Abstract
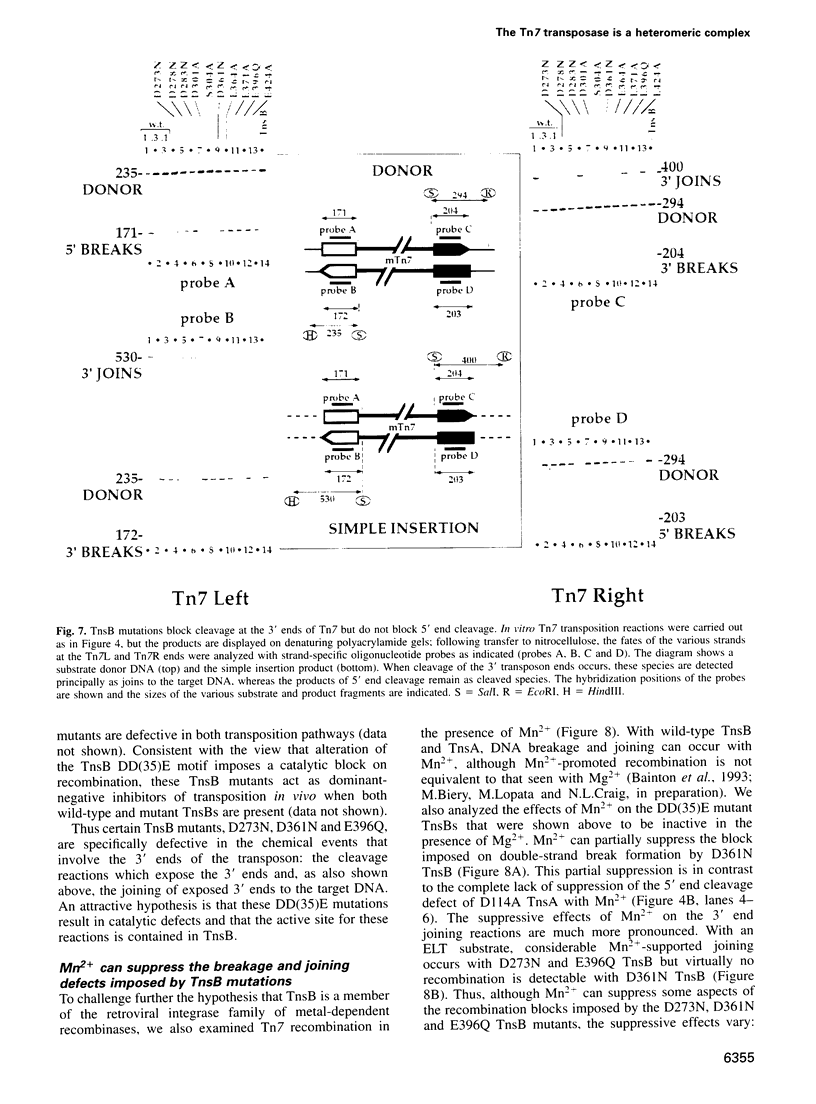
The bacterial transposon Tn7 translocates by a cut and paste mechanism: excision from the donor site results from double-strand breaks at each end of Tn7 and target insertion results from joining of the exposed 3' Tn7 tips to the target DNA. Through site-directed mutagenesis of the Tn7-encoded transposition proteins TnsA and TnsB, we demonstrate that the Tn7 transposase is a heteromeric complex of these proteins, each protein executing different DNA processing reactions. TnsA mediates DNA cleavage reactions at the 5' ends of Tn7, and TnsB mediates DNA breakage and joining reactions at the 3' ends of Tn7. Thus the double-strand breaks that underlie Tn7 excision result from a collaboration between two active sites, one in TnsA and one in TnsB; the same (or a closely related) active site in TnsB also mediates the subsequent joining of the 3' ends to the target. Both TnsA and TnsB appear to be members of the retroviral integrase superfamily: mutation of their putative DD(35)E motifs blocks catalytic activity. Recombinases of this class require a divalent metal cofactor that is thought to interact with these acidic residues. Through analysis of the metal ion specificity of a TnsA mutant containing a sulfur (cysteine) substitution, we provide evidence that a divalent metal actually interacts with these acidic amino acids.
Full text
PDF




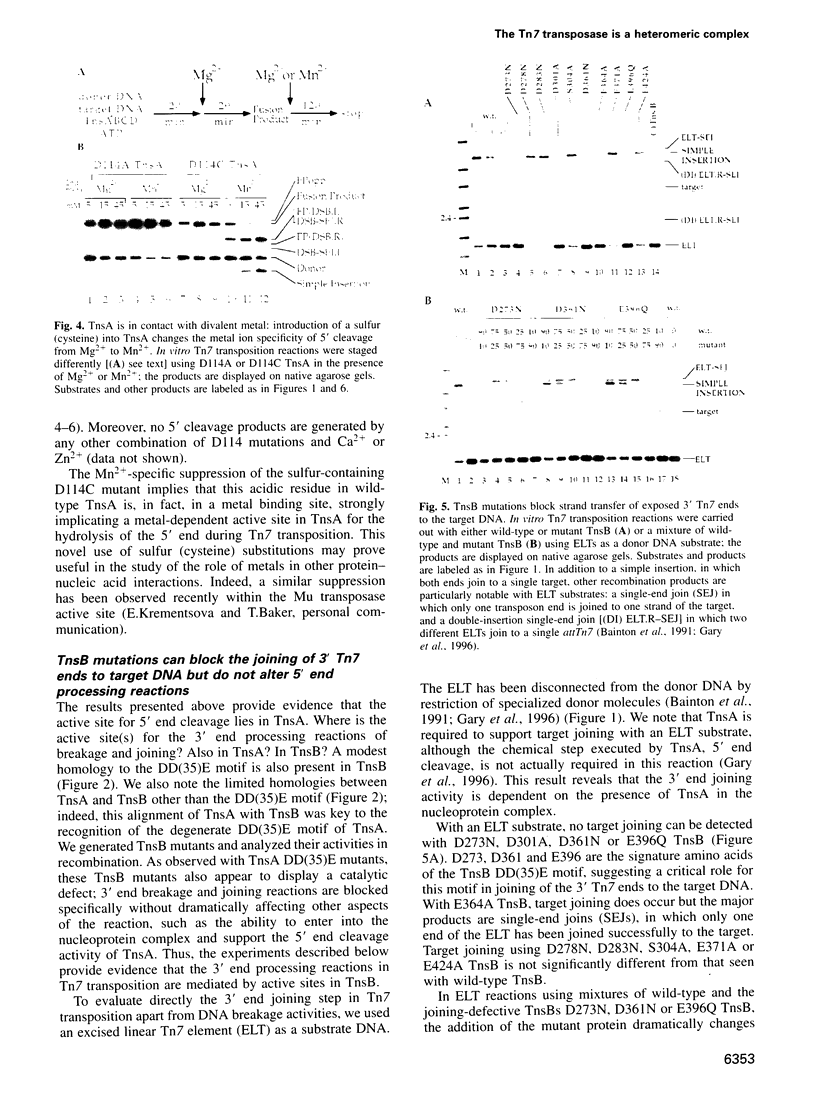







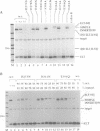
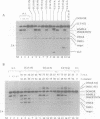
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldaz H., Schuster E., Baker T. A. The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell. 1996 Apr 19;85(2):257–269. doi: 10.1016/s0092-8674(00)81102-2. [DOI] [PubMed] [Google Scholar]
- Arciszewska L. K., Craig N. L. Interaction of the Tn7-encoded transposition protein TnsB with the ends of the transposon. Nucleic Acids Res. 1991 Sep 25;19(18):5021–5029. doi: 10.1093/nar/19.18.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arciszewska L. K., Drake D., Craig N. L. Transposon Tn7. cis-Acting sequences in transposition and transposition immunity. J Mol Biol. 1989 May 5;207(1):35–52. doi: 10.1016/0022-2836(89)90439-7. [DOI] [PubMed] [Google Scholar]
- Arciszewska L. K., McKown R. L., Craig N. L. Purification of TnsB, a transposition protein that binds to the ends of Tn7. J Biol Chem. 1991 Nov 15;266(32):21736–21744. [PubMed] [Google Scholar]
- Arciszewska L. K., Sherratt D. J. Xer site-specific recombination in vitro. EMBO J. 1995 May 1;14(9):2112–2120. doi: 10.1002/j.1460-2075.1995.tb07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur A., Sherratt D. Dissection of the transposition process: a transposon-encoded site-specific recombination system. Mol Gen Genet. 1979 Oct 1;175(3):267–274. doi: 10.1007/BF00397226. [DOI] [PubMed] [Google Scholar]
- Bainton R. J., Kubo K. M., Feng J. N., Craig N. L. Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993 Mar 26;72(6):931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- Bainton R., Gamas P., Craig N. L. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell. 1991 May 31;65(5):805–816. doi: 10.1016/0092-8674(91)90388-f. [DOI] [PubMed] [Google Scholar]
- Baker T. A., Kremenstova E., Luo L. Complete transposition requires four active monomers in the mu transposase tetramer. Genes Dev. 1994 Oct 15;8(20):2416–2428. doi: 10.1101/gad.8.20.2416. [DOI] [PubMed] [Google Scholar]
- Baker T. A., Luo L. Identification of residues in the Mu transposase essential for catalysis. Proc Natl Acad Sci U S A. 1994 Jul 5;91(14):6654–6658. doi: 10.1073/pnas.91.14.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. A., Mizuuchi K. DNA-promoted assembly of the active tetramer of the Mu transposase. Genes Dev. 1992 Nov;6(11):2221–2232. doi: 10.1101/gad.6.11.2221. [DOI] [PubMed] [Google Scholar]
- Baker T. A. Protein-DNA assemblies controlling lytic development of bacteriophage Mu. Curr Opin Genet Dev. 1993 Oct;3(5):708–712. doi: 10.1016/s0959-437x(05)80087-5. [DOI] [PubMed] [Google Scholar]
- Barth P. T., Datta N., Hedges R. W., Grinter N. J. Transposition of a deoxyribonucleic acid sequence encoding trimethoprim and streptomycin resistances from R483 to other replicons. J Bacteriol. 1976 Mar;125(3):800–810. doi: 10.1128/jb.125.3.800-810.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall E. L., Admon A., Rio D. C. A Drosophila protein homologous to the human p70 Ku autoimmune antigen interacts with the P transposable element inverted repeats. Proc Natl Acad Sci U S A. 1994 Dec 20;91(26):12681–12685. doi: 10.1073/pnas.91.26.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely G., May G., McCulloch R., Arciszewska L. K., Burke M., Lovett S. T., Sherratt D. J. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K12. Cell. 1993 Oct 22;75(2):351–361. doi: 10.1016/0092-8674(93)80076-q. [DOI] [PubMed] [Google Scholar]
- Bolland S., Kleckner N. The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell. 1996 Jan 26;84(2):223–233. doi: 10.1016/s0092-8674(00)80977-0. [DOI] [PubMed] [Google Scholar]
- Bujacz G., Jaskólski M., Alexandratos J., Wlodawer A., Merkel G., Katz R. A., Skalka A. M. The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations. Structure. 1996 Jan 15;4(1):89–96. doi: 10.1016/s0969-2126(96)00012-3. [DOI] [PubMed] [Google Scholar]
- Chalmers R. M., Kleckner N. Tn10/IS10 transposase purification, activation, and in vitro reaction. J Biol Chem. 1994 Mar 18;269(11):8029–8035. [PubMed] [Google Scholar]
- Colloms S. D., McCulloch R., Grant K., Neilson L., Sherratt D. J. Xer-mediated site-specific recombination in vitro. EMBO J. 1996 Mar 1;15(5):1172–1181. [PMC free article] [PubMed] [Google Scholar]
- Craig N. L. Transposon Tn7. Curr Top Microbiol Immunol. 1996;204:27–48. doi: 10.1007/978-3-642-79795-8_2. [DOI] [PubMed] [Google Scholar]
- Craig N. L. Unity in transposition reactions. Science. 1995 Oct 13;270(5234):253–254. doi: 10.1126/science.270.5234.253. [DOI] [PubMed] [Google Scholar]
- Craig N. L. V(D)J recombination and transposition: closer than expected. Science. 1996 Mar 15;271(5255):1512–1512. doi: 10.1126/science.271.5255.1512. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Mechanism of transposition of bacteriophage Mu: structure of a transposition intermediate. Cell. 1985 Jul;41(3):867–876. doi: 10.1016/s0092-8674(85)80067-2. [DOI] [PubMed] [Google Scholar]
- Craigie R. Quality control in Mu DNA transposition. Cell. 1996 Apr 19;85(2):137–140. doi: 10.1016/s0092-8674(00)81089-2. [DOI] [PubMed] [Google Scholar]
- Dahm S. C., Uhlenbeck O. C. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry. 1991 Oct 1;30(39):9464–9469. doi: 10.1021/bi00103a011. [DOI] [PubMed] [Google Scholar]
- Dyda F., Hickman A. B., Jenkins T. M., Engelman A., Craigie R., Davies D. R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994 Dec 23;266(5193):1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- Engelman A., Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992 Nov;66(11):6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A., Mizuuchi K., Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991 Dec 20;67(6):1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- Fayet O., Ramond P., Polard P., Prère M. F., Chandler M. Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol Microbiol. 1990 Oct;4(10):1771–1777. doi: 10.1111/j.1365-2958.1990.tb00555.x. [DOI] [PubMed] [Google Scholar]
- Flores C., Qadri M. I., Lichtenstein C. DNA sequence analysis of five genes; tnsA, B, C, D and E, required for Tn7 transposition. Nucleic Acids Res. 1990 Feb 25;18(4):901–911. doi: 10.1093/nar/18.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P., Craig N. L. Purification and characterization of TnsC, a Tn7 transposition protein that binds ATP and DNA. Nucleic Acids Res. 1992 May 25;20(10):2525–2532. doi: 10.1093/nar/20.10.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary P. A., Biery M. C., Bainton R. J., Craig N. L. Multiple DNA processing reactions underlie Tn7 transposition. J Mol Biol. 1996 Mar 29;257(2):301–316. doi: 10.1006/jmbi.1996.0164. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Leschziner A. E. DNA transposition: from a black box to a color monitor. Cell. 1995 Dec 29;83(7):1063–1066. doi: 10.1016/0092-8674(95)90132-9. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Benjamin H. W., Kleckner N. Kinetic and structural analysis of a cleaved donor intermediate and a strand transfer intermediate in Tn10 transposition. Cell. 1991 Jan 11;64(1):171–179. doi: 10.1016/0092-8674(91)90218-n. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Chelouche A. R., Kleckner N. A specific class of IS10 transposase mutants are blocked for target site interactions and promote formation of an excised transposon fragment. Cell. 1989 Oct 20;59(2):385–394. doi: 10.1016/0092-8674(89)90299-7. [DOI] [PubMed] [Google Scholar]
- Jayaram M. Phosphoryl transfer in Flp recombination: a template for strand transfer mechanisms. Trends Biochem Sci. 1994 Feb;19(2):78–82. doi: 10.1016/0968-0004(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- Kaufman P. D., Rio D. C. P element transposition in vitro proceeds by a cut-and-paste mechanism and uses GTP as a cofactor. Cell. 1992 Apr 3;69(1):27–39. doi: 10.1016/0092-8674(92)90116-t. [DOI] [PubMed] [Google Scholar]
- Kim K., Namgoong S. Y., Jayaram M., Harshey R. M. Step-arrest mutants of phage Mu transposase. Implications in DNA-protein assembly, Mu end cleavage, and strand transfer. J Biol Chem. 1995 Jan 20;270(3):1472–1479. doi: 10.1074/jbc.270.3.1472. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Chalmers R. M., Kwon D., Sakai J., Bolland S. Tn10 and IS10 transposition and chromosome rearrangements: mechanism and regulation in vivo and in vitro. Curr Top Microbiol Immunol. 1996;204:49–82. doi: 10.1007/978-3-642-79795-8_3. [DOI] [PubMed] [Google Scholar]
- Kruklitis R., Nakai H. Participation of the bacteriophage Mu A protein and host factors in the initiation of Mu DNA synthesis in vitro. J Biol Chem. 1994 Jun 10;269(23):16469–16477. [PubMed] [Google Scholar]
- Kubo K. M., Craig N. L. Bacterial transposon Tn7 utilizes two different classes of target sites. J Bacteriol. 1990 May;172(5):2774–2778. doi: 10.1128/jb.172.5.2774-2778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J., Jones K. S., Katz R. A., Mack J. P., Skalka A. M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992 May;12(5):2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B. D., Chaconas G. Transposition of phage Mu DNA. Curr Top Microbiol Immunol. 1996;204:83–102. doi: 10.1007/978-3-642-79795-8_4. [DOI] [PubMed] [Google Scholar]
- Leavitt A. D., Shiue L., Varmus H. E. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem. 1993 Jan 25;268(3):2113–2119. [PubMed] [Google Scholar]
- May E. W., Craig N. L. Switching from cut-and-paste to replicative Tn7 transposition. Science. 1996 Apr 19;272(5260):401–404. doi: 10.1126/science.272.5260.401. [DOI] [PubMed] [Google Scholar]
- McBlane J. F., van Gent D. C., Ramsden D. A., Romeo C., Cuomo C. A., Gellert M., Oettinger M. A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995 Nov 3;83(3):387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- McKown R. L., Orle K. A., Chen T., Craig N. L. Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. J Bacteriol. 1988 Jan;170(1):352–358. doi: 10.1128/jb.170.1.352-358.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Adzuma K. Inversion of the phosphate chirality at the target site of Mu DNA strand transfer: evidence for a one-step transesterification mechanism. Cell. 1991 Jul 12;66(1):129–140. doi: 10.1016/0092-8674(91)90145-o. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. Polynucleotidyl transfer reactions in transpositional DNA recombination. J Biol Chem. 1992 Oct 25;267(30):21273–21276. [PubMed] [Google Scholar]
- Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Zenilman M., Ohtsubo H., McCormick M., Machida C., Machida Y. Mechanism of insertion and cointegration mediated by IS1 and Tn3. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):283–295. doi: 10.1101/sqb.1981.045.01.041. [DOI] [PubMed] [Google Scholar]
- Orle K. A., Craig N. L. Identification of transposition proteins encoded by the bacterial transposon Tn7. Gene. 1991 Jul 31;104(1):125–131. doi: 10.1016/0378-1119(91)90478-t. [DOI] [PubMed] [Google Scholar]
- Pecoraro V. L., Hermes J. D., Cleland W. W. Stability constants of Mg2+ and Cd2+ complexes of adenine nucleotides and thionucleotides and rate constants for formation and dissociation of MgATP and MgADP. Biochemistry. 1984 Oct 23;23(22):5262–5271. doi: 10.1021/bi00317a026. [DOI] [PubMed] [Google Scholar]
- Piccirilli J. A., Vyle J. S., Caruthers M. H., Cech T. R. Metal ion catalysis in the Tetrahymena ribozyme reaction. Nature. 1993 Jan 7;361(6407):85–88. doi: 10.1038/361085a0. [DOI] [PubMed] [Google Scholar]
- Polard P., Chandler M. Bacterial transposases and retroviral integrases. Mol Microbiol. 1995 Jan;15(1):13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- Rice P., Mizuuchi K. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell. 1995 Jul 28;82(2):209–220. doi: 10.1016/0092-8674(95)90308-9. [DOI] [PubMed] [Google Scholar]
- Rogers M., Ekaterinaki N., Nimmo E., Sherratt D. Analysis of Tn7 transposition. Mol Gen Genet. 1986 Dec;205(3):550–556. doi: 10.1007/BF00338097. [DOI] [PubMed] [Google Scholar]
- Rowland S. J., Dyke K. G. Characterization of the staphylococcal beta-lactamase transposon Tn552. EMBO J. 1989 Sep;8(9):2761–2773. doi: 10.1002/j.1460-2075.1989.tb08418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland S. J., Dyke K. G. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol. 1990 Jun;4(6):961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- Rådström P., Sköld O., Swedberg G., Flensburg J., Roy P. H., Sundström L. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J Bacteriol. 1994 Jun;176(11):3257–3268. doi: 10.1128/jb.176.11.3257-3268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savilahti H., Mizuuchi K. Mu transpositional recombination: donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell. 1996 Apr 19;85(2):271–280. doi: 10.1016/s0092-8674(00)81103-4. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Lichtenstein C., Cotterill S. Purification and characterisation of the TnsB protein of Tn7: a transposition protein that binds to the ends of Tn7. Nucleic Acids Res. 1991 Jun 25;19(12):3395–3402. doi: 10.1093/nar/19.12.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermote C. L., Halford S. E. EcoRV restriction endonuclease: communication between catalytic metal ions and DNA recognition. Biochemistry. 1992 Jul 7;31(26):6082–6089. doi: 10.1021/bi00141a018. [DOI] [PubMed] [Google Scholar]
- Vink C., Yeheskiely E., van der Marel G. A., van Boom J. H., Plasterk R. H. Site-specific hydrolysis and alcoholysis of human immunodeficiency virus DNA termini mediated by the viral integrase protein. Nucleic Acids Res. 1991 Dec 25;19(24):6691–6698. doi: 10.1093/nar/19.24.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos J. C., De Baere I., Plasterk R. H. Transposase is the only nematode protein required for in vitro transposition of Tc1. Genes Dev. 1996 Mar 15;10(6):755–761. doi: 10.1101/gad.10.6.755. [DOI] [PubMed] [Google Scholar]
- Vos J. C., Plasterk R. H. Tc1 transposase of Caenorhabditis elegans is an endonuclease with a bipartite DNA binding domain. EMBO J. 1994 Dec 15;13(24):6125–6132. doi: 10.1002/j.1460-2075.1994.tb06959.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell C. S., Craig N. L. Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. Genes Dev. 1988 Feb;2(2):137–149. doi: 10.1101/gad.2.2.137. [DOI] [PubMed] [Google Scholar]
- Yang J. Y., Kim K., Jayaram M., Harshey R. M. A domain sharing model for active site assembly within the Mu A tetramer during transposition: the enhancer may specify domain contributions. EMBO J. 1995 May 15;14(10):2374–2384. doi: 10.1002/j.1460-2075.1995.tb07232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D. C., Groeneger A. A., Plasterk R. H. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D. C., Mizuuchi K., Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996 Mar 15;271(5255):1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- van Luenen H. G., Colloms S. D., Plasterk R. H. The mechanism of transposition of Tc3 in C. elegans. Cell. 1994 Oct 21;79(2):293–301. doi: 10.1016/0092-8674(94)90198-8. [DOI] [PubMed] [Google Scholar]