Abstract
In order to replicate, a retrovirus must integrate a DNA copy of the viral RNA genome into a chromosome of the host cell. The study of retroviral integration has advanced considerably in the last few years. Here we focus on host factor interactions and the linked area of integration targeting. Genome-wide screens for cellular factors affecting HIV replication have identified a series of host cell proteins that may mediate subcellular trafficking of integration complexes, nuclear import, and integration target site selection. The cell transcriptional co-activator protein LEDGF/p75 has been identified as a tethering factor important for HIV integration, and recently, BET proteins (Brd2, 4, and 4) have been identified as tethering factors for the gammaretroviruses. A new class of HIV inhibitors has been developed targeting the HIV-1 IN-LEDGF binding site, though surprisingly these inhibitors appear to block assembly late during replication and do not act at the integration step. Going forward, genome-wide studies of HIV-host interactions offer many new starting points to investigate HIV replication and identify potential new inhibitor targets.
Introduction
Retroviruses integrate a DNA copy of the viral genome into cellular DNA as an obligatory step in the viral replication cycle. Once integrated, the viral DNA is stably replicated with cellular DNA through cycles of DNA replication and cell division. The first clues regarding the mechanism of integration came from genetic experiments (1–3). Mutations at two locations within the viral genome resulted in a phenotype in which reverse transcription occured normally but the viral DNA failed to integrate. These mutations mapped to regions which we now know encode the viral integrase (IN) protein and the ends of the viral DNA sequence recognized by IN. The finding that viral DNA within extracts of infected cells efficiently integrated into exogenously added target DNA in vitro (4–6) facilitated biochemical studies of integration. This in vitro integration system enabled the DNA breaking and joining events to be unambiguously determined (6, 7). It also established that the viral DNA forms part of a large nucleoprotein complex termed the preintegration complex (PIC) (8). Later biochemical experiments showed that viral IN protein is necessary and sufficient to carry out the DNA cutting and joining steps of integration in the presence of a divalent metal ion (9–13). Subsequent studies established reaction conditions that facilitated efficient concerted integration of both viral DNA ends into the target DNA molecule in vitro (14–19).
This chapter focuses on mechanisms of targeting integration and the contributions of viral and cellular proteins. For structural information on nucleoprotein complexes involved in retroviral DNA integration see the chapter by Engelman and Cherepanov. For detailed discussions of the mechanisms of DNA transposition of related elements see other chapters in Mobile DNA III.
Composition of preintegration complexes
The composition and architecture of PICs remains poorly understood, largely due to their low abundance in extracts of infected cells; typically only a few PICs are present per cell. Knowledge of the protein composition is largely limited to immunoprecipitation studies which can identify protein components, but not their abundance, organization or functional roles. Even then, some identified components border on the limits of detection. Viral DNA within PICs is much easier to monitor because of highly sensitive assays such as Southern blotting and PCR. PICs have been valuable for studying the fate of the viral DNA in vitro and defining the DNA cutting and joining steps (6, 7).
A near-full complement of viral proteins have been detected within HIV-1 PICs including IN, RT, CA, NC, Vpr, and PR (20–27). Whereas IN, MA, RT, and Vpr are reported to be present in most studies, CA, NC, and PR are only detected in a few studies. These inconsistencies are likely due to differences in methods of preparation and sensitivity of the assays. Cellular proteins implied to be associated with PICs by biochemical studies include LEDGF/p75 (28), BAF (29), and HMGA1 (30). Mass spectroscopy screens have identified many additional cellular proteins potentially associated with HIV-1 PICs (31, 32). The functional role, if any, of the majority of these proteins is unknown.
The key viral protein within PICs is IN, which is responsible for the initial steps of covalent joining of viral DNA into host DNA. CA within PICs has been implicated in nuclear import (see below). Reverse transcriptase is also present in PICs but its unclear if it plays any functional role once reverse transcription is complete. Functional studies of HIV-1 and MLV PICs demonstrate a tight association of IN to the viral DNA--PICs remain active for in vitro DNA integration even after treatment with greater than 500 mM NaCl. A tetramer of HIV-1 IN forms a tight complex with viral DNA ends in vitro (33) and this likely bridges the viral DNA ends in the PIC. Footprinting of the viral DNA within MLV and HIV PICs (34–36) revealed extensive protection extending up to approximately 1 kb from the viral DNA ends. This protection is dependent on IN and is not observed in PICs derived from virus that lack IN. However, it is unclear if the protection is mediated by IN itself or recruitment of other factors by IN.
The viral DNA within PICs is sensitive to endonucleases suggesting that it adopts a relatively open structure (27). In contrast, it is not sensitive to digestion with exonucleases (27), as expected if the DNA ends are stably bridged by IN. A cellular factor termed barrier-to-autointegration factor (BAF) protects MLV PICs from autointegration in in vitro PIC integration assays (37). BAF is an essential cellular protein involved in DNA management within the nucleus (38, 39) which makes it difficult to study the effects of BAF depletion on integration in the cell. BAF is a DNA bridging protein that compacts DNA (40). Stripping BAF from MLV PICs by high-salt treatment leads to slower sedimentation in sucrose gradients (41), consistent with decompaction of the viral DNA. Addition of BAF to salt-stripped PICs restores the normal sedimentation behavior in parallel with restoring the preference for intermolecular integration and avoidance of autointegration. Consistent with these findings, treatment of MLV PICs with VRK1, the major kinase responsible for phosphorylation of BAF, promotes autointegration in a similar manner as salt- stripping (42). Since phosphorylation of BAF abrogates DNA binding (40, 43) it is expected that VRK1 would promote loss of BAF from the viral DNA within the PIC. A similar role of BAF in preventing autointegration of HIV-1 PICs has not been unambiguously demonstrated.
Nuclear import of PICs
The large size of PICs presents a formidable challenge to nuclear entry. In the case of gammaretroviruses, the chromosomes are accessed during mitosis when the nuclear enveloped is disassembled. However HIV-1 and other lentiviruses can integrate in non-dividing cells, so entry into the nucleus must be an active process involving the nuclear pores. HIV-1 PICs have comparable sedimentation properties to ribosomes (44) and an estimated Stokes radius of about 28 nM based on size exclusion chromatography (27). The mechanism has been the subject of a controversy that is still not completely resolved. Many of the protein components of the PIC possess nuclear localization signals that, when fused to normally cytoplasmic proteins result in their localization to the nucleus. MA (45);, IN (23, 46), and Vpr (24, 47, 48) have all been proposed to mediate nuclear localization of HIV-1 PICs, as has a DNA structure near the center of the viral DNA called the central flap (49). However, other studies have shown that each of these can be dispensible for nuclear entry under some circumstances. It is possible that the interpretation of the data may be complicated by redundancy of nuclear import mechanisms. More recent data strongly suggests an important role for CA in mediating nuclear entry (reviewed in (50)).
The involvement of CA in nuclear entry was suggested by the finding that infection by chimeric HIV-1 harboring an MLV CA is cell cycle dependent (51) and this conclusion has been further substantiated by subsequent studies. Certain point mutations in HIV-1 CA also confer a cell cycle dependent phenotype. Genome wide screens for cellular co-factors identified a large number proteins known to be involved in nuclear import and related functions (52–55). Of these, TNPO3, NUP358 and NUP153 are of particular interest. Knockdown of these proteins diminishes HIV-1 replication, but does not affect replication of MLV, suggesting the mechanism is specific for HIV-1 rather than an indirect effect on cellular function (53, 56–59) . Experiments with HIV-1/MLV chimeric viruses map the TNPO3 and NUP153 dependency to CA (56, 58).
Host factors affecting integration targeting
Intranuclear trafficking
Following import into the nucleus, the PIC must access the chromosomes of an infected cell. Though not fully clarified, several studies suggest that this may not be purely diffusion but rather a controlled process (60, 61). Imaging studies have suggested that integration may commonly be localized within the nucleus, particularly at the nuclear periphery (62). Consistent with this idea, data suggest that active genes may be enriched near the nuclear periphery in yeast (63); however, other data from studies in animal cells suggest that in fact active genes may be enriched more in the center of the nucleus (64–66).
A study of nuclear localization and integration targeting suggested a possible model in which retroviral integration complexes engaged a “railroad track” in the cytoplasm that transfer PICs to specific regions of the nucleus rich in favored sites of integration (57, 67). The CA mutant N74D, which alters the interaction with the cellular splicing factor CPSF6, also altered integration targeting, suggesting involvement of this system. Modulating the level or activity of the cyclophillin protein another protein that binds CA, also affected targeting. These findings point to greater involvement of CA in PIC functioning and targeting than was appreciated previously (57). Several additional nuclear-localized cell factors have been reported to influence integration (68–70) and could act at this step. More work is needed to clarify the mechanisms underlying the possible relationship between nuclear positioning of chromosomes and frequency of retroviral integration.
Favored target sites for retroviral DNA integration in cellular chromosomes
Studies mapping the distribution of integration sites in cellular chromosomes have led to a detailed picture of favored and disfavored target sites (for some previous reviews in this area see (33)(36)(71)). The first genome-wide mapping study, carried out shortly after the completion of the first draft human genome sequence, showed that HIV favored integration particularly strongly within transcription units of the SupT1 T-cell line(72). Comparison to microarray data quantifying transcriptional activity in the same cells showed that integration was particularly strongly favored in active transcription units. Subsequent studies have shown that this pattern is maintained in most cell lines studied (73, 74, 75, 76–80), with the exception of a few experimentally manipulated examples described below (Figure 1). A variety of features on the human genome correlate with active transcription units (81, 82) such as sites of histone modification including H3K4 mono and dimethylation, H3 K9 and K27 monomethylation, H3K36 trimethylation, DNAseI hypersensitive sites and sites of histone acetylation generally (67, 82, 83). In addition, gene-rich regions are associated with a syndrome of associated features, such as high G/C content, high Alu element content, low LINE element content, and Geimsa dark banding patterns, all of which are positively associated with integration (72).
Figure 1.
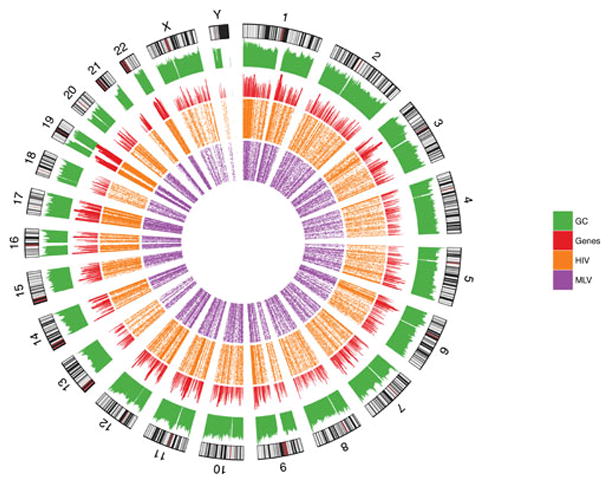
Sites of HIV DNA integration in chromosomes. The human chromosomes are shown in the outermost ring. The green histograms indicated relative G/C content; red indicates gene density, orange indicates density of HIV integration sites in T cells (78), and purple indicates MLV integration site density in T cells(115).
The above collection of favorable features characterizes all the lentiviruses studied to date, but other retroviral genera show different patterns. The gammaretroviruses, which include the MLVs and XMRV, show favored integration close to transcription start sites, rather than throughout transcription units as is seen with lentiviruses (74, 84). The alpha-retroviruses, which include RSV and the allied ALVs, show near random distributions (76, 85). A study of the viral determinants of integration targeting implicated IN as a major determinant.
Swapping the MLV (gammaretroviral) IN for that of HIV, in a background of an otherwise wild-type HIV provirus, resulted in chimeric viruses that showed the integration pattern expected for MLV (86). Although these viruses showed reduced ability to replicate, these studies suggested that IN was a major determinant of integration target site specificity. As is described below, targeting by the lentiviruses and gammaretroviruses can be understood as engagement of different cellular tethering factors that are distinctive for each group.
LEDGF/p75—a cellular tethering factor for HIV integration
The LEDGF/p75 protein (product of the PSIP1 gene) was first implicated in HIV replication when it was identified as a tight binder to HIV IN protein (87–89) These studies involved artificial systems, such as over-expression of IN in animal cells or yeast two-hybrid assays, leaving the significance of the interaction uncertain initially. However, subsequent experiments eventually documented that reduction of LEDGF/p75 function did have strong negative effects on viral replication (79, 90, 91), establishing the importance.
Mapping studies showed that LEDGF/p75 is comprised of several protein domains. Multiple N-terminal domains mediate chromatin binding, potentially in part to the H3 histone trimethylated at the K36 position (92, 93). The C-terminal domain binds IN (“integrase binding domain” or IBD). X-ray crystallography reveals that the LEDGF-IBD engages the catalytic and N-terminal domains of integrase. The most extensive interactions involve a pocket formed at the dimer interface of two monomers of the integrase catalytic domain and the tip of the IBD helical bundle structure (94, 95).
Given that LEDGF/p75 was a cellular protein implicated in both transcriptional regulation and HIV integration, it was of interest to test whether integration in active transcription units required LEDGF/p75. Knockdown studies showed that depleting LEDGF/p75 in fact caused a significant shift in integration targeting toward a more random pattern, consistent with the idea that the LEDGF/p75-IN interaction mediated targeting to transcription units (77, 79, 96, 97).
LEDGF/p75 is a member of a protein family, and the related protein HRP2 has also been shown to act as an integration tether. However, effects of HRP2 on HIV integration efficiency and targeting are only prominent in cells lacking LEDGF/p75, and the influence of HRP2 even in this setting is modest, supporting the idea that LEDGF/p75 is the more significant cofactor (98–101).
Evidence supporting the idea that LEDGF/p75 acts as a simple tether was provided by results with LEDGF/p75 chimeras (102–104). Several groups substituted new chromatin binding domains for the LEDGF/p75 N-terminal chromatin-binding domain. These chimeras, which retained the LEDGF/p75 IN binding C-terminal domain but grafted to new domains conferring new chromatin binding specificities, were introduced into cells depleted for wild-type LEDGF/p75, and analysis of HIV integration showed new favored locations. A variety of such novel tethers have now been reported, collectively supporting the idea that LEDGF/p75 acts as a tethering protein for PICs. Studies of the yeast Ty elements Ty1-5, described elsewhere in this book, have disclosed additional examples of targeting by tethering of integrase to cellular chromatin-binding proteins.
BET proteins—cellular factors directing gammaretroviral integration
The observation that HIV targeting is directed by tethering to LEDGF/p75 raised the question of whether other retroviral genera with different targeting preferences might dock with their own group-specific tethering factors. The gammaretroviruses favor integration near transcription start sites, and within the last year cellular BET proteins (Brd2, 3 and 4) have been identified as targeting factors (BET stands for “Bromodomain and extra terminal”, and Brd stands for “bromodomain containing”) (105–108) Brd2-4 are known to bind to sites of histone acetylation, explaining their association with transcription start regions. Initially these proteins were identified as binders to MLV IN in yeast two-hybrid screens (109), but the importance was uncertain. Later studies found that BET proteins also bound MLV IN in affinity capture-mass spec protocols, and that the addition of BET proteins could affect the activity of MLV IN in vitro (105–108).
Definitive data on the effects of BET proteins came with experiments modulating the activity of BET proteins in cells (105–108). Reducing BET protein activity was difficult with siRNA because three proteins were involved, and full elimination of BET activity was lethal to cells. A key breakthrough came with studies based on the anticancer drug JQ1, which binds to BET proteins and competes with binding of actetylated histone tails. This allowed activity of all BET proteins to be abrogated acutely, allowing experimental analysis of cells before the onset of lethality. Using JQ1, it was possible to show that the frequency of MLV integration was reduced in the region around transcription start sites. Effects could also be detected with siRNA knockdowns of the three genes, though with lower magnitude. In follow up experiments, hybrid tethering proteins were synthesized that contained the BET IN binding region and novel chromatin binding domains. In cells depleted for normal BET proteins but containing the chimeric tethers, integration sites were notably redistributed, again strongly supporting the tethering model.
Retroviral integration and nucleosomal association
Several studies have explored the association between nucleosome packaging and integration bias. Early studies suggesting a loose association of retroviral integration sites with chromosomal sites sensitive to DNAseI digestion (reviewed in (16, 19, 20, 34)), which led to the idea that packaging DNA in nucleosomes would inhibit integration. However, once assays were available that recapitulated integration in vitro, nucleosomal wrapping was found instead to favor integration (110–114). Subsequent studies in vivo have suggested that DNA predicted to be facing outward when wrapped on nucleosomal particles is favored for integration (82, 115). However, complicating the analysis, not all methods for calling nucleosome positions in vivo agree well, and experimental mapping of nucleosome positions is imperfect and fraught with lab-specific noise. Additional studies have investigated possible roles of cellular factors in modulating interactions with nucleosomes (116, 117). Different integrating elements, described elsewhere in this book, show a wide range of responses to nucleosome binding to integration target DNA.
Role of DNA repair enzymes in completing provirus formation
The initial steps in the integration reaction are catalyzed by the viral-encoded IN enzymes, but this only takes the reaction to the point of joining one DNA strand at each end of the provirus to host DNA. The integration intermediate produced by IN has DNA gaps at each host-virus DNA junction, requiring intervention of additional enzymes to generate the fully integrated provirus. Cocktails of host-encoded DNA repair enzymes have been shown to be able to carry out the needed polymerization, branch excision, and ligation steps in vitro on substrate DNAs modeling the expected gaps (118). These enzymes are ubiquitous in cell nuclei and quite active, so their involvement seems likely. However, another possibility is that the virus-encoded RT and IN enzymes actually complete gap repair. Some in vitro evidence supports this (119), although the observed enzymatic activities may not be adequately robust. So far no studies have succeeded in creating mutant combinations that result in production of stalled gapped intermediates inside cells, leaving the activities involved incompletely clarified. In addition, several further cellular DNA repair enzymes have been proposed to act on retroviral DNA and influence integration or replication (120–127).
Inhibitors of HIV integration
Inhibitors acting at the active site
Antiviral agents are now available that target many steps in the HIV replication cycle, greatly improving the quality of life of HIV-positive individuals, and IN is among the targets (128). Initial work improving assays for integration allowed high-throughput screening under relatively authentic conditions, yielding starting points for drug development (129–131). All IN inhibitors targeting the active site contain chemical groups that can bind both catalytic metal atoms together with pendant groups that contact other parts of the IN-DNA complex. Several chemical families were explored before the pyrimidinone group was identified. Extensive development eventually yielded raltegravir, which received FDA approval in 2007 (132).
Mechanistic studies show that raltegravir acts by blocking binding of the integration target DNA to the PIC (133). In addition, structural studies of raltegravir bound to the PFV IN/DNA complex show that the drug displaces the LTR 3′ end from its normal position in the active site, thereby obstructing the chemical steps of DNA transesterification (134).
As with all antiviral agents, resistance mutations have arisen in viruses in subjects treated with raltegravir. The effects of these amino acid substitutions can also be understood from the PFV structure as altering the drug binding site (see section by Engelman and Cherepanov). The observation of escape mutations has motivated a search for second generation IN inhibitors, resulting in promising new compounds such as dolutegravir, which shows high potency and favorable pharmacokinetic properties (135, 136).
Inhibitors acting at the LEDGF/p75 binding site on IN
Another site on HIV IN, the LEDGF/p75 binding site, was recently shown to be a promising target. The observation that removal of LEDGF/p75 from cells or overexpression of isolated LEDGF/p75 IBD could inhibit HIV-1 replication, motivated searches for compounds that obstructed the LEDGF/p75 interaction (79, 90, 91, 137). Further motivating this search was a report of a small molecule inhibitor of HIV integration bound to a site at the dimer interface of the HIV IN catalytic domain (138) that was later shown to also be the LEDGF/p75 IBD binding site. Several groups carried out large-scale screens and identified small molecules that bound at this site and blocked binding of the LEDGF/p75 IBD to HIV-1 IN (137, 139–143). Some of these molecules were found to have low nanomolar affinities, acceptable toxicity to cells, and inhibited virus infection quite potently. Structural studies using X-ray crystallography showed the drugs to be bound as expected in a pocket at the catalytic domain dimer interface (Figure 2). Growth of HIV in the presence of the drugs elicited escape mutations that mapped to regions encoding segments of IN near the drug binding site, genetically specifying IN as the drug target during viral replication.
Figure 2.
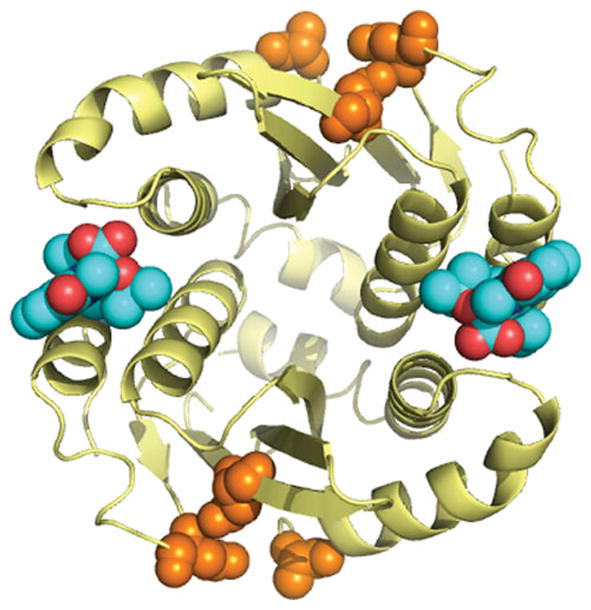
The inhibitor GSK1264 bound to the LEDGF-binding site on the HIV IN catalytic domain dimer. The alpha carbon backbone of the IN catalytic domain dimer is shown in gold. The GSK1264 compound is show in cyan (carbons) and red (oxygens). Active site residues are shown in orange. Details on the structure and function of GSK1264 are reported in (137).
Mechanistic studies of the steps affected in the viral replication cycle by these drugs yielded a surprise. Unexpectedly, the drugs were not particularly potent during the early phase of the viral replication cycle (entry through integration). In some cases the small molecules reduced targeting to transcription units, but this has been variable across the drug series. Compounds inhibited potently during the late steps (gene expression through assembly and release). It turned out that drugs of this class were promoting abnormal assembly of HIV polyprotein precursors and/or IN, so that viral particles had a disrupted structure and were fatally impaired during early steps of the next infectious cycle. LEDGF does not seem to be involved in normal assembly, because assembly proceeds normally, and the inhibitors are active in the absence of LEDGF. At the time of writing, large-scale efforts are under way in several pharmaceutical companies to complete development and bring this class of inhibitor to patients.
Controlling retroviral integration site selection with small molecules
As is described above, it is now possible to control the distributions of retroviral integration target sites by adding small molecules to cells during infections. For both lentiviruses and gammaretroviruses, addition of the drugs (some LEDGF site blockers (139–143) or JQ1 (105–108)) reduces targeting biases, so that integration site patterns more closely resemble random distributions. These molecules may have applications in human gene therapy, where targeting integration in or near transcription units is undesirable because of the risk of genotoxicity. If the efficiency of gene expression after integration is acceptable at these new locations, and if the small molecules are not too toxic to primary cells, these small molecules may be useful additives during cell transduction prior to transplantation of modified cells into patients.
Integration and HIV latency: implications for cure research
Recent work has focused attention on the possible influence of the chromosomal environment of the integrated proviruses on HIV latency. The formation of latent (transcriptionally silent) proviruses represents a major block to eradicating HIV from infected cells (144, 145). Studies show that HIV+ subjects who have viral replication suppressed by anti-retroviral therapy to below the limit of detection nevertheless harbor rare cells containing integrated proviruses that are transcriptionally silent. Some of these are known to be resting memory CD4+ T-cells, which, upon exposure to antigen, can resume active transcription that allows proviral induction. As a result, renewed viral replication can restart active infection.
The observation that several individuals may have been cured of HIV has resulted in intense interest in developing generalizable means of eradicating HIV from infected people (146–148). One idea holds that cells harboring latent viruses might be selectively killed if latent proviruses could be coaxed to resume transcription, provided that the host was equiped to destroy these cells efficiently once the latent cells were disclosed to the immune system. A recent clinical trial tested this by treating well-suppressed HIV+ subjects with the histone deacetylase inhibitor SAHA (vorinostat) with the goal of altering the chromatin environment in latent cells to promote HIV transcription (149). Studies to date have reported an increase in cell-associated HIV RNA following SAHA treatment, though no increase in circulating virus, providing an early suggestion that the shock-and-kill strategy might be feasible.
The idea that SAHA would boost transcription presupposes that a lack of histone acetylation is the limiting factor for transcription of the HIV provirus. This assumption can be addressed experimentally using cell-based models of latency and proviral induction (150). In such studies, cells are infected with HIV or an HIV-based vector. Cells are then fractionated based on whether or not they express the HIV provirus. The non-expressing cells can then be induced to express HIV with an agent that activates T-cells. Those cells that respond and transcribe the proviral genome are operationally defined as the inducibly latent population.
Several studies have explored integration site distributions in these cell populations (150–152). In general, inducible proviruses are found in locations similar to from typical HIV favored integration sites. A recent meta-analysis showed that no single feature characterized latent cells studied in five different cell models (150). Significant trends were seen within each model, but the chromosomal features near favored sites were mostly dissimilar between models. The most prominent shared feature of latent integration sites was proximity to cellular alphoid repeats, which was observed to be positively associated with latency in four of five models. Alphoid repeats are enriched near centromeres, so the silencing detected may resembles position effect variagation seen in early studies in Drosophila, where apposition of an active gene to centromeric heterochromatin by inversion resulted in reduction in expression (153). However, latent sites near alphoid repeats comprised only a minority of all inducible integration sites. SAHA shock-and-kill protocols predicted a negative association between latent sites and histone acetylation, but this association was not consistently observed. Latency in these models was not associated with a single integration site pattern, indicating that transcriptional latency in HIV may have many molecular mechanisms, a conclusions also reached with other experimental approaches (154). Unfortunately these findings complicate efforts to expose and remove cells harboring latent HIV.
Other studies suggest additional possible roles for integration targeting in HIV latency. Ikeda et al. reported in 2007 that rare cells from HIV+ patients on long term successful drug therapy showed clustering of integration sites (155), and recent reports have developed this picture further (156–158). Some of these integration sites were in genes related to cell growth or persistence, such as the transcription factor BACH2, which is a lymphoid cancer-related gene. These results raise the possibility that gene disruption by HIV integration may promote growth or persistence of latently infected cells. An example of lentiviral integration correlated with outgrowth of a cell clone has been reported in a gene therapy trial. In this case, the vector integrated in the expanded cells was located in the proto-oncogene HMGA2, providing a possible analogy for clonal outgrowth during latency (159).
Genome-wide studies to identify candidate host factors
Numerous genome-wide screens have been carried out to identify host factors important for HIV replication (collected in (160–162)). A wide variety of methods have been applied. In a few cases proteins have been identified that are candidates for affecting the integration step. Loss-of-function screens have been carried out using siRNA knockdowns, in which siRNAs targeting most human genes are assayed individually for their ability to either reduce or increase HIV infection (14)(53, 54). Gain of function studies have been carried out by overexpressing cDNAs and assessing whether they increased or decreased HIV infection (163). Focused screens have queried interferon-inducible genes, a particularly rich source of anti-viral factors (164, 165).
Another approach involves genome-wide studies to identify host proteins that bind to viral proteins (32). In these protocols, HIV proteins are expressed in cells, then captured using appropriate tagging systems. Bound proteins are then eluted and analyzed by mass spectrometry approaches. Recent comprehensive studies have identified many more cellular proteins binding to the HIV proteins, in some cases with strong statistical support for specificity. Another approach involves yeast two-hybrid assays, in which candidate interactors are expressed together as fusions in yeast, and interactions scored by effects on yeast transcription. For both LEDGF/p75 and BET proteins, early studies used yeast two-hybrid assays to document binding.
Given the gigantic size of the genome-wide data sets, the development of useful summaries and convenient web browers becomes as important as generating the data to begin with. Table 1 summarizes some of web-enabled sites for investingating these data. The GuavaH web site allows one to query HIV and human genotypes and their relationship to phenotypes (166). The GeneOverlapper site can be used to query lists of genes in groups to find genes in common (162). Over 40 lists are included, which can be queried in over 200 trillion combinations, so a blog site is included for readers to post comments on their findings, allowing “crowd sourcing” of discovery. The NCBI keeps a list of HIV proteins and their interactions. So far, with respect to integration biology, these large data sets have helped identify tethering factors and a variety of cellular factors implicated in PIC trafficking and nuclear import. A rich array of additional cellular proteins have been implicated, and some bind IN, offering many new starting points for investigating the roles of cellular proteins in integration.
Table 1.
Resources for working with genome-wide data on HIV.
Table 1. Some resources for working with genome-wide data from HIV research
| Name of resource | Link |
|---|---|
| GuavaH | http://www.GuavaH.org |
| NCBI | http://www.ncbi.nlm.nih.gov/ |
| Los Alamos HIV Databases | www.hiv.lanl.gov/ |
| UNAIDS | http://www.unaids.org/en/ |
| Stanford University HIV Drug Resistance Database | http://hivdb.stanford.edu/index.html |
| GPS-prot | http://www.gpsprot.org |
| HIV Replication Cycle | http://www.hivsystemsbiology.org/wiki/index.php/Introduction |
| Gene Overlapper | http://www.hivsystemsbiology.org/GeneListOverlapper/ |
Other fates of the viral DNA
The normal fate of retroviral DNA is integration into the host genome. However, forms of viral DNA are also found that are thought to be abortive products not on the integration pathway (167, 168). Some viral DNA is found in the form of 2-LTR circles that likely arise by non-homologous end joining of the linear viral DNA after preintegration complexes have fallen apart (125). 1-LTR circles appear to be the product of homologous recombination between the two LTRs in the linear viral DNA. Other types of aberrant product, including full length circular viral DNA with a rearrangement and smaller circles with deletion, are the result of autointegration.
Several models have suggested that the unrepaired integration intermediate and/or unintegrated forms of viral DNA may trigger pathways leading to cell death. Retroviral infection was reported by some to induce cell death in NHEJ-deficient cell lines (120, 121, 169). Cell death in these cell lines was reported to be dependent on the presence of functional IN, suggesting that the integration intermediate is recognized as DNA damage. Consistent with this idea, inhibition of DNA-PK by wortmannin makes normal cells sensitive to killing by retroviral vectors with an active IN, but not by vectors with an inactive IN. Results with these models have varied (125, 170). In another study, infection of activated primary CD4+ lymphocytes by HIV-1 also induced cell killing (171). Killing was not observed in the absence of active IN or the presence of IN inhibitors, suggesting a requirement for integration. However, in a recent study from another group, unintegrated viral DNA was implicated as the signal in killing of CD4+ lymphocytes (172). Knockdown of the interferon–γ-inducible protein 16 (IFI16), a DNA sensor, abrogated cell killing in this model, suggesting that the integrated viral DNA triggers an innate immune response that leads to caspase 1 activation and pyroptosis. Killing was still observed with virus with an active site mutation in IN, but not in the presence of a nonnucleoside reverse transcriptase inhibitor, demonstrating a requirement for DNA synthesis but not integration. Further work is required to reconcile and advance work in this area.
Conclusions
Retroviral host factors and their roles in integration targeting has been a very active area of study. Genome-wide screens have yielded many new starting points for investigating HIV biology, including HIV integration. The discoveries of tethering factors for HIV and MLV provide a simple mechanism. In contrast, the finding that drugs targeting the LEDGF/p75 binding site in fact disrupt assembly in a fashion that is mostly independent of LEDGF/p75 was not at all expected, and focuses attention on the apparent role of IN in late replication steps. The role of integration in latency, and the role of replication intermediates in cell toxicity and immunodepletion are further areas where additional investigation would be valuable.
Acknowledgments
We are grateful to Karen Ocwieja, Scott Sherrill-Mix, Alan Engelman and Peter Cherepanov for comments on the manuscript. This work was supported by the Intramural Program of NIDDK, NIH and by the AIDS Targeted Antiviral Program of the Office of the Director of the NIH RC), AI 052845 and AI 090935 (FDB), and the HINT collaboratory.
References
- 1.Donehower LA, Varmus HE. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc Natl Acad Sci USA. 1984;81:6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panganiban AT, Temin HM. The retrovirus pol gene encodes a product required for DNA integration: Identification of a retrovirus int locus. Proc Natl Acad Sci USA. 1984;81:7885–7889. doi: 10.1073/pnas.81.24.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartzberg P, Colecilli J, Goff SP. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984;37:1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- 4.Brown PO, Bowerman B, Varmus HE, Bishop JM. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 5.Farnet CM, Haseltine WA. Integration of human immunodeficiency virus type 1 DNA in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 7.Brown PO, Bowerman B, Varmus HE, Bishop JM. Retroviral integration: structure of the initial covalent complex and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci U S A. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowerman B, Brown PO, Bishop JM, Varmus HE. A nucleoprotein complex mediates the integration of retroviral DNA. Genes and Development. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 9.Bushman FD, Craigie R. Sequence requirements for integration of Moloney murine leukemia virus DNA in vitro. J Virol. 1990;64:5645–5648. doi: 10.1128/jvi.64.11.5645-5648.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 11.Bushman FD, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 12.Sherman PA, Fyfe JA. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katzman M, Katz RA, Skalka AM, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka AM, Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 avain sarcoma virus concerted DNA integration in vitro. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M, Craigie R. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. The Journal of biological chemistry. 2005;280:29334–29339. doi: 10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinha S, Grandgenett DP. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. Journal of virology. 2005;79:8208–8216. doi: 10.1128/JVI.79.13.8208-8216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sinha S, Pursley MH, Grandgenett DP. Efficient concerted integration by recombinant human immunodeficiency virus type 1 integrase without cellular or viral cofactors. Journal of virology. 2002;76:3105–3113. doi: 10.1128/JVI.76.7.3105-3113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valkov E, Gupta SS, Hare S, Helander A, Roversi P, McClure M, Cherepanov P. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37:243–255. doi: 10.1093/nar/gkn938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carteau S, Gorelick R, Bushman FD. Coupled integration of human immunodeficiency virus cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iordanskiy S, Berro R, Altieri M, Kashanchi F, Bukrinsky M. Intracytoplasmic maturation of the human immunodeficiency virus type 1 reverse transcription complexes determines their capacity to integrate into chromatin. Retrovirology. 2006;3:4. doi: 10.1186/1742-4690-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bukrinsky MI, Sharova N, McDonald TL, Pushkarskaya T, Tarpley GW, Stevenson M. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc Natl Acad Sci USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV Nuclear Import is Governed by the Phosphotyrosine-mediated Binding of Matrix to the Core Domain of Integrase. Cell. 1995;17:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 23.Gallay P, Hope T, Chin D, Trono D. HIV-1 Infection of Nondividing Cells Through the Recognition of Integrase by the Importin/Karyopherin Pathway. Proc Natl Acad Sci USA. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinzinger NK, Bukrinsky MI, Haggerty SA, Ragland AM, VK, Lee MA, Gendelman HE, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karageorgos L, Li P, Burrell C. Characterization of HIV replication complexes early after cell-to-cell infection. AIDS Res Human Retrovir. 1993;9:817–823. doi: 10.1089/aid.1993.9.817. [DOI] [PubMed] [Google Scholar]
- 26.Farnet CM, Haseltine WA. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller MD, Farnet CM, Bushman FD. Human Immunodeficiency Virus Type 1 Preintegration Complexes: Studies of Organization and Composition. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farnet C, Bushman FD. HIV-1 cDNA Integration: Requirement of HMG I(Y) Protein for Function of Preintegration Complexes In Vitro. Cell. 1997;88:1–20. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 31.Raghavendra NK, Shkriabai N, Graham R, Hess S, Kvaratskhelia M, Wu L. Identification of host proteins associated with HIV-1 preintegration complexes isolated from infected CD4+ cells. Retrovirology. 2010;7:66. doi: 10.1186/1742-4690-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D’Orso I, Fernandes J, Fahey M, Mahon C, O’Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: reaction pathway and critical intermeidates. Embo J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei SQ, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei SQ, Mizuuchi K, Craigie R. Footprints of the viral DNA ends in Moloney murine leukemia virus preintegration complexes reflect a specific association with integrase. Proc Natl Acad Sci USA. 1998;95:10535–10540. doi: 10.1073/pnas.95.18.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H, Wei SQ, Engelman A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J Biol Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- 37.Lee MS, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng R, Ghirlando R, Lee MS, Mizuuchi K, Krause M, Craigie R. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc Natl Acad Sci U S A. 2000;97:8997–9002. doi: 10.1073/pnas.150240197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Margalit A, Segura-Totten M, Gruenbaum Y, Wilson KL. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc Natl Acad Sci U S A. 2005;102:3290–3295. doi: 10.1073/pnas.0408364102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skoko D, Li M, Huang Y, Mizuuchi M, Cai M, Bradley CM, Pease PJ, Xiao B, Marko JF, Craigie R, Mizuuchi K. Barrier-to-autointegration factor (BAF) condenses DNA by looping. Proc Natl Acad Sci U S A. 2009;106:16610–16615. doi: 10.1073/pnas.0909077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki Y, Craigie R. Regulatory Mechanisms by Which Barrier-to-Autointegration Factor Blocks Autointegration and Stimulates Intermolecular Integration of Moloney Murine Leukemia Virus Preintegration Complexes. J Virol. 2002;76:12376–12380. doi: 10.1128/JVI.76.23.12376-12380.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki Y, Ogawa K, Koyanagi Y, Suzuki Y. Functional disruption of the moloney murine leukemia virus preintegration complex by vaccinia-related kinases. J Biol Chem. 2010;285:24032–24043. doi: 10.1074/jbc.M110.116640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nichols RJ, Wiebe MS, Traktman P. The vaccinia-related kinases phosphorylate the N’ terminus of BAF, regulating its interaction with DNA and its retention in the nucleus. Molecular biology of the cell. 2006;17:2451–2464. doi: 10.1091/mbc.E05-12-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bushman FD. Targeting Retroviral Integration. Science. 1995;267:1443–1444. doi: 10.1126/science.7878462. [DOI] [PubMed] [Google Scholar]
- 45.Bukrinsky MI, Haggerty S, Dempsey MP, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouyac-Bertoia M, Dvorin JD, Fouchier RA, Jenkins Y, Meyer BE, Wu LI, Emerman M, Malim MH. HIV-1 infection requires a functional integrase NLS. Molecular cell. 2001;7:1025–1035. doi: 10.1016/s1097-2765(01)00240-4. [DOI] [PubMed] [Google Scholar]
- 47.Fouchier RA, Meyer BE, Simon JH, Fischer U, Albright AV, Gonzalez-Scarano F, Malim MH. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. Journal of virology. 1998;72:6004–6013. doi: 10.1128/jvi.72.7.6004-6013.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vodicka MA, Koepp DM, Silver PA, Emerman M. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes & development. 1998;12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 50.Matreyek KA, Engelman A. Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes. Viruses. 2013;5:2483–2511. doi: 10.3390/v5102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamashita M, Emerman M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol. 2004;78:5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science (New York, NY ) 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 53.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. Genome-Scale RNAi Screen for Host Factors Required for HIV Replication. Cell host & microbe. 2008 doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 55.Yeung ML, Houzet L, Yedavalli VS, Jeang KT. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J Biol Chem. 2009;284:19463–19473. doi: 10.1074/jbc.M109.010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krishnan L, Matreyek KA, Oztop I, Lee K, Tipper CH, Li X, Dar MJ, Kewalramani VN, Engelman A. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. Journal of virology. 2010;84:397–406. doi: 10.1128/JVI.01899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hue S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS pathogens. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matreyek KA, Engelman A. The requirement for nucleoporin NUP153 during human immunodeficiency virus type 1 infection is determined by the viral capsid. Journal of virology. 2011;85:7818–7827. doi: 10.1128/JVI.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thys W, De Houwer S, Demeulemeester J, Taltynov O, Vancraenenbroeck R, Gerard M, De Rijck J, Gijsbers R, Christ F, Debyser Z. Interplay between HIV entry and transportin-SR2 dependency. Retrovirology. 2011;8:7. doi: 10.1186/1742-4690-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Greene WC, Peterlin BM. Charting HIV’s remarkable voyage through the cell: Basic science as a passport to future therapy. Nature medicine. 2002;8:673–680. doi: 10.1038/nm0702-673. [DOI] [PubMed] [Google Scholar]
- 61.Campbell EM, Hope TJ. Live cell imaging of the HIV-1 life cycle. Trends in microbiology. 2008;16:580–587. doi: 10.1016/j.tim.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di Primio C, Quercioli V, Allouch A, Gijsbers R, Christ F, Debyser Z, Arosio D, Cereseto A. Single-cell imaging of HIV-1 provirus (SCIP) Proc Natl Acad Sci U S A. 2013;110:5636–5641. doi: 10.1073/pnas.1216254110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 64.Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- 65.Chubb JR, Bickmore WA. Considering nuclear compartmentalization in light of nuclear dynamics. Cell. 2003;112:403–406. doi: 10.1016/s0092-8674(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 66.Wei Z, Huang D, Gao F, Chang WH, An W, Coetzee GA, Wang K, Lu W. Biological implications and regulatory mechanisms of long-range chromosomal interactions. J Biol Chem. 2013;288:22369–22377. doi: 10.1074/jbc.R113.485292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ocwieja KE, Brady TL, Ronen K, Huegel A, Roth SL, Schaller T, James LC, Towers GJ, Young JA, Chanda SK, Konig R, Malani N, Berry CC, Bushman FD. HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS pathogens. 2011;7:e1001313. doi: 10.1371/journal.ppat.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, Lusic M, Marcello A, Giacca M. Acetylation of HIV-1 integrase by p300 regulates viral integration. Embo J. 2005;24:3070–3081. doi: 10.1038/sj.emboj.7600770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, Giacca M, Cereseto A. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe. 2011;9:484–495. doi: 10.1016/j.chom.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 70.Kalpana GV, Goff SP. Genetic Analysis of Homomeric Interactions of Human Immunodeficiency Virus Type 1 Integrase Using the Yeast Two-Hybrid System. Proc Natl Acad Sci USA. 1993;90:10593–10597. doi: 10.1073/pnas.90.22.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lewinski M, Bushman FD. Retroviral DNA integration--mechanism and consequences. Adv Genet. 2005;55:147–181. doi: 10.1016/S0065-2660(05)55005-3. [DOI] [PubMed] [Google Scholar]
- 72.Schroder ARW, Shinn P, Chen HM, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 73.Mitchell R, Chiang C, Berry C, Bushman FD. Global effects on cellular transcription following infection with an HIV-based vector. Mol Ther. 2003;8:674–687. doi: 10.1016/s1525-0016(03)00215-6. [DOI] [PubMed] [Google Scholar]
- 74.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science (New York, NY ) 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 75.Barr SD, Ciuffi A, Leipzig J, Shinn P, Ecker JR, Bushman FD. HIV integration site selection: targeting in macrophages and the effects of different routes of viral entry. Molecular therapy : the journal of the American Society of Gene Therapy. 2006;14:218–225. doi: 10.1016/j.ymthe.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 76.Barr SD, Leipzig J, Shinn P, Ecker JR, Bushman FD. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J Virol. 2005;79:12035–12044. doi: 10.1128/JVI.79.18.12035-12044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, Bickmore W, Poeschla E, Bushman FD. Role of PSIP1/LEDGF/p75 in Lentiviral Infectivity and Integration Targeting. PLoS One. 2007:2. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berry CC, Ocwieja KE, Malani N, Bushman FD. Comparing DNA integration site clusters with scan statistics. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes & development. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Han Y, Lassen K, Monie D, Sedaghat AR, Shimoji S, Liu S, Pierson TC, Margolick JB, Siliciano RF, Siliciano JD. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected inndividuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol. 2004;78:6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS computational biology. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome research. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ciuffi A, Ronen K, Brady T, Malani N, Wang G, Berry CC, Bushman FD. Methods for integration site distribution analyses in animal cell genomes. Methods (San Diego, Calif ) 2009;47:261–268. doi: 10.1016/j.ymeth.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mitchell RS, Beitzel BF, Schroder ARW, Shinn P, Chen HM, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:1127–1137. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C, Skalka AM, Katz RA. Genome-wide analyses of avain sarcoma virus integration sites. J Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, Hannenhalli S, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS pathogens. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 Integrase Forms Stable Tetramers and Associates with LEDGF/p75 Protein in Human Cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 88.Turlure F, Devroe E, Silver PA, Engelman A. Human cell proteins and human immunodeficiency virus DNA integration. Front Biosci. 2004;9:3187–3208. doi: 10.2741/1472. [DOI] [PubMed] [Google Scholar]
- 89.Emiliani S, Mousnier A, Busschots K, Maroun M, Van Maele B, Tempe D, Vandekerckhove L, Moisant F, Ben-Slama L, Witvrouw M, Christ F, Rain JC, Dargemont C, Debyser Z, Benarous R. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J Biol Chem. 2005;280:25517–25523. doi: 10.1074/jbc.M501378200. [DOI] [PubMed] [Google Scholar]
- 90.Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo WL, Poeschla EM. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 91.De Rijck J, Vandekerckhove L, Gijsbers R, Hombrouck A, Hendrix J, Vercammen J, Engelborghs Y, Christ F, Debyser Z. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. Journal of virology. 2006;80:11498–11509. doi: 10.1128/JVI.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eidahl JO, Crowe BL, North JA, McKee CJ, Shkriabai N, Feng L, Plumb M, Graham RL, Gorelick RJ, Hess S, Poirier MG, Foster MP, Kvaratskhelia M. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 2013;41:3924–3936. doi: 10.1093/nar/gkt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van Nuland R, van Schaik FM, Simonis M, van Heesch S, Cuppen E, Boelens R, Timmers HM, van Ingen H. Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain. Epigenetics & chromatin. 2013;6:12. doi: 10.1186/1756-8935-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci U S A. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hare S, Shun MC, Gupta SS, Valkov E, Engelman A, Cherepanov P. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS pathogens. 2009;5:e1000259. doi: 10.1371/journal.ppat.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 97.Ciuffi A, Diamond T, Hwang Y, Marshall H, Bushman FD. Fusions of LEDGF/p75 to lambda repressor promote HIV DNA integration near lambda operators in vitro. Hum Gene Ther. 2006;17:960-967–960-967. doi: 10.1089/hum.2006.17.960. [DOI] [PubMed] [Google Scholar]
- 98.Vandegraaff N, Devroe E, Turlure F, Silver PA, Engelman A. Biochemical and genetic analyses of integrase-interacting protein lens epithelium-derived growth factor (LEDGF)/p75 and hepatoma-derived growth factor related protein 2 (HRP2) in preintegration complex function and HIV-1 replication. Virology. 2006;346:415–426. doi: 10.1016/j.virol.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 99.Schrijvers R, Vets S, De Rijck J, Malani N, Bushman FD, Debyser Z, Gijsbers R. HRP-2 determines HIV-1 integration site selection in LEDGF/p75 depleted cells. Retrovirology. 2012;9:84. doi: 10.1186/1742-4690-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vandegraaff N, Devroe E, Turlure F, Silver PA, Engelman A. Biochemical and genetic analysis of integrase-interacting protein lens epithelium-derived growth factor (LEDGF)/p75 and hepatoma-derived growth factor related protein 2 (HRP2) in preintegration complex function and HIV-1 replication. Virology Epub. 2005 doi: 10.1016/j.virol.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 101.Schrijvers R, De Rijck J, Demeulemeester J, Adachi N, Vets S, Ronen K, Christ F, Bushman FD, Debyser Z, Gijsbers R. LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs. PLoS pathogens. 2012;8:e1002558. doi: 10.1371/journal.ppat.1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Silvers RM, Smith JA, Schowalter M, Litwin S, Liang Z, Geary K, Daniel R. Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Hum Gene Ther. 2010;21:337–349. doi: 10.1089/hum.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gijsbers R, Ronen K, Vets S, Malani N, De Rijck J, McNeely M, Bushman FD, Debyser Z. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol Ther. 2010;18:552–560. doi: 10.1038/mt.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, Wang GG, Shun MC, Allis CD, Engelman A, Hughes SH. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc Natl Acad Sci U S A. 2010;107:3135–3140. doi: 10.1073/pnas.0914142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De Rijck J, de Kogel C, Demeulemeester J, Vets S, El Ashkar S, Malani N, Bushman FD, Landuyt B, Husson SJ, Busschots K, Gijsbers R, Debyser Z. The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell reports. 2013;5:886–894. doi: 10.1016/j.celrep.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sharma A, Larue RC, Plumb MR, Malani N, Male F, Slaughter A, Kessl JJ, Shkriabai N, Coward E, Aiyer SS, Green PL, Wu L, Roth MJ, Bushman FD, Kvaratskhelia M. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci U S A. 2013;110:12036–12041. doi: 10.1073/pnas.1307157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Larue RC, Plumb MR, Crowe BL, Shkriabai N, Sharma A, Difiore J, Malani N, Aiyer SS, Roth MJ, Bushman FD, Foster MP, Kvaratskhelia M. Bimodal high-affinity association of Brd4 with murine leukemia virus integrase and mononucleosomes. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gupta SS, Maetzig T, Maertens GN, Sharif A, Rothe M, Weidner-Glunde M, Galla M, Schambach A, Cherepanov P, Schulz TF. Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration. Journal of virology. 2013;87:12721–12736. doi: 10.1128/JVI.01942-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Studamire B, Goff SP. Host proteins interacting with the Moloney murine leukemia virus integrase: multiple transcriptional regulators and chromatin binding factors. Retrovirology. 2008;5:48. doi: 10.1186/1742-4690-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pryciak PM, Varmus HE. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 111.Pryciak PM, Sil A, Varmus HE. Retroviral integration into minichromosomes in vitro. EMBO J. 1992;11:291–303. doi: 10.1002/j.1460-2075.1992.tb05052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pryciak P, Muller HP, Varmus HE. Simian Virus 40 minichromosomes as targets for retroviral integration in vivo. Proc Natl Acad Sci USA. 1992;89:9237–9241. doi: 10.1073/pnas.89.19.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pruss D, Bushman FD, Wolffe AP. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc Natl Acad Sci USA. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pruss D, Reeves R, Bushman FD, Wolffe AP. The Influence of DNA and Nucleosome Structure on Integration Events Directed by HIV Integrase. J Biol Chem. 1994;269:25031–25041. [PubMed] [Google Scholar]
- 115.Roth SL, Malani N, Bushman FD. Gammaretroviral integration into nucleosomal target DNA in vivo. Journal of virology. 2011;85:7393–7401. doi: 10.1128/JVI.00635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lesbats P, Botbol Y, Chevereau G, Vaillant C, Calmels C, Arneodo A, Andreola ML, Lavigne M, Parissi V. Functional coupling between HIV-1 integrase and the SWI/SNF chromatin remodeling complex for efficient in vitro integration into stable nucleosomes. PLoS pathogens. 2011;7:e1001280. doi: 10.1371/journal.ppat.1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Botbol Y, Raghavendra NK, Rahman S, Engelman A, Lavigne M. Chromatinized templates reveal the requirement for the LEDGF/p75 PWWP domain during HIV-1 integration in vitro. Nucleic Acids Res. 2008;36:1237–1246. doi: 10.1093/nar/gkm1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yoder K, Bushman FD. Repair of gaps in retroviral DNA integration intermediates. J Virol. 2000;74:11191–11200. doi: 10.1128/jvi.74.23.11191-11200.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chow SA, Vincent KA, Ellison V, Brown PO. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 120.Daniel R, Katz RA, Skalka AM. A Role for DNA-PK in Retroviral DNA Integration. Science. 1999;284:644–647. doi: 10.1126/science.284.5414.644. [DOI] [PubMed] [Google Scholar]
- 121.Daniel R, Katz RA, Merkel G, Hittle JC, Yen TJ, Skalka AM. Wortmannin potentiates integrase-mediated killing of lymphocytes and reduces the efficency of stable transduction by retroviruses. Mol Cell Biol. 2001;21:1164–1172. doi: 10.1128/MCB.21.4.1164-1172.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lau A, Kanaar R, Jackson SP, O’Connor MJ. Suppression of retroviral infection by the RAD52 DNA repair protein. EMBO J. 2004;23:3421–3429. doi: 10.1038/sj.emboj.7600348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Daniel R, Kao G, Taganov K, Greger JG, Favorova O, Merkel G, Yen TJ, Katz RA, Skalka AM. Evidence that the retroviral DNA integration process triggers an ATR-dependent DNA damage response. Proc Natl Acad Sci U S A. 2003;100:4778–4783. doi: 10.1073/pnas.0730887100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yoder K, Sarasin A, Kraemer K, McIlhatton M, Bushman F, Fishel R. The DNA repair genes XPB and XPD defend cells from retroviral infection. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4622–4627. doi: 10.1073/pnas.0509828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li L, Olvera JM, Yoder K, Mitchell RS, Butler SL, Lieber MR, Martin SL, Bushman FD. Role of the Non-Homologous DNA End Joining Pathway in Retroviral Infection. EMBO J. 2001;20:3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Espeseth AS, Fishel R, Hazuda D, Huang Q, Xu M, Yoder K, Zhou H. siRNA screening of a targeted library of DNA repair factors in HIV infection reveals a role for base excision repair in HIV integration. PLoS One. 2011;6:e17612. doi: 10.1371/journal.pone.0017612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yoder KE, Espeseth A, Wang XH, Fang Q, Russo MT, Lloyd RS, Hazuda D, Sobol RW, Fishel R. The base excision repair pathway is required for efficient lentivirus integration. PLoS One. 2011;6:e17862. doi: 10.1371/journal.pone.0017862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Arts EJ, Hazuda DJ. HIV-1 antiretroviral drug therapy. Cold Spring Harbor perspectives in medicine. 2012;2:a007161. doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hazuda DJ, Hastings JC, Wolfe AL, Emini EA. A novel assay for the DNA strand-transfer reaction of HIV-1 integrase. Nuc Acids Res. 1994;22:1121–1122. doi: 10.1093/nar/22.6.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. Inhibitors of Strand Transfer That Prevent Integration and Inhibit HIV-1 Replication in Cells. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 131.Hazuda DJ, Anthony NJ, Gomez RP, Jolly SM, Wai JS, Zhuang L, Fisher TE, Embrey M, PGJ, Egbertson MS, Vacca JP, Huff JR, Felock PJ, Witmer MV, Stillmock KA, Danovich R, Grobler J, Miller MD, Espeseth AS, Jin L, Chen IW, Lin JH, Kassahun K, Ellis JD, Wong BK, Xu W, Pearson PG, Schleif WA, Cortese R, Emini E, VS, Holloway MK, Young SD. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proc Natl Acad Sci U S A. 2004;101:11233–11238. doi: 10.1073/pnas.0402357101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Metifiot M, Marchand C, Pommier Y. HIV integrase inhibitors: 20-year landmark and challenges. Advances in pharmacology. 2013;67:75–105. doi: 10.1016/B978-0-12-405880-4.00003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Espeseth AS, Felock P, Wolfe A, Witmer M, Grobler J, Anthony N, Egbertson M, Melamed JY, Young S, Hamill T, Cole JL, Hazuda DJ. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc Natl Acad Sci U S A. 2000;97:11244–11249. doi: 10.1073/pnas.200139397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Krishnan L, Li X, Naraharisetty HL, Hare S, Cherepanov P, Engelman A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc Natl Acad Sci U S A. 2010;107:15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Min S, Sloan L, DeJesus E, Hawkins T, McCurdy L, Song I, Stroder R, Chen S, Underwood M, Fujiwara T, Piscitelli S, Lalezari J. Antiviral activity, safety, and pharmacokinetics/pharmacodynamics of dolutegravir as 10-day monotherapy in HIV-1-infected adults. Aids. 2011;25:1737–1745. doi: 10.1097/QAD.0b013e32834a1dd9. [DOI] [PubMed] [Google Scholar]
- 136.Walmsley SL, Antela A, Clumeck N, Duiculescu D, Eberhard A, Gutierrez F, Hocqueloux L, Maggiolo F, Sandkovsky U, Granier C, Pappa K, Wynne B, Min S, Nichols G, Investigators S. Dolutegravir plus abacavir-lamivudine for the treatment of HIV-1 infection. The New England journal of medicine. 2013;369:1807–1818. doi: 10.1056/NEJMoa1215541. [DOI] [PubMed] [Google Scholar]
- 137.Gupta K, Brady T, Dyer BM, Malani N, Hwang Y, Male F, Nolte RT, Wang L, Velthuisen E, Jeffrey J, Van Duyne GD, Bushman FD. Allosteric inhibition of human immunodeficiency virus integrase: late block during viral replication and abnormal multimerization involving specific protein domains. J Biol Chem. 2014 doi: 10.1074/jbc.M114.551119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Molteni V, Greenwald J, Rhodes D, Hwang Y, Kwiatkowski W, Bushman FD, Siegel JS, Choe S. Identification of a small molecule binding site at the dimer interface of the HIV integrase catalytic domain. Acta Crystallogr D Biol Crystallogr. 2001;57:536–544. doi: 10.1107/s0907444901001652. [DOI] [PubMed] [Google Scholar]
- 139.Le Rouzic E, Bonnard D, Chasset S, Bruneau JM, Chevreuil F, Le Strat F, Nguyen J, Beauvoir R, Amadori C, Brias J, Vomscheid S, Eiler S, Levy N, Delelis O, Deprez E, Saib A, Zamborlini A, Emiliani S, Ruff M, Ledoussal B, Moreau F, Benarous R. Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage. Retrovirology. 2013;10:144. doi: 10.1186/1742-4690-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jurado KA, Wang H, Slaughter A, Feng L, Kessl JJ, Koh Y, Wang W, Ballandras-Colas A, Patel PA, Fuchs JR, Kvaratskhelia M, Engelman A. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc Natl Acad Sci U S A. 2013;110:8690–8695. doi: 10.1073/pnas.1300703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Feng L, Sharma A, Slaughter A, Jena N, Koh Y, Shkriabai N, Larue RC, Patel PA, Mitsuya H, Kessl JJ, Engelman A, Fuchs JR, Kvaratskhelia M. The A128T resistance mutation reveals aberrant protein multimerization as the primary mechanism of action of allosteric HIV-1 integrase inhibitors. J Biol Chem. 2013 doi: 10.1074/jbc.M112.443390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Engelman A, Kessl JJ, Kvaratskhelia M. Allosteric inhibition of HIV-1 integrase activity. Current opinion in chemical biology. 2013 doi: 10.1016/j.cbpa.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Desimmie BA, Schrijvers R, Demeulemeester J, Borrenberghs D, Weydert C, Thys W, Vets S, Van Remoortel B, Hofkens J, De Rijck J, Hendrix J, Bannert N, Gijsbers R, Christ F, Debyser Z. LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology. 2013;10:57. doi: 10.1186/1742-4690-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. Identification of a Reservoir for HIV-1 in Patients on Highly Active Antiretroviral Therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 145.Siliciano RF, Greene WC. HIV Latency. Cold Spring Harbor perspectives in medicine. 2011;1:a007096. doi: 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, Blau IW, Hofmann WK, Thiel E. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. The New England journal of medicine. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 147.Xing S, Siliciano RF. Targeting HIV latency: pharmacologic strategies toward eradication. Drug discovery today. 2013;18:541–551. doi: 10.1016/j.drudis.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Saez-Cirion A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, Potard V, Versmisse P, Melard A, Prazuck T, Descours B, Guergnon J, Viard JP, Boufassa F, Lambotte O, Goujard C, Meyer L, Costagliola D, Venet A, Pancino G, Autran B, Rouzioux C, Group AVS. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS pathogens. 2013;9:e1003211. doi: 10.1371/journal.ppat.1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sherrill-Mix S, Lewinski MK, Famiglietti M, Bosque A, Malani N, Ocwieja KE, Berry CC, Looney D, Shan L, Agosto LM, Pace MJ, Siliciano RF, O’Doherty U, Guatelli J, Planelles V, Bushman FD. HIV latency and integration site placement in five cell-based models. Retrovirology. 2013;10:90. doi: 10.1186/1742-4690-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lewinski MK, Bisgrove D, Shinn P, Chen H, Hoffmann C, Hannenhalli S, Verdin E, Berry CC, Ecker JR, Bushman FD. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. Journal of virology. 2005;79:6610–6619. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Han Y, Lin YB, An W, Xu J, Yang HC, O’Connell K, Dordai D, Boeke JD, Siliciano JD, Siliciano RF. Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell host & microbe. 2008;4:134–146. doi: 10.1016/j.chom.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Elgin SC, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harbor perspectives in biology. 2013;5:a017780. doi: 10.1101/cshperspect.a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Spina CA, Anderson J, Archin NM, Bosque A, Chan J, Famiglietti M, Greene WC, Kashuba A, Lewin SR, Margolis DM, Mau M, Ruelas D, Saleh S, Shirakawa K, Siliciano RF, Singhania A, Soto PC, Terry VH, Verdin E, Woelk C, Wooden S, Xing S, Planelles V. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS pathogens. 2013;9:e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ikeda T, Shibata J, Yoshimura K, Koito A, Matsushita S. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. The Journal of infectious diseases. 2007;195:716–725. doi: 10.1086/510915. [DOI] [PubMed] [Google Scholar]
- 156.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, Huang HC, Edlefsen PT, Mullins JI, Frenkel LM. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Margolis D, Bushman F. HIV/AIDS. Persistence by proliferation? Science. 2014;345:143–144. doi: 10.1126/science.1257426. [DOI] [PubMed] [Google Scholar]
- 159.Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, Down J, Denaro M, Brady T, Westerman K, Cavallesco R, Gillet-Legrand B, Caccavelli L, Sgarra R, Maouche-Chretien L, Bernaudin F, Girot R, Dorazio R, Mulder GJ, Polack A, Bank A, Soulier J, Larghero J, Kabbara N, Dalle B, Gourmel B, Socie G, Chretien S, Cartier N, Aubourg P, Fischer A, Cornetta K, Galacteros F, Beuzard Y, Gluckman E, Bushman F, Hacein-Bey-Abina S, Leboulch P. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Goff SP. Knockdown screens to knockout HIV-1. Cell. 2008;135:417–420. doi: 10.1016/j.cell.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bushman FD, Malani N, Fernandes J, D’Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, Bandyopadhyay S, Ideker T, Goff SP, Krogan NJ, Frankel AD, Young JA, Chanda SK. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS pathogens. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bushman FD, Barton S, Bailey A, Greig C, Malani N, Bandyopadhyay S, Young J, Chanda S, Krogan N. Bringing it all together: big data and HIV research. Aids. 2013;27:835–838. doi: 10.1097/QAD.0b013e32835cb785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nguyen DG, Yin H, Zhou Y, Wolff KC, Kuhen KL, Caldwell JS. Identification of novel therapeutic targets for HIV infection through functional genomic cDNA screening. Virology. 2007;362:16–25. doi: 10.1016/j.virol.2006.11.036. [DOI] [PubMed] [Google Scholar]
- 164.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, Schoggins JW, Rice CM, Yamashita M, Hatziioannou T, Bieniasz PD. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. 2013;502:563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Goujon C, Moncorge O, Bauby H, Doyle T, Ward CC, Schaller T, Hue S, Barclay WS, Schulz R, Malim MH. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bartha I, McLaren PJ, Ciuffi A, Fellay J, Telenti A. GuavaH: a compendium of host genomic data in HIV biology and disease. Retrovirology. 2014;11:6. doi: 10.1186/1742-4690-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1997. [PubMed] [Google Scholar]
- 168.Farnet CM, Haseltine WA. Circularization of Human Immunodeficiency Virus Type 1 DNA In Vitro. J Virol. 1991;65:6942–6952. doi: 10.1128/jvi.65.12.6942-6952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Daniel R, Greger JG, Katz RA, Taganov KD, Wu X, Kappes JC, Skalka AM. Evidence that stable retroviral transduction and cell survival following DNA integration depend on components of the nonhomologous end joining repair pathway. Journal of virology. 2004;78:8573–8581. doi: 10.1128/JVI.78.16.8573-8581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ariumi Y, Turelli P, Masutani M, Trono D. DNA damage sensors ATM, ATR, DNA-PKcs, and PARP-1 are dispensable for human immunodeficiency virus type 1 integration. Journal of virology. 2005;79:2973–2978. doi: 10.1128/JVI.79.5.2973-2978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cooper A, Garcia M, Petrovas C, Yamamoto T, Koup RA, Nabel GJ. HIV-1 causes CD4 cell death through DNA-dependent protein kinase during viral integration. Nature. 2013;498:376–379. doi: 10.1038/nature12274. [DOI] [PubMed] [Google Scholar]
- 172.Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, Greene WC. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]


