Abstract
Background
Microbial constituents of the gut microbiome interact with each other and the host to alter the luminal environment and impact development, motility, and homeostasis of the gut. Powerful methods are becoming available to investigate connections between the gut microbiome and human health. While high-throughput sequencing of 16S rRNA genes can be used to identify and enumerate microbes in the gut, advances in several techniques (e.g., metagenomics, metatranscriptomics, metabolomics and metaproteomics) are providing a clearer view as to the specific activities of the microbiota in the context of functional host-microbial interactions. Testing emergent hypotheses regarding microbial effects on host biology, which arise from analyses of “Big Data” generated from massive parallel high-throughput sequencing technology and spectroscopic techniques, to guide translational research is an important goal for the future. Insights regarding the fundamental operating principles of the gut microbiota should lay the foundation for rational manipulation of the microbiota to promote human health.
Purpose
In this review we provide an overview of current research on the gut microbiome emphasizing current state-of-the-art technologies, approaches and directions for improvement of our understanding of the impact of the gut microbiota with specific focus on gastrointestinal motility disorders.
Keywords: enteric, genomics, germ-free, humanized, physiology, transcriptomics
Introduction
Deciphering the functional impact of gastrointestinal (GI) microbes on human hosts is critical for a better understanding of GI form and function. The assemblage of microbial symbionts in a typical adult includes representatives from all domains of life, performs important roles in health, and exhibits the properties of a dynamic organ that is capable of more varied biochemical transformations than any other system in the body (1). Mutualistic microbes perform important functions in the gut including synthesis of vitamins and cofactors (2), fermentation of complex polysaccharides to short chain fatty acids (SCFA; e.g., butyrate, the main nutritional substrate for colonic epithelial cells (3)), and modulation of GI motility (4-6). Gut microbes also play key roles in the development, and perhaps evolution, of adaptive immunity (7). Although bacteria, Archaea, microeukaryotes, and viruses are all important constituents of the gut microbiome, in this review, we will describe technologies for understanding the microbiota with a focus on the impact of bacteria.
As molecular techniques have been developed to enumerate and characterize constituents of the human microbiome, the totality of microbial life within and upon us, our understanding of “self” is extending to better appreciate the microbial factors and metabolic capacities that shape human biology (8). The development of novel computational tools, along with decreasing costs associated with DNA sequencing, brought about a revolution in microbial ecology that allowed unprecedented, cultivation-independent surveys of microbial biodiversity in the human gut and many other environments. As sequencing and computational capabilities have expanded, increasingly sophisticated investigations have been possible, and relatively straightforward nucleic acid-based enumeration studies have given way to coordinate, high-throughput analyses of macromolecular and metabolic flux within the GI tract.
As interdisciplinary research on the microbiome progresses, so will our ability to integrate principles of microbial ecology with functional studies to describe the mechanistic interactions through which gut microbes influence human health. The development and application of tools to assess the influence of the microbiome on health and disease should reveal novel metabolic and signaling pathways, and facilitate identification of therapeutic targets for treating disease.
FROM PHYLOTYPES TO FUNCTIONAL NETWORKS
Phylotyping: where we are and where we need to go
The term phylotype is a taxon-neutral description of an evolutionarily related group initially coined to describe a bacterial variant found in oceanic hydrothermal vents (9). It can be used to refer to a group defined by Linnean taxonomy or to an operational taxonomic unit (OTU) defined as a group sharing some high percentage (often ≥97%) of DNA sequence identity which are often referred to as species (10). Although several genetic elements have been proposed as indicators of evolutionary history, variable regions in the 16S rRNA gene have been the most widely used, due largely to contributions from Woese and Fox (11). 16S rRNA is required for protein synthesis, forms part of the scaffold for ribosomal proteins, and is present in all bacteria and Archaea. Importantly, genes encoding 16S rRNA have conserved regions, which are useful for broad-range PCR where multiple targets may be detected by a single primer set, and variable regions that exhibit properties of a “molecular clock.” As there is little evidence for horizontal gene transfer or recombination of rRNA genes, amplification and sequencing of the variable regions with degenerate primers can be used to identify the species present within microbial habitats.
The advent of high-throughput massively parallel next generation sequencing platforms, such as 454 Pyrosequencing, Illumina and SOLiD (Sequencing by Oligonucleotide Ligation and Detection), over the last decade has allowed characterization of entire microbial communities using partial 16S amplicons. Use of these methods has allowed us to effectively characterize the biodiversity in the gut and other body sites far surpassing molecular and cultivation-based methods which only captures about 12% of all phylotypes (12). Large-scale projects such as the European Metagenomics of the Human Intestinal Tract (MetaHIT; http://www.metahit.eu) and the NIH Human Microbiome Project (HMP; http://www.hmpdacc.org) utilizing next generation sequencing platforms have made substantial progress in defining normal, healthy microbiota. Several potentially important concepts have emerged from these efforts. For example, the term enterotype has been used to refer to gut microbiomes characterized by clusters dominated by a particular bacterial genus (e.g., Bacteroides, Prevotella, or Ruminococcus) as determined in an analysis of 33 metagenomes (13). However, enterotypes are not immutable and have been shown to shift from one type to another (14), whether due to dietary change, environmental effects, or other factors. Furthermore, in most body habitats, smooth abundance gradients of key genera are observed, with the characterization of discrete clusters depending greatly on computational approach and other technical variables (15). While reports have described distribution of enterotypes in certain populations (13, 16), interestingly, a study including over 300 adults from the United States, Venezuela, and Malawi did not corroborate discrete divisions between the three previously reported enterotypes (17). However, it did reveal exclusivity with respect to variation in Prevotella versus Bacteroides lineages (i.e., individuals with an abundance of Prevotella spp. tend not to have an abundance of Bacteroides spp. and vice versa), suggesting important ecological roles for these groups in the human gut.
Over the past decade, phylotyping has allowed us to understand the taxonomic distribution and diversity of enteric microbial communities in health and disease. 16S rRNA-based microbial community analysis has further allowed us to identify factors that drive normal variation. Age is an important contributor to microbial diversity, with a fairly chaotic microbiota at birth that increases in diversity and stability over the first three years of life, eventually reaching a stable, species-rich state; however, the diversity again declines in the elderly (18, 19). The effect of host genetics, environment (e.g., sanitation, hygiene, geography and climate), and diet on the microbiota is evident by studies investigating the gut microbial composition of individuals from different countries (Burkina Faso, Italy, Venezuela, Malawi, and US) (17, 20). Genetic influence is further apparent from similarities in the gut microbiota between twins and mother-daughter pairs (21). The effect of diet on the ratio of two main genera in the gut, Bacteroides and Prevotella, within the US population, highlights the strong independent effect of diet on gut microbiota (16). In addition to factors driving normal variation, phylotyping has helped define deviation from normal microbial states (dysbiosis) in diseases associated with the GI tract such as inflammatory bowel disease, irritable bowel syndrome, diarrheal states including Clostridium difficile-associated disease, as well as systemic diseases such as obesity, multiple sclerosis, and rheumatoid arthritis (22-29).
Although research has begun to define disease-associated dysbioses, study designs that use phylotyping to investigate longitudinal changes associated with disease states are lacking. Studies involving regular sampling to assess changes in individuals and their microbiota (e.g., prior to disease onset and with its onset and progression) will become increasingly valuable and should provide insights regarding changes in gut microbial communities that may precede clinical manifestations, providing both disease biomarkers and therapeutic targets. While this has been done in healthy adults (30), during pregnancy (31), and during infancy (19), there is a dearth of data regarding disease states and the interactions among factors known to influence variation of the microbiota in disease. Furthermore, there is a clear lack of studies addressing the composite effect of individual factors on gut microbial composition. For example, while studies have shown independent effects of diet and genotype on gut microbiota, it is unclear whether the same diet given in individuals with two different genotypes has the same effect on the gut microbiota. As a result, environmental factors, host diet, and genotype may introduce significant heterogeneity or biases in phylotyping studies addressing the effect of any single factor, given the strong influence of other factors on the gut microbiota (15, 16, 20, 32, 33).
Advances in bioinformatics are beginning to allow us to derive more information from 16S rRNA-based data sets in order to estimate protein-coding and metabolic capacities of intestinal populations based on extant microbial genome sequences, but such approaches require large inferential steps and do not typically measure the abundance of non-16S genes. Software tools (e.g., PICRUST: http://picrust.sourceforge.net) are being developed in an attempt to predict functional metagenomic content from 16S rRNA surveys using information from genomic databases, but it is unclear how they will compare with in-depth surveys of microbial gene content.
In order to understand true impact of gut microbes in health and disease, we need to move beyond phylotyping using complimentary -omics approaches (i.e., metagenomics, metatranscriptomics, metabolomics, and metaproteomics) to decipher functional changes in the microbiome. Well-defined animal models, including germ-free (GF), gnotobiotic (GF mice with defined microbial populations), and humanized (GF mice colonized with human-derived microbes) mice, will be an invaluable complement to studies of naturally occurring populations for understanding microbiota-dependent changes in host physiology. Integrating these technologies with disease-specific measurements of host physiology (e.g., markers of inflammation, changes in GI motility, behavioral studies) will provide further mechanistic insight into role of gut microbes.
Beyond phylotyping: predicting and testing functional networks
Metagenomics
Metagenomics is the study of collective genomes present in an ecosystem and was first described in 1998 (34). Early metagenomic studies utilized large-insert DNA libraries and were relatively expensive and difficult, due to the paucity of available sequencing platforms and bioinformatics tools. However, with newer generations of sequencing technologies, rapidly declining costs, and sophisticated bioinformatic tools, metagenomics has become a powerful tool supplementing 16S-based analyses. While 16S-based compositional analysis helps characterize the microbial members of the community, it currently provides limited insight into functionality of this community. Shotgun sequencing and assembly of the entire microbiome allows reconstruction of microbial genomes and pathways within a given habitat, as well as gene counting/quantitative metagenomic methods. Microbial functionality can be inferred from annotation of genes identified by shotgun sequencing, as well as by sequencing of inserts used in functional metagenomic libraries. Metagenomic analyses in large population-based studies of healthy individuals performed by the MetaHIT and HMP consortia have revealed similarities in functional gene profiles among individuals despite significant taxonomic diversity (1, 35). This highlights the presence among individuals of a “functional core” microbiome with conserved molecular activities rather than a “taxonomic core” microbiota consisting of a strictly conserved assemblage of phylotypes. Understanding the distribution of microbial phylotypes and their gene content in the gut facilitates comparisons between individuals and elucidates the functional relationships that characterize “healthy” microbial populations in humans. Furthermore, it facilitates the generation of hypotheses regarding trophic structures (nutrient and energy transfer) and deeper aspects of host-microbial interaction in the gut with health impacts in humans. These hypotheses must, of course, be tested to reveal how functional microbial mechanisms are integrated to establish or disrupt homeostasis in the gut.
Metatranscriptomics
Global gene expression studies of microbes have traditionally utilized cDNA microarrays, which utilize immobilized single-stranded, gene-specific DNA probes. Microarray-based technology works well for surveys of individual species, however, it doesn't perform as well when looking at interactions among multiple microbes or between the host and resident microbes. Moreover, to analyze the entire transcriptome including non-coding regions, hundreds of millions of probes are required. The advent of next generation sequencing has revolutionized the field of transcriptomics with introduction of RNA-seq (36). RNA-seq represents massively parallel sequencing of cDNA generated from all RNA molecules in a sample. Strand-specific RNA-seq preserves information about transcript directionality, which is invaluable for analyzing non-coding and antisense RNAs, as well as for defining operons in bacteria. RNA-seq holds several advantages over array-based transcriptomics in that it possesses a higher dynamic range and provides full-genome coverage, as well as the potential to identify novel transcripts and analyze transcriptomes of both the host and its microbial inhabitants at the same time (dual RNA-seq (37)). Although dual RNA-seq poses challenges given the significant heterogeneity of RNA present in eukaryotic and bacterial transcriptomes, differences in methodology to extract RNA, use of rRNA depletion methods to increase information content (mRNA), and difficulties in estimating the sequencing depth, it still has the potential to further revolutionize the study of host-microbial interactions.
Metatranscriptomics is the study of RNA from all of the microbes in a given environment (e.g., GI tract, soil, or aquatic habitats). Metatranscriptomic analysis of the human gut microbiota from 10 healthy volunteers revealed major functional roles of gut microbes in carbohydrate metabolism, energy production, and synthesis of cellular components, while certain activities such as lipid transport metabolism were underrepresented (38). The benefit of metatranscriptomics is demonstrated by a study of microbial populations in the oligotrophic surface waters of the Pacific Ocean, where transcriptomic analysis of microbial communities revealed gene expression patterns specific to particular taxa, as well as gene categories undetected in earlier DNA-based metagenomic surveys (39). Thus, complementary -omics approaches can be used to extract additional ecological, genetic, and biochemical information from various microbial habitats.
Metatranscriptomics adds to metagenomic data sets by providing information about which genes in a microbial community are expressed, and can also be used to investigate host transcriptional changes that occur in response to microbes. In order to further extend our understanding of host-microbe interactions, we need to profile low molecular weight metabolites (metabolomics), peptides, and proteins produced by the host, as well as associated microbes (metaproteomics), in response to differential gene expression in health and disease. Small molecules including nutritional substrates and chemical messengers may have profound effects on shaping the activities gut microbial communities (e.g., through trophic effects on dietary fermentation, quorum sensing mechanisms, or the production of bacteriocins and other toxins). Furthermore, low molecular weight compounds may modulate the crosstalk that occurs between the resident microbiota and host epithelial and immune cells.
Metabolomics
Metabolomics or metabolic profiling of biological fluids, such as urine, serum, or fecal water, is usually accomplished by spectroscopic techniques, which can be used for global analyses of metabolites (untargeted approach) or measurement of selected metabolites of interest (targeted approach). Mass spectrometry (MS)-based methods help to discriminate metabolites based on their mass to charge (m/z) ratio and are often preceded by separation techniques such as gas chromatography or high-performance/ultra-performance liquid chromatography. Identification of metabolites is then achieved using existing databases of m/z values such as METLIN (40). On the other hand, 1H nuclear magnetic resonance spectroscopy helps to identify chemical structures based on the chemical shift (i.e., the resonant frequencies of atomic nuclei relative to a reference standard) following perturbation with radiofrequency pulses. This technique can also integrate information about the measured influence of atomic nuclei on each other (spin-spin coupling). Given the minimal sample preparation required, 1H NMR is better suited than MS-based methods for high-throughput studies (41).
Metabolomic profiles, especially those of exclusively microbial origin, can be integrated with metagenomics and metatranscriptomics using multivariate computational modeling to provide insight regarding microbial functionality in dynamic environmental conditions. In gnotobiotic mice colonized with a 15-species model human gut microbiota, introduction of a fermented milk product containing five sequenced bacterial strains did not result in marked changes to the gut microbial community structure; however, metatranscriptomic analysis of fecal samples and MS-based analyses of urinary metabolites revealed potentially important effects on the expression of microbial enzymes involved in carbohydrate metabolism (42). Observed discordance between effects on community structure relative to effects on gene expression and metabolism is evidence that complementary approaches beyond 16S-based enumeration and metagenomics are required to elucidate certain biological phenomena. Data from the human vaginal microbiome further show that changes in metabolite profiles can be observed even in the absence of significant alterations in microbial community structure (43).
Metaproteomics
A developing approach for high-throughput, non-targeted identification of thousands of proteins from complex, microbial habitats (shotgun metaproteomics) involves cellular lysis and subsequent enzymatic digestion of all accessible proteins in a given sample. The resultant peptide fragments are separated by liquid chromatography and subjected to tandem mass spectrometry (MS/MS). Peptide masses and spectra are quantified and compared to reference protein databases predicted from genomic sequence information. This method requires no gel-based separation nor de novo protein sequencing, and has been applied to gut microbial communities both in healthy individuals and patients with inflammatory bowel disease (44). Although it will require significant effort to characterize the entire metaproteome, it appears to be well within our reach. A recent study of a single individual before, during, and after antibiotic therapy highlights how four -omic technologies can be used to provide more comprehensive information on the microbiome (45).
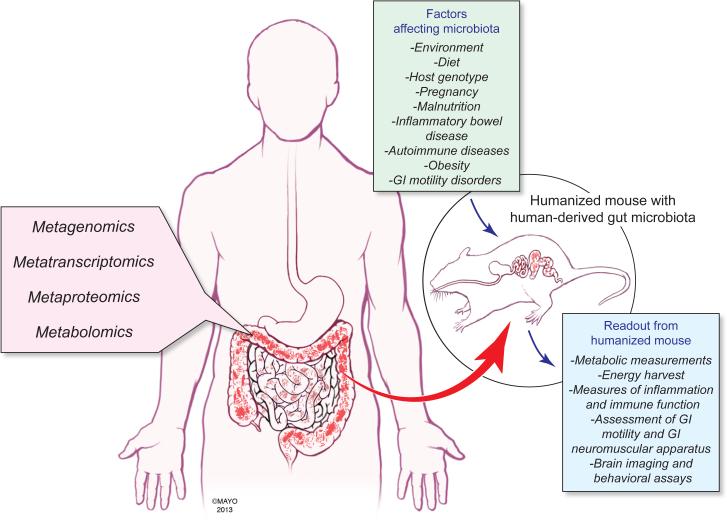
The term multi-omics can be used to refer to approaches that combine two or more high-throughput -omic technologies (e.g., metagenomics together with metatranscriptomics or metabolomics or metaproteomics). Multi-omic platforms can help predict, define and confirm various microbial metabolic pathways, as well as provide insight into microbe-host interactions (Fig. 1). Combining multi-omics platforms with 16S-based phylotyping can significantly improve our understanding of the taxonomic and molecular characteristics of the microbiome. A recent study investigating effects of gut microbes on the luminal metabolome clearly demonstrates the power of linking phylotyping analysis with metabolomics and defining microbial functionality by determining metabolites produced in vivo by specific bacterial groups (46). To further compliment these platforms, we need a model host that will provide a highly controllable experimental system with reduced complexity to deconstruct interactions between the host and its resident microbiota and test emergent hypotheses generated by multi-omics platforms.
Figure 1. Multi-omic and gnotobiotic approaches to host-microbial interaction.
High-throughput analyses of host and microbial DNA, RNA, proteins and metabolites can be performed using metagenomics, metatranscriptomics, metaproteomics and metabolomics, respectively, to investigate interrelationships between host and microbial factors. Hypotheses that emerge from multi-omic studies can be tested using gnotobiotics through colonization of germ-free mice with characterized microbial consortia or human gut microbiota. This approach will help to elucidate effects on host biology of factors that modulate the gut microbiota including environment (e.g., geography, hygiene), diet, host genotype, physiological states (e.g., pregnancy) and disease states.
Gnotobiotics
Gnotobiotic approaches (47) use controlled procedures to establish, breed, and study animals in the absence of microbes (GF) or in the presence of intentional communities of one or more defined microbial species used to colonize GF mice. Gnotobiotic techniques have been effectively employed to study response to microbes in several vertebrate model organisms (47, 48). These techniques provide a controlled setting with regard to environment (housing), host genetics, and microbial exposure to assess the impact of microbes and their molecular products on host biology. They also facilitate the study of alterations in the microbiome in response to perturbations including dietary interventions and pharmacologic treatments (49).
Despite the apparent complexity of the human intestinal microbiota, which contains ~1000 bacterial species (1), just four phyla dominate. The Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria constitute >95% of the bacterial residents in a typical, healthy adult gut (50). This composition of a restricted number of related bacterial groups suggests that it can be modeled in a simplified, experimentally tractable manner. An investigation of species abundance and microbial gene expression in a model consortium of 10 sequenced human gut-derived bacteria introduced into gnotobiotic mice identified responses to randomized perturbations of four defined dietary components. In turn, this allowed for development of a statistical algorithm capable of predicting 60% of the variation in microbial species abundance in response to specific dietary perturbations (49). This study highlights that the functionality of a healthy human gut microbiome can be modeled in gnotobiotic mice harboring this 10-member community of cultivatable microbes.
GF mice can also be colonized with microbial collections from human donors (e.g., samples derived from intestinal contents or feces) to generate “humanized” mice (51, 52). Humanized mice represent a model for testing effects of human-derived microbial consortia in vivo in a surrogate mammalian host (Fig. 1). Studies in humanized mice have helped define the role of gut microbiota in the physiological changes observed in diet-induced obesity (53). Mice colonized with human-derived fecal microbiota and fed a high-fat, high-sugar (Western) diet exhibit increased adiposity compared to GF mice fed the same diet (53). Furthermore, transplantation of the microbiota from humanized mice that had received the Western diet into GF recipients led to increased adiposity in these mice compared to GF mice that received microbiota transplanted from lean controls (53). A study using humanized mice to model the influence of microbes during pregnancy revealed that an altered gut microbiome was, at least in part, responsible for increased weight gain and inflammation, and decreased insulin sensitivity during the third trimester (31).
Integration of multi-omics platforms with animal models such as humanized mice can be useful for defining the impact of gut microbiota in maintaining health and causing disease (Fig. 1), as well as for conducting pre-clinical trials to test the effects of environmental and genetic factors on the gut microbiota and host physiology. To this point, a recent study that implicates gut microbiota in the development of kwashiorkor, a form of acute and severe malnutrition, used a combination of metagenomics and metabolomics to identify microbial perturbations and metabolic changes that exacerbate malnutrition in this population (40). Moreover, the authors were able to recapitulate this phenotype in humanized mice, which could be transiently ameliorated by using a customized dietary formulation.
The scientific community has made significant progress in understanding our microbial co-conspirators over the past decade through the development of new sequencing technologies, novel bioinformatics tools to handle “Big Data”, and efforts by the NIH Human Microbiome Project. However, we now need to shift our focus towards applying technologies to address basic biological questions at a mechanistic level. This will require combining -omic technologies with the measurement of changes in host physiology (Fig. 1). While the measures of host physiology will be different for particular disease states, we will focus on the role of gut microbiota on GI motility.
A variety of techniques in animal models are available to study GI motility and associated cellular processes, including ex vivo techniques such as single-cell electrophysiology, contractility recordings and spatiotemporal mapping of intestinal segments in organ baths, as well as in vivo methods including gastric emptying measurements, whole gut transit assays using colored dyes, and colonic manometry using miniature pressure transducers.
GUT MICROBIOTA AND GASTROINTESTINAL MOTILITY
Interdependence of gut microbes and gastrointestinal motility
GI motility and the enteric microbiota are interrelated and can exert reciprocal effects on each other. For example, experiments in GF mice revealed that gut microbes stimulate small bowel motility (5, 6, 54). Small bowel bacterial overgrowth is a clinical syndrome often seen in patients with impaired GI motility, indicating that changes in gut motility can also alter resident microbial communities (55). Furthermore, the gut microbiome is altered in GI disorders (56) which can be associated with changes in GI motility (e.g., IBD (24), IBS (28), and infectious diarrhea (29)). Because the gut microbiome responds to changes in environmental factors, abnormal GI motility likely shapes the organizational structure of the microbiota by changing the relative fitness of gut microbes depending on the “flow rate” of intestinal contents; likewise, microbial gene expression is also likely to change in response to intraluminal conditions depending on GI transit time.
We recently demonstrated that introducing complex, fecal microbiota from a healthy human into GF mice (humanized mice) significantly shortened GI transit time and increased colonic contractility (57). Interestingly, the magnitude and directionality of this effect depends on the type of carbohydrates provided in diet, suggesting that microbial influence on the GI tract is partially dependent on diet. Conversely, abundance of gut microbial communities was altered by perturbations resulting from changes in host GI transit. Accelerating or decelerating GI transit using PEG or loperamide, respectively, led to characterized differences in the structure of the gut microbiome that were reversed upon return to a normal rate of GI transit (57).
Recent studies further support an interdependent relationship between gut microbiota and motility. Electrophysiological studies have revealed abnormal function of intrinsic primary afferent neurons isolated from the jejuna of GF mice, with reduced excitability in the myenteric afterhyperpolarization neurons and prolonged slow afterhyperpolarization after action potentials, compared to conventional mice (58). Furthermore, spatiotemporal mapping and intraluminal pressure recordings have been used to assess jejunal and colonic migrating motor complex (MMC) frequency and velocity in response to luminal application of two purportedly beneficial strains of Lactobacilli (59). Intriguingly, this study found that luminal L. rhamnosus significantly increased MMC velocity in the jejunum, but decreased MMC velocity in the colon, suggesting region-specific responses to neuroactive gut microbes. While these studies highlight effect of microbes on GI motility, it has also been demonstrated, in a mouse model (Ednrb−/−) of intestinal aganglionosis, that the enteric nervous system is an important determinant of microbiome composition, with Ednrb−/− mice possessing abnormally high Bacteroidetes:Firmicutes ratios and altered fecal metabolite profiles (60).
Mechanisms of microbial regulation of gastrointestinal motility
While it is clear that microbiota and motility are related, the mechanisms by which microbes mediate this effect are still not well defined. A recent study demonstrated that microbial products—most likely bacterial lipopolysaccharide (LPS)—promote GI motility by signaling through TLR4/Myd88 in derivatives of the neural crest (4), which gives rise to several cell types in the gut, including enteric neurons and glial cells. Toll-like receptors (TLRs) comprise a large family of host receptors involved in metazoan development and microbial defense, and most TLR signaling occurs through the Myd88 adaptor protein (61). Compared to wild-type mice, mice lacking either Myd88 or Tlr4 exhibited longer GI transit times and possessed reduced numbers of colonic nitrergic neurons (4). GF mice were also found to possess significantly lower number of colonic nitrergic neurons than conventionally raised mice, suggesting that both intact TLR4/Myd88 signaling in the host and luminal LPS are required for maintenance of these neuronal populations in the colon (4). Knockdown of the indigenous microbiota by a 12-week regimen of broad-spectrum antibiotics resulted in a significant delay in GI motility (4). Interestingly, the authors reported that incubation of enteric neuronal cells with subtoxic concentrations of LPS led to activation of the transcription factor nuclear factor (NF)-κB and decreased apoptosis, suggesting that signaling downstream of LPS reception promotes survival of the enteric neurons that promote gut motility.
In our recent study, we integrated 16S-rRNA based microbial community analysis and metabolomics to address the mechanisms by which gut microbiota regulate GI motility (57). We demonstrated that a diet composed primarily of fructo-oligosaccharide (FOS) led to a marked increase in GI transit time compared to a standard diet in humanized mice; this change was accompanied by alterations in microbial community structure, with a significant decrease in the abundance of members of the family Lachnospiraceae. Furthermore, metabolic profiles revealed significantly lower fecal concentrations of all detectable SCFA as well as the serotonin (5-HT) metabolite 5-hydroxyindoleacetic acid (5-HIAA), compared to mice fed a standard diet (57). These findings suggest the possible involvement of the 5-HT pathway in the modulation of GI motility by gut microbiota which was further supported by a significant delay in GI transit in humanized mice, but not in GF mice, following intraperitoneal administration of SDZ205-557 (5-HT3/4 antagonist).
Role of microbial metabolites in gastrointestinal motility
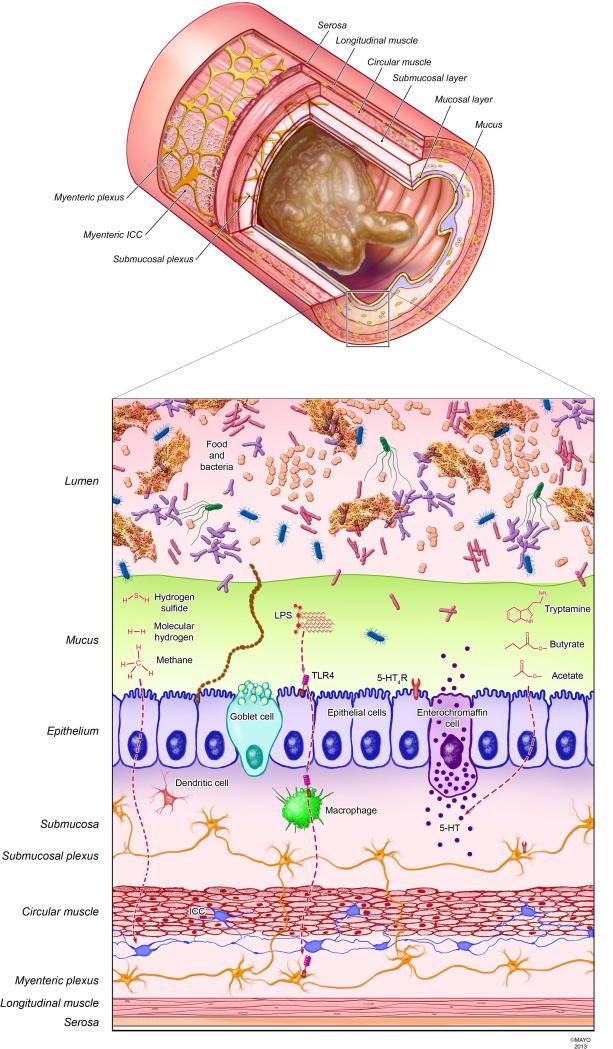
The majority of microbes are separated from the epithelium by the mucus layer and hence bioactive microbial products, which can diffuse through the mucus layers, are the likely mediators of effects of gut microbiota on the host (Fig. 2). Several microbial products have been suggested to influence GI motility. Experimentally administered methane (CH4) has been demonstrated to delay small intestinal transit in guinea pig and canine models (62), possibly by an effect on smooth muscle contractility. Production of methane in humans and other mammals is thought to arise exclusively from the actions of gut-dwelling members of the domain Archaea (e.g., Methanobrevibacter spp.) and a relatively small subset of bacteria capable of methanogenesis (e.g., Bacteroides and Clostridium spp. (63)).
Figure 2. Microbial products with putative effects on GI motility.
GI motility depends on the complex interaction of multiple cell types including enteric neurons, interstitial cells of Cajal, smooth muscle, and immune cells (e.g., macrophages). Luminal gases mediate GI motility; methane and hydrogen sulfide have inhibitory effects on GI transit potentially due to an effect on smooth muscle cells, and molecular hydrogen increases colonic motility by an unidentified mechanism. Lipopolysaccharide produced by Gram-negative bacteria has been suggested to promote survival of enteric nitrergic neurons and motility through Toll-like receptor 4 signaling. The bacterial metabolite, tryptamine, mimics stimulatory effects on motility of serotonin. Luminal short chain fatty acids such as butyrate and acetate promote GI motility through several mechanisms including direct effects on smooth muscle and production of mucosal 5-HT. Production of microbial products regulating motility depends on the availability of specific dietary compounds. 5-HT, serotonin; 5-HT4R, serotonin receptor; TLR4, Toll-like receptor 4; ICC, interstitial cells of Cajal; LPS, lipopolysaccharide.
Hydrogen sulfide (H2S) is another compound with documented effects on GI motor function. Unlike methane, however, H2S is produced endogenously in tissues by regulated enzymatic activity as well as in the gut lumen by resident sulfate-reducing bacteria. Endogenous H2S has been shown to inhibit the contractile activity of smooth muscle in the mouse colon (64) and rat ileum (65), an effect reported to be dependent on K+ channels (66). H2S also has a variety of effects on interstitial cells of Cajal, enteric and extrinsic neurons, and can modulate synaptic transmission (67); elucidating the functional integration of these signals to regulate motility remains an important area of research. Regarding a role for bacterially derived H2S in GI motility, however, there is little evidence that luminal H2S survives detoxification pathways present in the colonic mucosa (67). Although it has been proposed that impaired H2S degradation could promote inflammatory disease in the colon (68), a causative link has not been proven. Nonetheless, as disruption of barrier function is known to occur in inflamed GI mucosa and there is evidence that hydrogen sulfide can augment signaling through TLR4 (69), reinvestigation of bacterially mediated effects of H2S on motility may be warranted, especially in inflammatory disease.
Along with CH4 and H2S, molecular hydrogen (H2) is among the gases formed as byproducts of microbial metabolism in the gut. Exogenously administered H2 has been reported to significantly shorten transit times in the proximal and distal colon (70). This is interesting in light of the fact that H2 is consumed during microbial production of both H2S by sulfate-reducing bacteria and CH4 by methanogens. One possibility is that some of the influence of H2 on gut motility is mediated indirectly through effects on these bacteria. It has been shown in vitro that sulfate-reducing bacteria outcompete methanogens for H2, but only if sulfate concentrations are high enough (71); thus, dietary sulfate may be an important factor to consider in vivo. Although osmotic diarrhea resulting from high-levels of unabsorbed sulfate has been reported (72), it is unknown whether susceptibility depends on aspects of the gut microbiome.
Metabolites produced by the microbiota, including both primary (e.g., the fermentative end-products, butyrate and acetate) and secondary (e.g., derivatives of aromatic amino acids), can also alter GI transit (Fig. 2). In conscious rats, intraluminal administration of a mixture of the SCFA was reported to stimulate colonic transit by triggering of 5-HT release, as this response was abolished by intraluminal pretreatment with lidocaine and a 5-HT3 receptor antagonist (73). 5-HT release, as well as ascending contraction and descending relaxation of the rat colon induced by mechanical stimulation, was also shown to be significantly augmented by acetate alone (1 mM) (74). 5-HT release from the mucosa depends largely on the expression of tryptophan hydroxylase 1, the rate-limiting enzyme in 5-HT synthesis. Interestingly, a butyrate-inducible transcription factor, ZBP-89, has recently been shown to regulate Tph1 expression in vivo (75).
Tryptamine is a secondary metabolite resulting from the transformation of the aromatic amino acid tryptophan and has been previously attributed to microbial metabolism (76). Like 5-HT, tryptamine has been shown to increase contractility in ex vivo preparations of guinea pig ileum, indicating that this metabolite may be one of the microbial mediators of host GI motility (77).
Additional unidentified compounds produced by gut microbes may also influence GI motility. For example, there is some evidence that gut bacteria synthesize metabolic precursors to ligands of the GABAA receptor (78), which has well-documented suppressive effects on GI motility. Additional motility-related molecules that are profoundly dependent on the gut microbiota include bile acid metabolites (79). Although the introduction of bile acids directly into the sigmoid colon and rectum has been shown to stimulate colonic motility in humans (80), the effects of differential bacterial metabolism of bile acids on GI motility remain to be elucidated.
Future investigations of the interrelationships among diet, the microbiome, and GI motility should enhance our understanding of GI function as well as modalities for treating disease. By utilizing multi-omic technologies, we can identify how changes in gut microbiota composition and function influence the synthesis of bioactive bacterial products that alter host physiology under different environmental conditions, such as dietary alteration. While individual nutrients including dietary fermentable poly- and oligosaccharides, tryptophan, and sulfate can affect production of specific metabolites (Fig. 2) such as SCFA, tryptamine, and hydrogen sulfide, respectively, the complex signaling among GI microbes and the microbial food web that exists in the gut make it challenging to predict the effect of any single nutrient that is introduced. Systems biology approaches can make important predictions using mathematical models of these changes, which can be subsequently tested in gnotobiotic animals. These studies are needed desperately to understand the mechanisms by which gut microbiota alter motility and to develop more directed therapeutic strategies targeting the gut microbiota.
Microbiome-gut-brain axis
Although the gut microbiota has important immediate effects in the GI tract, its effects on the central nervous system (CNS) may also modulate aspects of GI physiology, such as nociception and motility. The microbiome-gut-brain axis is now well established as bidirectional and provides a conceptual model that can be used to further investigate and, ultimately, to treat diseases with currently poorly defined etiologies, including irritable bowel syndrome, and perhaps even psychiatric and autoimmune diseases (81, 82). The effect of gut microbes on behavior is highlighted by a recent study investigating anxiety and locomotor activity in gnotobiotic mice (83). This study indicated that ex-GF mice, colonized with normal specific pathogen-free microbiota, were less anxious and active than GF mice, and that this phenotype was accompanied by higher turnover rates of norepinephrine, dopamine, and serotonin in most regions of the brain, suggesting modulation of monoaminergic neurotransmission by the gut microbiota. Interestingly, in earlier studies, GF mice of different genetic backgrounds exhibited increased motor activity and reduced anxiety, suggesting microbial effects on behavior may be dependent on host genetics (84). Furthermore, a probiotic strain (Bifidobacterium longum NCC3001) has been shown to decrease anxiety-like behavior and hippocampal brain derived neurotrophic factor (BDNF) in mice with infectious colitis (85), highlighting the potential for use of select microbes as therapeutic agents. As the field evolves, we will be better able to understand the mechanisms by which constituents of the gut microbiota affect CNS function.
Consistent with the bidirectional nature of the microbiome-gut-brain axis, signals from the CNS can alter composition of the gut microbiota. In two different models of stress in mice (grid floor-induced and water avoidance stress), the gut microbiome was found to be significantly altered compared to that of non-stressed mice, suggesting that CNS-derived signals can act to shape the gut microbiota (86, 87). Exposure to water avoidance stress in mice led to inhibition of the NLRP6 inflammasome with altered the composition of the microbiota (86). Given the complexity of the microbiome-gut-brain axis, much work will be needed to elucidate how these systems are interrelated and regulated, and what aspects are important for determining health in humans.
PROSPECTUS FOR GASTROINTESTINAL MOTILITY AND FUNCTIONAL GASTROINTESTINAL DISORDERS
As we move beyond characterizing gut microbial configurations, and identify specific changes in microbial functionality and reciprocal changes in host function, it will be important to define mechanisms through which gut microbes alter host cells.
Microbiota-driven host epigenetic changes
One important phenomenon, which remains to be completely elucidated, is the alteration of epigenetic status in host cells by effector microbes in the gut. Gnotobiotic techniques offer a powerful platform upon which to test the effects of monocultures or groups of bacteria identified using the various -omics platforms on epigenetic changes in the host. Epigenetic research involves the study of transcriptional modulation that occurs through chemical modifications to DNA or associated histone proteins that act as localized signals and are retained by chromatin through progressive cell divisions. Epigenetic changes include DNA methylation and various histone modifications that alter the accessibility of DNA to RNA polymerase and transcription factors. These changes are added or removed enzymatically, in response to environmental factors, and can alter cellular function in a variety of ways. While it is still unclear how microbes mediate epigenetic changes, a few studies have described these phenomena in response to both commensal and pathogenic bacteria.
For example, infection by Helicobacter pylori is now well-accepted as a risk factor for developing gastric ulcer and cancer in humans (88). These bacteria have been shown to cause hypermethylation of the FOXD3 promoter in the progression of H. pylori-associated gastric tumors (89). However, the normal microbiota also affects epigenetic status in host cells. Commensal microbes in mice have been shown to reduce the accumulation of invariant natural killer T cell (iNKT) in the lungs and colon via epigenetic changes in the chemokine, CXCL16. Association of GF mice with a conventional microbiota early in life acts to decrease hypermethylation of the Cxcl16 gene, facilitating its normal expression (90). However, this is not observed when adult GF mice are similarly colonized, implicating CXCL16 in modulating susceptibility to pulmonary and colonic inflammation by regulating the abundance of iNKT cells in an age- and tissue-specific manner (90). This finding has important implications with respect to early associations between microbes and the developing host, especially if other cell types are similarly influenced by microbial interactions. Interactions with commensals and other microbes could influence heritable, chromatin-level changes that predispose individuals to functional GI disorders through effects on the gut neuromuscular apparatus or associated immune cells. Several functional GI disorders, including those of gut motility, can be passed to offspring but lack an identified causative genetic component. Some of these may involve altered transcriptional control and display a pedigree consistent with epigenetic inheritance (91). However, testing host epigenetic responses to microbial groups, microbial end-products or other metabolites in animal models will likely require a different approach than typical pharmacological dose-response assays. Relevant host epigenetic, cellular and physiological responses may not be elicited immediately following introduction of microbes or microbially derived compounds; responses may require some chronic exposure to bacterial products, integration of an immunological response, or exposure during an age-dependent timeframe. Experiments in gnotobiotic animals will be essential to look effects of complex experimental communities on the host in a controlled setting where the presence or absence of a particular strain or luminal compound can be tested systematically.
Quorum sensing
Many bacteria use diffusible, small molecules as messengers to communicate with members of their own (and potentially other) species to mediate symbiotic interactions and to regulate the production of catabolic enzymes, pathogenic virulence factors, and other functional molecules. The process, known as quorum sensing (QS), is widespread among bacteria and has been identified as a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7 (92), and is required for biofilm production in both Acinetobacter, which can cause a variety of opportunistic, nosocomial infections, and Clostridium difficile (93), which can also form biofilms during infection (94). One important class of QS signals includes the N-acyl-homoserine lactones (AHL), which are used by Gram-negative bacteria to coordinate gene expression in response to accumulation of these molecules in the environment; Gram-positive bacteria typically use other compounds, including secreted peptides, to regulate density-dependent community behavior.
We are beginning to appreciate the potential for recognition of prokaryotic QS molecules by the eukaryotic cells of colonized hosts. Although further study is necessary, it is possible that such trans-domain interactions act to modify aspects of microbial mutualism and pathogenesis in humans. Host responses to bacterial QS signals appear to include alterations in growth responses, immune response and defense, as well as hormone production (95). P. aeruginosa-derived homoserine lactones have been shown to interact with the host immune system to attenuate LPS-induced inflammatory responses by means of an unidentified mechanism that may promote chronic infection in the lung (96); similar phenomena may be relevant in diseases of the GI tract and can be explored integrating multi-omic technologies. These findings reinforce our understanding and prompt inquiry regarding the interdependent nature of physiology, disease, and the activities of the microbiome; diligent investigation of QS mechanisms in the GI tract may provide further revelations regarding our symbiotic functionality and the mechanisms underlying microbiota-associated disease. Furthermore, by integrating knowledge of the gut microbiota with understanding of effects on the host, we will profoundly expand the repertoire of potential therapeutic targets (within the gut ecosystem) for the treatment of disease by rational manipulation of the microbiome.
Identification of novel metabolites produced by specific subsets of bacteria is now feasible by using an integrated multi-omics approach correlating data obtained from 16S rRNA-based community analysis with metatranscriptomics, metabolomics and metaproteomics. Furthermore, UPLC or HPLC (ultra-performance liquid chromatography or high-performance liquid chromatography) individual fractions from mass spectrometric methods with specific retention times can be collected and tested in in vitro and in vivo systems allowing discovery of novel bacterial peptides with effects on host physiology. Again, gnotobiotics offers a powerful tool to test these hypotheses. Identification of novel peptides produced by specific bacteria and the genes responsible for these metabolites will allow targeted therapeutics by engineering commensal gut-adapted bacteria to produce metabolites of interest for administration as probiotics.
SUMMARY
A wealth of information is emerging about interactions within the microbiome and their impact on human health, but numerous questions remain as to how composition of the intestinal microbiota shapes human biology. As we learn more, it is hoped that we will be able to assemble more accurate models of the interdependence of genetic, environmental, and key microbial products and activities related to host response. Understanding beneficial and deleterious molecular mechanisms linking the microbiome to host physiology will enable more effective therapeutic approaches and provide the basis for targeted manipulation of the microbiota. In conclusion, bringing together studies of host and microbe using traditional methodologies and newly developed techniques for high-throughput multi-omics analysis will allow us to build and test mathematical models of microbial activity, interaction, and effects on human health as we move beyond phylotyping.
ACKNOWLEDGEMENTS
We would like to thank Kristy Zodrow for secretarial assistance and Michael King for illustrations.
FUNDING
CSR is supported by a post-doctoral fellowship from the Center for Individualized Medicine, Mayo Clinic, Rochester, MN. This work was supported by pilot and feasibility awards to PCK from the Center for Individualized Medicine and Center for Cell Signaling, Mayo Clinic, Rochester, MN.
Footnotes
AUTHOR CONTRIBUTION
PCK was responsible for the overall organization of the review. CSR and PCK contributed to the research, writing of this review and designing of the figures.
DISCLOSURES
The authors have no competing interests.
REFERENCES
- 1.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 2.Resta SC. Effects of probiotics and commensals on intestinal epithelial physiology: implications for nutrient handling. The Journal of physiology. 2009;587:4169–4174. doi: 10.1113/jphysiol.2009.176370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donohoe DR, Garge N, Zhang X, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell metabolism. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anitha M, Vijay-Kumar M, Sitaraman SV, Gewirtz AT, Srinivasan S. Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology. 2012;143:1006–1016. e1004. doi: 10.1053/j.gastro.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Husebye E, Hellstrom PM, Midtvedt T. Intestinal microflora stimulates myoelectric activity of rat small intestine by promoting cyclic initiation and aboral propagation of migrating myoelectric complex. Digestive diseases and sciences. 1994;39:946–956. doi: 10.1007/BF02087542. [DOI] [PubMed] [Google Scholar]
- 6.Husebye E, Hellstrom PM, Sundler F, Chen J, Midtvedt T. Influence of microbial species on small intestinal myoelectric activity and transit in germ-free rats. American journal of physiology Gastrointestinal and liver physiology. 2001;280:G368–380. doi: 10.1152/ajpgi.2001.280.3.G368. [DOI] [PubMed] [Google Scholar]
- 7.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon JI. Honor thy gut symbionts redux. Science. 2012;336:1251–1253. doi: 10.1126/science.1224686. [DOI] [PubMed] [Google Scholar]
- 9.Polz MF, Cavanaugh CM. Dominance of one bacterial phylotype at a Mid-Atlantic Ridge hydrothermal vent site. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7232–7236. doi: 10.1073/pnas.92.16.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hein JJ. An optimal algorithm to reconstruct trees from additive distance data. Bulletin of mathematical biology. 1989;51:597–603. doi: 10.1007/BF02459968. [DOI] [PubMed] [Google Scholar]
- 11.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proceedings of the National Academy of Sciences of the United States of America. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- 13.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zupancic ML, Cantarel BL, Liu Z, et al. Analysis of the gut microbiota in the old order Amish and its relation to the metabolic syndrome. PloS one. 2012;7:e43052. doi: 10.1371/journal.pone.0043052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koren O, Knights D, Gonzalez A, et al. A Guide to Enterotypes across the Human Body: Meta-Analysis of Microbial Community Structures in Human Microbiome Datasets. PLoS computational biology. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claesson MJ, Cusack S, O'Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong-Si T, Weber J, Liu YB, et al. Increased prevalence of and gene transcription by Chlamydia pneumoniae in cerebrospinal fluid of patients with relapsing-remitting multiple sclerosis. Journal of neurology. 2004;251:542–547. doi: 10.1007/s00415-004-0360-0. [DOI] [PubMed] [Google Scholar]
- 23.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 24.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nature reviews Gastroenterology & hepatology. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 25.Rea MC, O'Sullivan O, Shanahan F, et al. Clostridium difficile carriage in elderly subjects and associated changes in the intestinal microbiota. Journal of clinical microbiology. 2012;50:867–875. doi: 10.1128/JCM.05176-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rousseau C, Levenez F, Fouqueray C, Dore J, Collignon A, Lepage P. Clostridium difficile colonization in early infancy is accompanied by changes in intestinal microbiota composition. Journal of clinical microbiology. 2011;49:858–865. doi: 10.1128/JCM.01507-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. The Journal of rheumatology. 2008;35:1500–1505. [PubMed] [Google Scholar]
- 28.Verdu EF. Differences in intestinal microbial composition in children with IBS-what does it all mean? The American journal of gastroenterology. 2012;107:1752–1754. doi: 10.1038/ajg.2012.291. [DOI] [PubMed] [Google Scholar]
- 29.Verma AK, Verma R, Ahuja V, Paul J. Real-time analysis of gut flora in Entamoeba histolytica infected patients of Northern India. BMC microbiology. 2012;12:183. doi: 10.1186/1471-2180-12-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, Lauber CL, Costello EK, et al. Moving pictures of the human microbiome. Genome biology. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koren O, Goodrich JK, Cullender TC, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanski I, von Hertzen L, Fyhrquist N, et al. Environmental biodiversity, human microbiota, and allergy are interrelated. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8334–8339. doi: 10.1073/pnas.1205624109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nature reviews Microbiology. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 34.Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chemistry & biology. 1998;5:R245–249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 35.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westermann AJ, Gorski SA, Vogel J. Dual RNA-seq of pathogen and host. Nature reviews Microbiology. 2012;10:618–630. doi: 10.1038/nrmicro2852. [DOI] [PubMed] [Google Scholar]
- 38.Gosalbes MJ, Durban A, Pignatelli M, et al. Metatranscriptomic approach to analyze the functional human gut microbiota. PloS one. 2011;6:e17447. doi: 10.1371/journal.pone.0017447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frias-Lopez J, Shi Y, Tyson GW, et al. Microbial community gene expression in ocean surface waters. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith CA, O'Maille G, Want EJ, et al. METLIN: a metabolite mass spectral database. Therapeutic drug monitoring. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 41.Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends in microbiology. 2011;19:349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 42.McNulty NP, Yatsunenko T, Hsiao A, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Science translational medicine. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gajer P, Brotman RM, Bai G, et al. Temporal dynamics of the human vaginal microbiota. Science translational medicine. 2012;4:132ra152. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verberkmoes NC, Russell AL, Shah M, et al. Shotgun metaproteomics of the human distal gut microbiota. The ISME journal. 2009;3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 45.Perez-Cobas AE, Gosalbes MJ, Friedrichs A, et al. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut. 2012 doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsumoto M, Kibe R, Ooga T, et al. Impact of intestinal microbiota on intestinal luminal metabolome. Scientific reports. 2012;2:233. doi: 10.1038/srep00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiology and molecular biology reviews : MMBR. 1998;62:1157–1170. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung H, Pamp SJ, Hill JA, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodman AL, Kallstrom G, Faith JJ, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science translational medicine. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barbara G, Stanghellini V, Brandi G, et al. Interactions between commensal bacteria and gut sensorimotor function in health and disease. The American journal of gastroenterology. 2005;100:2560–2568. doi: 10.1111/j.1572-0241.2005.00230.x. [DOI] [PubMed] [Google Scholar]
- 55.Toskes PP. Bacterial overgrowth of the gastrointestinal tract. Advances in internal medicine. 1993;38:387–407. [PubMed] [Google Scholar]
- 56.Simren M, Barbara G, Flint HJ, et al. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62:159–176. doi: 10.1136/gutjnl-2012-302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kashyap PC, Marcobal A, Ursell LK, et al. Complex Interactions Among Diet, Gastrointestinal Transit, and Gut Microbiota in Humanized Mice. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McVey Neufeld KA, Mao YK, Bienenstock J, Foster JA, Kunze WA. The microbiome is essential for normal gut intrinsic primary afferent neuron excitability in the mouse. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2013;25:183–e188. doi: 10.1111/nmo.12049. [DOI] [PubMed] [Google Scholar]
- 59.Wu RY, Pasyk M, Wang B, et al. Spatiotemporal maps reveal regional differences in the effects on gut motility for Lactobacillus reuteri and rhamnosus strains. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2013 doi: 10.1111/nmo.12072. [DOI] [PubMed] [Google Scholar]
- 60.Ward NL, Pieretti A, Dowd SE, Cox SB, Goldstein AM. Intestinal aganglionosis is associated with early and sustained disruption of the colonic microbiome. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2012;24:874–e400. doi: 10.1111/j.1365-2982.2012.01937.x. [DOI] [PubMed] [Google Scholar]
- 61.Carvalho FA, Aitken JD, Vijay-Kumar M, Gewirtz AT. Toll-like receptor-gut microbiota interactions: perturb at your own risk! Annual review of physiology. 2012;74:177–198. doi: 10.1146/annurev-physiol-020911-153330. [DOI] [PubMed] [Google Scholar]
- 62.Pimentel M, Lin HC, Enayati P, et al. Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. American journal of physiology Gastrointestinal and liver physiology. 2006;290:G1089–1095. doi: 10.1152/ajpgi.00574.2004. [DOI] [PubMed] [Google Scholar]
- 63.McKay LF, Holbrook WP, Eastwood MA. Methane and hydrogen production by human intestinal anaerobic bacteria. Acta pathologica, microbiologica, et immunologica Scandinavica Section B, Microbiology. 1982;90:257–260. doi: 10.1111/j.1699-0463.1982.tb00114.x. [DOI] [PubMed] [Google Scholar]
- 64.Dhaese I, Van Colen I, Lefebvre RA. Mechanisms of action of hydrogen sulfide in relaxation of mouse distal colonic smooth muscle. European journal of pharmacology. 2010;628:179–186. doi: 10.1016/j.ejphar.2009.11.024. [DOI] [PubMed] [Google Scholar]
- 65.Nagao M, Linden DR, Duenes JA, Sarr MG. Mechanisms of action of the gasotransmitter hydrogen sulfide in modulating contractile activity of longitudinal muscle of rat ileum. Journal of gastrointestinal surgery : official journal of the Society for Surgery of the Alimentary Tract. 2011;15:12–22. doi: 10.1007/s11605-010-1306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gallego D, Clave P, Donovan J, et al. The gaseous mediator, hydrogen sulphide, inhibits in vitro motor patterns in the human, rat and mouse colon and jejunum. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2008;20:1306–1316. doi: 10.1111/j.1365-2982.2008.01201.x. [DOI] [PubMed] [Google Scholar]
- 67.Linden DR, Levitt MD, Farrugia G, Szurszewski JH. Endogenous production of H2S in the gastrointestinal tract: still in search of a physiologic function. Antioxidants & redox signaling. 2010;12:1135–1146. doi: 10.1089/ars.2009.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Picton R, Eggo MC, Langman MJ, Singh S. Impaired detoxication of hydrogen sulfide in ulcerative colitis? Digestive diseases and sciences. 2007;52:373–378. doi: 10.1007/s10620-006-9529-y. [DOI] [PubMed] [Google Scholar]
- 69.Irie K, Ekuni D, Tomofuji T, et al. Combined effects of hydrogen sulfide and lipopolysaccharide on osteoclast differentiation in rats. Journal of periodontology. 2012;83:522–527. doi: 10.1902/jop.2011.110315. [DOI] [PubMed] [Google Scholar]
- 70.Jahng J, Jung IS, Choi EJ, Conklin JL, Park H. The effects of methane and hydrogen gases produced by enteric bacteria on ileal motility and colonic transit time. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2012;24:185–190. e192. doi: 10.1111/j.1365-2982.2011.01819.x. [DOI] [PubMed] [Google Scholar]
- 71.Gibson GR, Cummings JH, Macfarlane GT. Competition for hydrogen between sulphate-reducing bacteria and methanogenic bacteria from the human large intestine. The Journal of applied bacteriology. 1988;65:241–247. doi: 10.1111/j.1365-2672.1988.tb01891.x. [DOI] [PubMed] [Google Scholar]
- 72.Institute of Medicine (U.S.) Panel on Dietary Reference Intakes for Electrolytes and Water. DRI, dietary reference intakes for water, potassium, sodium, chloride, and sulfate. National Academies Press; Washington, D.C.: 2005. [Google Scholar]
- 73.Fukumoto S, Tatewaki M, Yamada T, et al. Short-chain fatty acids stimulate colonic transit via intraluminal 5-HT release in rats. American journal of physiology Regulatory, integrative and comparative physiology. 2003;284:R1269–1276. doi: 10.1152/ajpregu.00442.2002. [DOI] [PubMed] [Google Scholar]
- 74.Grider JR, Piland BE. The peristaltic reflex induced by short-chain fatty acids is mediated by sequential release of 5-HT and neuronal CGRP but not BDNF. American journal of physiology Gastrointestinal and liver physiology. 2007;292:G429–437. doi: 10.1152/ajpgi.00376.2006. [DOI] [PubMed] [Google Scholar]
- 75.Essien B, Grasberger H, Romain RD, et al. ZBP-89 Regulates Expression of Tryptophan Hydroxylase I and Mucosal Defense Against Salmonella Typhimurium in Mice. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martin FP, Wang Y, Sprenger N, et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Molecular systems biology. 2008;4:157. doi: 10.1038/msb4100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takaki M, Mawe GM, Barasch JM, Gershon MD, Gershon MD. Physiological responses of guinea-pig myenteric neurons secondary to the release of endogenous serotonin by tryptamine. Neuroscience. 1985;16:223–240. doi: 10.1016/0306-4522(85)90059-4. [DOI] [PubMed] [Google Scholar]
- 78.Yurdaydin C, Walsh TJ, Engler HD, et al. Gut bacteria provide precursors of benzodiazepine receptor ligands in a rat model of hepatic encephalopathy. Brain research. 1995;679:42–48. doi: 10.1016/0006-8993(95)00241-h. [DOI] [PubMed] [Google Scholar]
- 79.Sayin SI, Wahlstrom A, Felin J, et al. Gut Microbiota Regulates Bile Acid Metabolism by Reducing the Levels of Tauro-beta-muricholic Acid, a Naturally Occurring FXR Antagonist. Cell metabolism. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 80.Kirwan WO, Smith AN, Mitchell WD, Falconer JD, Eastwood MA. Bile acids and colonic motility in the rabbit and the human. Gut. 1975;16:894–902. doi: 10.1136/gut.16.11.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nature reviews Microbiology. 2012;10:735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- 82.Cryan JF, O'Mahony SM. The microbiome-gut-brain axis: from bowel to behavior. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2011;23:187–192. doi: 10.1111/j.1365-2982.2010.01664.x. [DOI] [PubMed] [Google Scholar]
- 83.Nishino R, Mikami K, Takahashi H, et al. Commensal microbiota modulate murine behaviors in a strictly contamination-free environment confirmed by culture-based methods. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2013 doi: 10.1111/nmo.12110. [DOI] [PubMed] [Google Scholar]
- 84.Diaz Heijtz R, Wang S, Anuar F, et al. Normal gut microbiota modulates brain development and behavior. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bercik P, Park AJ, Sinclair D, et al. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2011;23:1132–1139. doi: 10.1111/j.1365-2982.2011.01796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun Y, Zhang M, Chen CC, et al. Stress-Induced Corticotropin-Releasing Hormone-Mediated NLRP6 Inflammasome Inhibition and Transmissible Enteritis in Mice. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bangsgaard Bendtsen KM, Krych L, Sorensen DB, et al. Gut microbiota composition is correlated to grid floor induced stress and behavior in the BALB/c mouse. PloS one. 2012;7:e46231. doi: 10.1371/journal.pone.0046231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clinical microbiology reviews. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheng AS, Li MS, Kang W, et al. Helicobacter pylori Causes Epigenetic Dysregulation of FOXD3 to Promote Gastric Carcinogenesis. Gastroenterology. 2013;144:122–133. e129. doi: 10.1053/j.gastro.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 90.Olszak T, An D, Zeissig S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ordog T, Syed SA, Hayashi Y, Asuzu DT. Epigenetics and chromatin dynamics: a review and a paradigm for functional disorders. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2012;24:1054–1068. doi: 10.1111/nmo.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sperandio V, Torres AG, Giron JA, Kaper JB. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. Journal of bacteriology. 2001;183:5187–5197. doi: 10.1128/JB.183.17.5187-5197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Carter GP, Purdy D, Williams P, Minton NP. Quorum sensing in Clostridium difficile: analysis of a luxS-type signalling system. Journal of medical microbiology. 2005;54:119–127. doi: 10.1099/jmm.0.45817-0. [DOI] [PubMed] [Google Scholar]
- 94.Anbazhagan D, Mansor M, Yan GO, Md Yusof MY, Hassan H, Sekaran SD. Detection of quorum sensing signal molecules and identification of an autoinducer synthase gene among biofilm forming clinical isolates of Acinetobacter spp. PloS one. 2012;7:e36696. doi: 10.1371/journal.pone.0036696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hartmann A, Schikora A. Quorum sensing of bacteria and trans-kingdom interactions of N-acyl homoserine lactones with eukaryotes. Journal of chemical ecology. 2012;38:704–713. doi: 10.1007/s10886-012-0141-7. [DOI] [PubMed] [Google Scholar]
- 96.Kravchenko VV, Kaufmann GF, Mathison JC, et al. Modulation of gene expression via disruption of NF-kappaB signaling by a bacterial small molecule. Science. 2008;321:259–263. doi: 10.1126/science.1156499. [DOI] [PubMed] [Google Scholar]