Abstract
The conserved RNA-binding protein Musashi1 (MSI1) has emerged as a key oncogenic factor in numerous solid tumors, including glioblastoma. However, its mechanism of action has not yet been established comprehensively. To identify its target genes comprehensively and determine the main routes by which it influences glioblastoma phenotypes, we conducted individual-nucleotide resolution cross-linking and immunoprecipitation (iCLIP) experiments. We confirmed that MSI1 has a preference for UAG sequences contained in a particular structural context, especially in 3′ untranslated regions. Although numerous binding sites were also identified in intronic sequences, our RNA transcriptome sequencing analysis does not favor the idea that MSI1 is a major regulator of splicing in glioblastoma cells. MSI1 target mRNAs encode proteins that function in multiple pathways of cell proliferation and cell adhesion. Since these associations indicate potentially new roles for MSI1, we investigated its impact on glioblastoma cell adhesion, morphology, migration, and invasion. These processes are known to underpin the spread and relapse of glioblastoma, in contrast to other tumors where metastasis is the main driver of recurrence and progression.
INTRODUCTION
Glioblastoma multiforme (GBM) is the most malignant form of brain cancer and carries a poor prognosis. Patients receive a combined treatment that includes surgical resection, followed by concurrent chemotherapy with the oral alkylating agent temozolomide and radiation therapy. Tumor relapse is very frequent, and patients have a median survival of 14.6 months. Many clinical trials have been carried out recently, but none have demonstrated improvement in overall survival according to the current standard of care (1, 2). In recent years, many studies have attempted to elucidate the genomics of glioblastoma using high-density microarrays and high-throughput sequencing. In a comprehensive genomic study of glioblastoma by The Cancer Genome Atlas (TCGA) project, 206 glioblastoma tumors were analyzed for DNA copy number, DNA methylation, and gene expression (3). Comparable results were revealed by other similar studies into the genomic landscape of glioblastoma (4). These studies highlighted the role of a common core of signaling pathways that are activated in GBM—the p53 pathway, the Rb pathway, and the RTK pathway (3, 4)—suggesting that these underlying pathways promote cell proliferation and enhanced cell survival in GBM. A follow-up study from the TCGA project described a robust molecular classification system to establish subtypes of glioblastoma (proneural, neural, classical, and mesenchymal) based on patterns of somatic mutations and DNA copy number that have profound implications on tumor behavior and response to therapy (5). The contribution of these studies to the understanding of glioblastoma development is unquestionable. However, they explore just a subset of alterations that lead to gliomagenesis. Co- and posttranscriptional events such as splicing, polyadenylation, mRNA decay, and translation are important components of gene expression, and alterations to these are known to be involved in many human diseases, including cancer. These processes are controlled by RNA-binding proteins, which are commonly deregulated in tumors (6).
Many RNA-binding proteins have important roles in the initiation and progression of glioblastoma, including LIN28 (7), PCBP2 (8), HuR (9–11), Quaking (12), and Musashi1 (MSI1) (13–21). Among these, MSI1 stands out due to its evolutionary conservation, connection to neural stem cells, and role in balancing self-renewal and differentiation (18, 22–26). High expression levels of MSI1 have a prognostic value in glioma (21) and in the pediatric malignant brain cancer medulloblastoma (27).
Previous literature has suggested a potentially critical role for MSI1 in glioblastoma based on a small number of MSI1 target mRNAs (19). We sought to identify the complete collection of MSI1 target mRNAs by using individual-nucleotide resolution cross-linking and immunoprecipitation (iCLIP) methodology coupled with RNA transcriptome sequencing (RNA-Seq). Our findings uncover a role for MSI1 as a central regulator of cell adhesion pathways that contribute to GBM by influencing cell adhesion, morphology, migration, and invasion.
MATERIALS AND METHODS
Cell culture.
U251 and U343 glioblastoma cells were obtained from the American Type Culture Collection (Manassas, VA) and cultured in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin.
Cells were transiently transfected with control siRNAs or MSI1 small interfering RNA (siRNA) using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) and harvested after 48 to 72 h for assays described below. siRNA was obtained from the Invitrogen Stealth RNAi collection (MSI1HSS106732, MSI1HSS106733, and MSI1HSS106734) and used in an equimolar pool. Invitrogen RNAi siRNA negative control, medium GC content was used as a control. Knockdown levels of MSI1 using siRNAs reproducibly exceeded 90% compared to the control siRNA transfection, as measured by quantitative reverse transcription-PCR (qRT-PCR).
Cell proliferation.
Transfected cells (siRNA control or MSI1) were harvested by using trypsin and replated with 104 cells per well in a 96-well tissue culture-grade plate. Cells were counted with the Countess automated cell counter (Invitrogen) using trypan blue exclusion for visualization. The cells were counted every day for three consecutive days.
Cell viability.
Cells were previously synchronized by serum starvation for 48 h, replated with 104 cells per well in a 96-well tissue culture-grade plate, and then transfected with siRNA (control or MSI1). After 72 h of transfection, cell viability was measured using CellTiter-Glo (Promega [Madison, CA]; catalog no. G7570) according to the manufacturer's instructions.
Cell cycle analysis.
Cells were transfected as described above. Cell cycle stages were analyzed by propidium iodide staining of DNA content. In brief, cells were harvested using trypsin and washed with phosphate-buffered saline (PBS). PBS-resuspended cells were added dropwise to 200-proof ethanol for fixation and incubated on ice for at least 30 min. The cells were then pelleted via centrifugation at 1,000 rpm for 5 min. The cell pellet was resuspended in a PBS solution containing 20 mg of RNase A/ml, 0.1% (vol/vol) Triton X-100, and 20 μg of propidium iodide/ml. The solution was incubated at 37°C for 15 min. The cells were then analyzed via fluorescence-activated cell sorting on the BD FACSCanto flow cytometry machine (BD Biosciences, San Jose, CA). Flow cytometry data were analyzed using Modfit LT (Verity Software House, Topsham, ME) to calculate cell cycle distribution.
Annexin V staining.
Transfected cells were analyzed for annexin V staining using an annexin V-phycoerythrin (PE) apoptosis detection kit I (BD Pharmingen [San Diego, CA]; catalog no. 559763). In brief, cells were treated with trypsin to prevent further apoptosis, washed with ice-cold PBS, and resuspended in 1× binding buffer. The PE-annexin V staining solution and 7-aminoactinomycin, used to detect cell viability, were added to the resuspended cell solution, followed by incubation at room temperature for 15 min. The annexin V staining was then analyzed on a BD FACSCanto flow cytometry machine (BD Biosciences).
Caspase-3/-7 activity assay.
In U251 and U343 transfected cells, the activated caspase-3/-7 activity was measured with Caspase-Glo 3/7 assay (Promega; catalog no. G8091) according to the manufacturer's instructions, and luminescence was measured with a Molecular Devices SpectraMax M5 microplate reader.
In vitro scratch assay.
Glioblastoma cells were grown in a 96-well ImageLock cell culture plate (Essen BioScience, Ann Arbor, MI) in a standard CO2 incubator. The cells were transfected at a low density with control or MSI1 siRNAs for 48 h to ca. 100% confluence. The floor of the wells was left uncoated. A 96-pin WoundMaker (Essen BioScience) was used to create precise and reproducible wounds in the 96-well plate. The assay plates were then equilibrated for 15 min within the IncuCyte automated microscope system (Essen BioScience) before the first scan. The software was set to scan every 15 min for 4 days. The data were analyzed by using the relative wound density integrated metric. At the end of the experiment, the data were inspected using analysis of variance and displayed as means ± the standard deviations.
Cell migration.
For single cell migration analyses, glioblastoma cells were seeded on a collagen coated 24-well plate (3 × 103/500 μl). A single cell migration was monitored over 10 h by time-lapse analyses (1 frame/5 min) using a Leica TCS SP5 confocal microscope equipped with a Ludin cube incubation chamber with a ×20 objective lens. The speed of cell migration was determined as the mean displacement (in micrometers per minute) of tracked cells using the “manual tracking” plugin (http://rsbweb.nih.gov/ij/plugins/track/track.html) for ImageJ. Statistical significance was determined by using a Student t test.
Cell morphology.
U343 and U251 cells were grown on glass coverslips, fixed, and processed for indirect immunostaining with the following antibodies: anti-MSI1 antibody (Merck-Millipore [Darmstadt, Germany]; catalog no. 04-1041), DAPI (4′,6′-diamidino-2-phenylindole; Sigma [St. Louis, MO]; catalog no. D9542) and phalloidin-TRITC (Sigma; catalog no. P1951). Image acquisition was performed on a Leica TCS SP5 confocal microscope using a ×63 Plan Apo oil objective lens.
Cell invasion assay.
Cell invasion was measured in vitro with a Boyden chamber assay using a CytoSelect 24-well cell invasion assay (Cell Biolabs, San Diego, CA). U251- and U343-transfected cells were harvested using trypsin and counted. Next, 0.5 × 106 cells were resuspended in serum-free medium. A total of 500 μl of medium containing 10% fetal bovine serum was added to the lower chamber, and 300 μl of resuspended cells was placed in the upper chamber. The assay mixture was incubated at 37°C and an 5% CO2 atmosphere for 24 h. The plate was removed, and the medium inside the insert was aspirated. The chamber was stained using cell stain solution and washed with deionized water. A microscopic image of the insert was taken with a Nikon Eclipse TS100 inverted microscope equipped with a DS-L2 camera control unit (Nikon Instruments, Inc., Melville, NY) at ×20 magnification. The stained cells were then extracted using 200 μl of extraction solution. Finally, 100 μl of the extracted solution was added to a 96-well microtiter plate, and the optical density at 560 nm was measured with a BioTek Synergy HT microplate reader (BioTek, Winooski, VT).
Fibronectin cell adhesion assay.
Ninety-six-well plates were coated with 0, 10, and 20 μg of human fibronectin (BD Biosciences; catalog no. 354008)/ml. The wells were washed with PBS and blocked with 3% bovine serum albumin for 60 min. At 72 h after control siRNA or MSI1 siRNA transfection (as described above), 5 × 105 cells were added to each well. The plates were incubated for 30 min at 37°C and 5% CO2 and then washed with PBS. Paraformaldehyde solution (4%) was used to fix adherent cells for 15 min at room temperature, followed by washes with PBS. Adherent cells were detected by crystal violet staining (5 mg/ml in 2% ethanol; Sigma-Aldrich [St. Louis, MO]; catalog no. C3886) for 10 min. The plates were extensively washed with water and allowed to dry completely. Then, 2% sodium dodecyl sulfate (SDS) was used to lyse the cells for 30 min, and the absorbance was read at 550 nm.
Western blotting.
Cells were resuspended and sonicated in Laemmli sample buffer, separated on a 10% SDS-PAGE gel, and transferred. After transfer, the membrane was blocked in Tris-buffered saline with Tween 20 and 5% milk. The membranes were probed with a collection of different antibodies, including anti-MSI1 antibody (Abcam [Cambridge, MA]; catalog no. ab52865; dilution, 1:2,000), anti-IGF1R antibody (Santa Cruz Biotechnology [Santa Cruz, CA]; catalog no. sc-713; dilution, 1:500), anti-epidermal growth factor receptor (anti-EGFR) antibody (Cell Signaling Technology [Danvers, MA]; catalog no. 2646; dilution, 1:200), anti-PDGFRα antibody (Cell Signaling Technology; catalog no. 5241; dilution, 1:1,000), and anti-α-tubulin antibody (Sigma; catalog no. T5168; dilution, 1:4,000). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (Santa Cruz Biotechnology) or HRP-conjugated goat anti-mouse antibody (Zymed Laboratories, Carlsbad, CA) was used as a secondary antibody. Electrochemiluminescence was used to detect the selected proteins using Immobilon Western chemiluminescence substrate (Millipore, Billerica, MA).
Polysomal gradient and analysis.
Cells were incubated for 10 min with 0.1 mg of cycloheximide (Sigma-Aldrich)/ml and then harvested in 1 ml of polysome extraction buffer (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 5 mM MgCl2, 0.3% Igepal CA-630, and 0.1 mg of cycloheximide/ml) on ice for 10 min. Cytoplasmic lysates were fractionated by centrifugation through 10 to 50% linear sucrose gradients. The eluted fractions were prepared with a fraction collector and their quality monitored at 254 nm using a UV-6 detector (Teledyne Isco, Lincoln, NE). RNA from each of the 12 fractions was extracted using TRIzol (Invitrogen; catalog no. 15596-026) and used for qRT-PCR analysis to determine the distributions of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), EGFR, and insulin-like growth factor 1 receptor (IGF1R) mRNAs.
Alternative splicing analysis.
Total RNA was extracted using the TRIzol reagent according to manufacturer's instructions. Reverse transcription of messenger RNAs was performed using a high-capacity cDNA reverse transcription kit (Applied Biosystems [Warrington, WA]; catalog no. 4368814) with random priming. For mRNA analysis, quantitative PCR was performed using the primers listed below and Power SYBR green PCR master mix (Applied Biosystems; catalog no. 4309155). Real-time PCRs were performed on a 7500 real-time PCR system (Applied Biosystems). The data were acquired using the SDS 2.0.1 software package (Applied Biosystems) and analyzed using the 2−ΔΔCT method with GAPDH as an endogenous control. The primers used were as follows: GAPDH forward (CCCCTGGCCAAGGTCATCCA), GAPDH reverse (ACAGCCTTGGCAGCGCCAGT), ZAK control forward (TCAGTGTTGGGAAGCTGATG), ZAK control reverse (CAGGAAGGCTCGTGTCATTT), ZAK short forward (GCATTCTGGGATGCAGATAAAC), ZAK short reverse (TCATCATCATCGTCACCTTCTG) ZAK long forward (CTCAGACAGAAGCAGGAACAA), and ZAK long reverse (GGAGTGGAATGCTGACTCTTT).
iCLIP experiments.
U251 cells were plated on 100-mm dishes and cross-linked when semiconfluent in 6 ml of PBS with a Spectrolinker XL-1500 two times at 100 mJ/cm2. Cells were scraped, pelleted, and snap-frozen. iCLIP was performed as described previously (28) using Abcam rabbit anti-MSI1 monoclonal antibody (catalog no. ab52865). Samples were amplified, purified with Beckman Genomics Ampure XP beads (Beckman Coulter Genomics, Danvers, MA), and quantitated with an 2100 Bioanalyzer and DNA 1000 chips (Agilent Technologies, Santa Clara, CA). Three biologic replicates were combined, and 50SE sequencing was performed on a Hi-Seq 2000 (Illumina, Inc., San Diego, CA).
Mapping of iCLIP sites and RNA-Seq data analysis.
To map high-throughput sequence data, we constructed a version of the hg18 genome in which the regions outside the genes were masked out. The transcriptome used was RefSeq (29), downloaded from the UCSC genome browser, which was also used for visualization (30, 31). RMAP (32) was used for read mapping. Peak identification from mapped iCLIP data was performed using Piranha (33). For binding site characterization, we used the union of all significant peaks from all replicates to enhance sensitivity. Identification of differential gene expression in RNA-Seq was performed using EdgeR (34), and differential usage of exons was calculated using the Fisher exact test. Enrichment of trinucleotides around iCLIP sites was conducted by calculating an observed over expected ratio, where the expected occurrences (stratified by region type: 3′ untranslated region [3′ UTR], 5′ UTR, coding sequence, and intronic) were determined from a collection of public iCLIP data sets for other RBPs (for full details, see the supplemental material). Computational prediction of RNA secondary structure (base-pairing probability) was conducted using an implementation of McCaskill's algorithm (35).
The Musashi1 targets used in gene ontology and pathway analyses consist of genes containing at least one significant iCLIP peak in either the 3′ UTR or the 5′ UTR in at least two replicates. Enrichment for the KEGG pathways was performed with DAVID (36) using a background of the expressed genes, as determined by our RNA-Seq data (defined as the genes with at least a single read in all total RNA replicates). (Protein-protein interactions were extracted from iRefIndex [37] and visualized with Circos (http://circos.ca/.) Full details of the computational methods used are provided in the supplemental material.
MSI1 gene expression analysis using TCGA GBM RNA-Seq data.
RNA sequencing raw reads of 167 GBM samples (43 classical, 40 proneural, 28 neural, and 56 mesenchymal) from TCGA (http://cancergenome.nih.gov/) were downloaded from Cancer Genomics Hub (https://browser.cghub.ucsc.edu). Normal brain samples were downloaded from the SRA database (www.ncbi.nlm.nih.gov/sra/ [SRP028705]). Reads were mapped against the human genome (version hg19/GRCh37; UCSC Genome Browser [http://genome.ucsc.edu]) using GSNAP (38). Mapped reads presenting a quality (Q) of ≥20 (Phred scale) were selected using Samtools (39). Read counts per gene were defined using HT-Seq (40) and Gencode (v19 [http://www.gencodegenes.org/]) as reference transcriptomes. Expression analysis was performed using DESeq2 (41) comparing tumor samples from each GBM subtype to normal samples. MSI1 expression is presented as log2 fold change.
Microarray data accession number.
Next-generation sequencing data, gene expression data, and iCLIP peak locations have been submitted to GEO (GSE68800).
RESULTS
iCLIP analysis reveals novel aspects of MSI1 binding site.
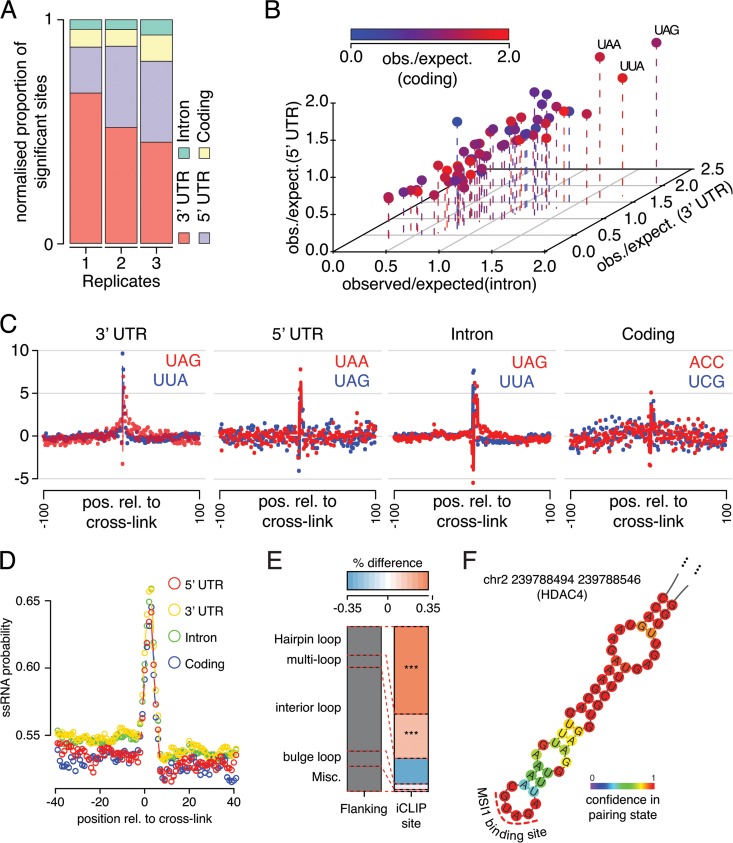
RNA-binding proteins affect gene expression in multiple ways and can regulate hundreds of targets in a given cell type (42). MSI1 is an important player in glioblastoma and displays high expression in all subgroups according to the TCGA data sets (see Fig. S1 in the supplemental material). We used iCLIP (28) to map MSI1 binding sites in the U251 glioblastoma line. This assay uses UV light to cross-link protein-RNA interactions, followed by immunoprecipitation of MSI1-interacting ribonucleoproteins (RNPs) (see Fig. S2 in the supplemental material), digestion, and finally high-throughput sequencing of the RNAs from these RNP complexes. The resultant next-generation sequencing reads are then mapped to the transcriptome. By exploiting the tendency of reverse transcriptase to truncate at the cross-link location, iCLIP provides us with single-nucleotide identification of MSI1-RNA cross-link locations. After the appropriate normalization of sequencing results, significant MSI1 iCLIP sites (P < 0.05) (see Materials and Methods for the iCLIP analysis) were enriched in 3′ UTRs and 5′ UTRs, as expected for a protein primarily known for its role in translation (Fig. 1A; see Fig. S3A in the supplemental material; Table S1 in the supplemental material contains complete sets of identified sites).
FIG 1.
Novel characteristics of the Musashi1 binding site revealed by iCLIP analysis. (A) Proportion of significant iCLIP sites falling within introns, coding regions, 3′ UTRs, and 5′ UTRs. The proportions are normalized to the total region size. (B) Observed/expected ratio for all trinucleotides around iCLIP sites stratified by region type where the site is located. (C) Z-scores for the number of occurrences of the top two trinucleotides in each region from panel B shown relative to the iCLIP cross-link site. (D) Average probability of nucleotide adopting single-stranded RNA (ssRNA) conformation for all nucleotides relative to iCLIP cross-link location, stratified by the site's location. (E) Proportion of bases predicted to be in each of the five possible ssRNA conformations within iCLIP sites compared to flanking regions (***, the P < 0.001 odds ratio for a given conformation between flanking and iCLIP sites is >1 [Fisher exact test]). (F) Representative structure of an iCLIP target site in the gene HDAC4, showing presence of the predicted MSI1 binding site in the hairpin loop.
The UAG trinucleotide, which has been previously reported as the core of the MSI1 binding site (43), was generally enriched around MSI1 iCLIP sites (Fig. 1B and C). Interestingly, however, UAG enrichment was not present in coding regions. As discussed further below, binding to UAGs that function as stop codons was rarely detected.
Nuclear magnetic resonance structures have suggested that the two RNA recognition motif (RRM) domains of MSI1 act independently to recognize RNA motifs (43). We investigated the tendency of UAG/GUAG motif pairs, in contrast to isolated UAG or GUAG sequences, to occur at iCLIP sites compared to flanking regions in different contexts. Interestingly, there was a marked increase in paired versus unpaired motifs around the 3′-UTR iCLIP sites (roughly 2× greater, P < 2.5 × 10−11 [Fisher exact test]) (see Fig. S3B and C in the supplemental material) but little discernible trend in 5′ UTRs, introns, or coding sequences (see Fig. S3D to F in the supplemental material). A comparative analysis between different contexts revealed that 3′-UTR sites have a significantly greater proportion of paired motifs than sites in other transcript areas (P < 6.74 × 10−14). Our data suggest the existence of different binding site characteristics based on context, but more detailed studies are required to determine whether these relate to functional differences.
MSI1 iCLIP sites showed a strong preference toward single-stranded regions (Fig. 1D) and, in particular, were contained in stem-loops (Fig. 1E). Interestingly, there was little discernible difference in structural preference between UTR, intronic, and coding sites, unlike the sequence preference (which, as mentioned above, avoids UAG and generally displays less sequence specificity in coding regions [Fig. 1C]). Representative structures are shown in Fig. 1F and in Fig. S4 in the supplemental material, where the MSI1 binding site occurs in the hairpin loop.
Since UAG forms the core of the expected MSI1 binding site, we looked for evidence of MSI1 binding to UAG stop codons, speculating that such binding may interfere with translation termination. However, we saw no preference for iCLIP sites around these locations (see Fig. S3G in the supplemental material). Computational prediction of the structure around human UAG stop codons indicated a region of double-stranded RNA preference encroaching on the UAG (see Fig. S3H in the supplemental material). Previous experimental findings have suggested strong structural signals at stop codons in yeast (44), and we speculate that the secondary structure at these sites is not conducive to MSI1 binding, explaining the lack of enrichment for iCLIP sites near stop codons, despite the apparently favorable sequence.
Musashi1 has modest impact on alternative splicing.
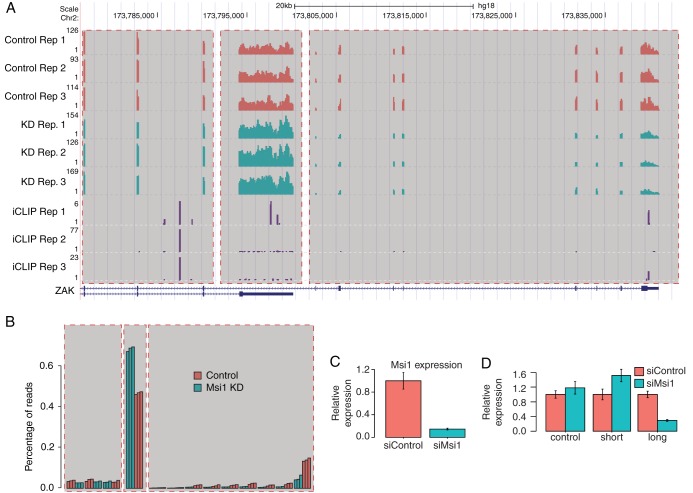
Since we identified a substantial number of MSI1 iCLIP sites in intronic locations (see Fig. S3A in the supplemental material), we sought to determine whether MSI1 is involved in regulating alternative splicing. We identified 26 genes with significant changes in exon usage upon MSI1 knockdown (see Table S3 in the supplemental material); 7 of the 26 genes also contain MSI1 iCLIP sites in proximity to the exons with changed inclusions rates in all three replicates, a number that is more than three times greater than that expected by chance (P < 6.9 × 10−3 [Fisher exact test]), and this hints at a direct involvement of MSI1 in these changes. In some cases, the affected exons are located in the coding sequence, whereas some genes show changed read profiles in 3′-UTR exons (Fig. 2; also see Fig. S5A to D in the supplemental material).
FIG 2.
Musashi1 impact on splicing. (A) UCSC genome-browser image showing read coverage for RNA-Seq and iCLIP replicates in the gene ZAK. Region α highlights exons with no significant change in inclusion ratio, region β (short isoform) highlights exons with an increased inclusion ratio upon MSI1 knockdown, and region γ (long isoform) highlights exons with a decreased inclusion ratio upon Musashi1 knockdown. MSI1 knockdown shows a shift in usage between the two isoforms. (B) Schematic representation of panel A, showing the proportion of reads for ZAK falling within each of its exons in each replicate. (C) Relative mRNA levels for MSI1 after knockdown for qRT-PCR validation of ZAK splicing event show successful knockdown. (D) Relative mRNA levels as measured by qRT-PCR of the short ZAK isoform (containing region β), the long ZAK isoform (containing region γ), and a control upon knockdown of MSI1 confirm the same change in usage as observed in the RNA-Seq data.
Although MSI1 iCLIP sites are not enriched in coding or intronic regions, in absolute terms we did observe a large number of such sites (see Fig. S3A in the supplemental material). The presence of only a small number of genes displaying changes in exon usage and alternative polyadenylation in spite of this motivated us to examine the distribution of iCLIP sites around splice junctions and polyadenylation signal sites more closely. The majority of intronic and coding MSI1 iCLIP sites are localized more than 200 nucleotides from splice sites (see Fig. S5E in the supplemental material); we observed only a modest enrichment of iCLIP sites in coding regions near splice sites, only marginally more than that expected by chance under the assumption of a uniform random distribution. This suggests that MSI1, at least in U251 cells, plays at most a minor role in regulating alternative splicing. With respect to polyadenylation, interestingly, MSI1 iCLIP sites are strongly depleted around polyadenylation signal sites (see Fig. S5F in the supplemental material), suggesting that if MSI1 does regulate polyadenylation in this cell system, the mechanism is unlikely to be via blockade of the signal site. It is worth mentioning that MSI has been shown in a different system to regulate polyadenylation and the signals involved to be conserved across evolution. In the Xenopus oocyte, polyadenylation of DNMT1, a gene critical for epigenetic control of various genes and for genome stability, requires MSI function. It was later shown that MSI interacts with poly(A) polymerase germ line development defective 2 (GLD2) protein, suggesting that coupling of MSI to the polyadenylation apparatus is a conserved mechanism to promote target mRNA translation (45, 46).
Musashi1 functions as a central regulator of cell adhesion pathways.
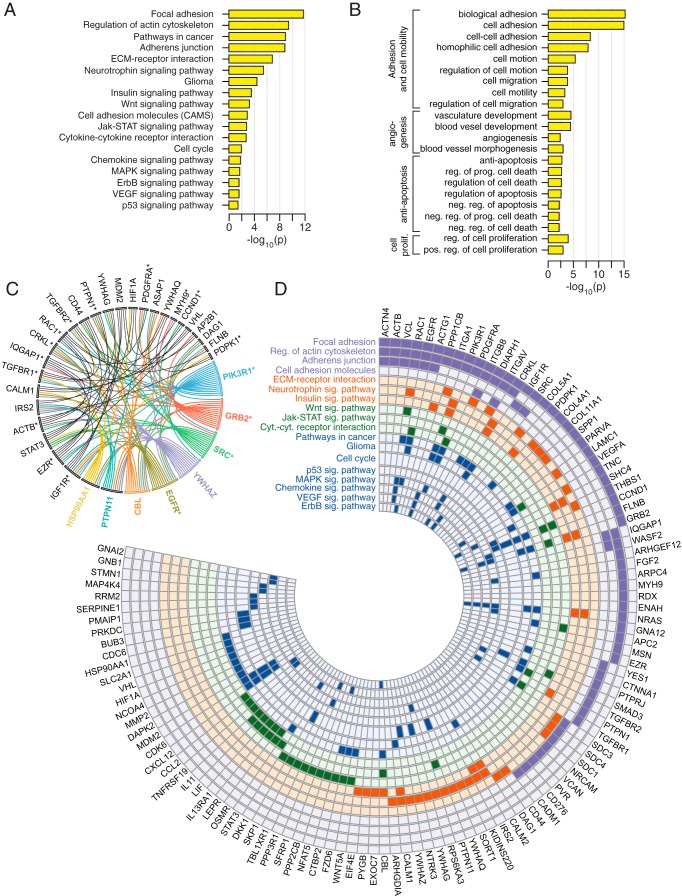
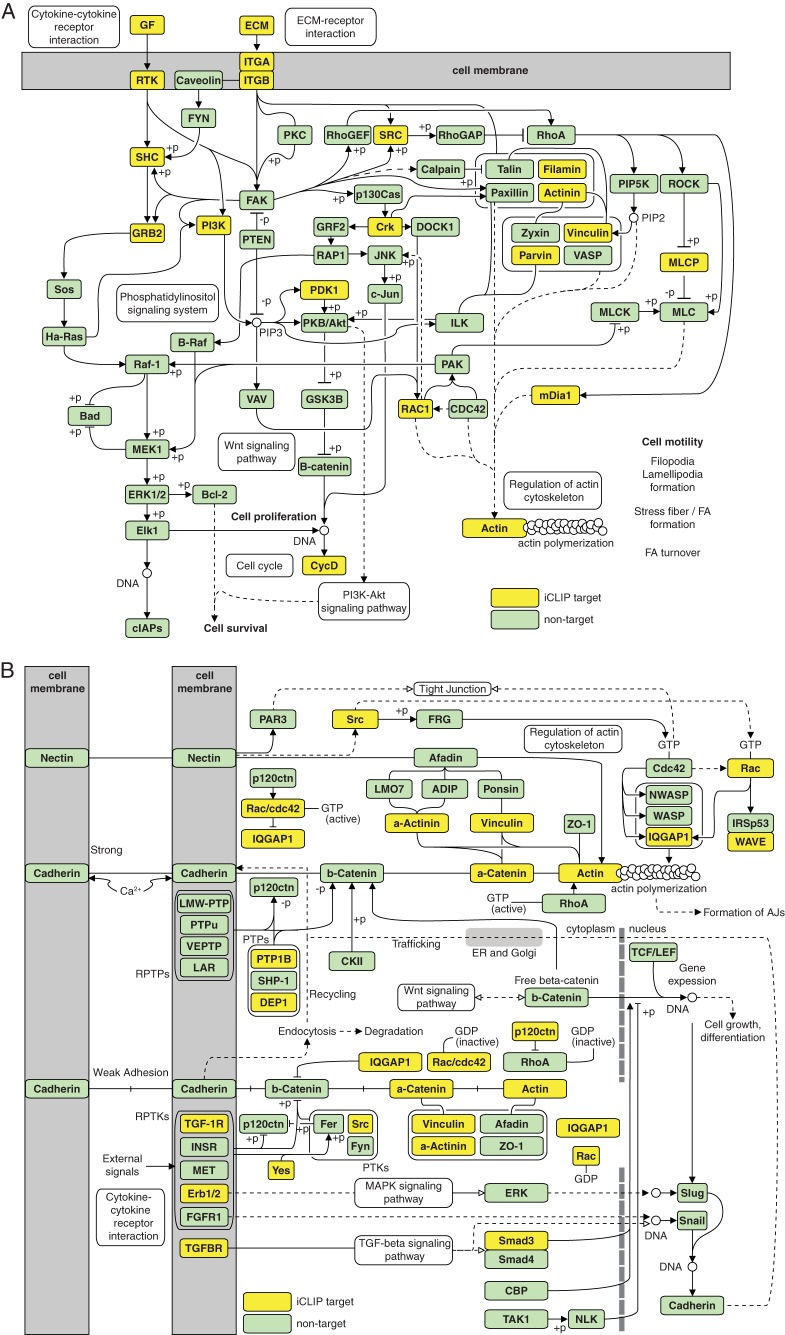
Enrichment analysis done with KEGG pathways and biological processes (corrected P < 0.05) revealed that mRNAs with MSI1 iCLIP tags in the 3′ UTRs encode proteins strongly associated with glioblastoma development (Fig. 3A and B; for a complete list, see Tables S4 and S5 in the supplemental material). We observed an extensive enrichment in adhesion-related pathways and processes, particularly the focal adhesion pathway, which impacts cell mobility and invasiveness. The distribution of MSI1 targets in the focal adhesion and adherens junction pathways is shown in Fig. 4. Other enriched pathways associated with glioblastoma growth include cytokines, chemokines, ErbB, p53, Wnt, insulin, vascular endothelial growth factor (VEGF), and mitogen-activated protein kinase (MAPK). Further, we selected the MSI1 iCLIP targets in the enriched KEGG pathways and constructed an interaction network using iRefIndex (shown in Fig. 3C). This software relies on protein-protein interaction information to build the associations. The main nodes based on the number of connections are the genes for PTPN11, IGF1R, HSP90AA1, PIK3R1, GRB2, SRC, YWHAZ, EGFR, and CBL, all of which have been implicated in tumorigenesis. This analysis also shows that target genes implicated in adhesion are highly interconnected (Fig. 3C).
FIG 3.
Gene ontology and pathway analyses for Musashi1 target sets. (A) Enrichment levels (P values) for KEGG pathways significantly enriched (corrected P < 0.05) in MSI1 iCLIP target genes. (B) Same as in panel A, but for biological processes. (C) Protein-protein interaction network for MSI1 iCLIP target genes present in enriched KEGG pathways. Shown are genes with at least three interactions in the network. Genes associated with adhesion pathways are indicated with an asterisk. (D) Distribution of MSI1 iCLIP targets in enriched KEGG pathways from panel A.
FIG 4.
MSI1 target distribution in the focal adhesion (A) and adherens junction (AJ) (B) pathways.
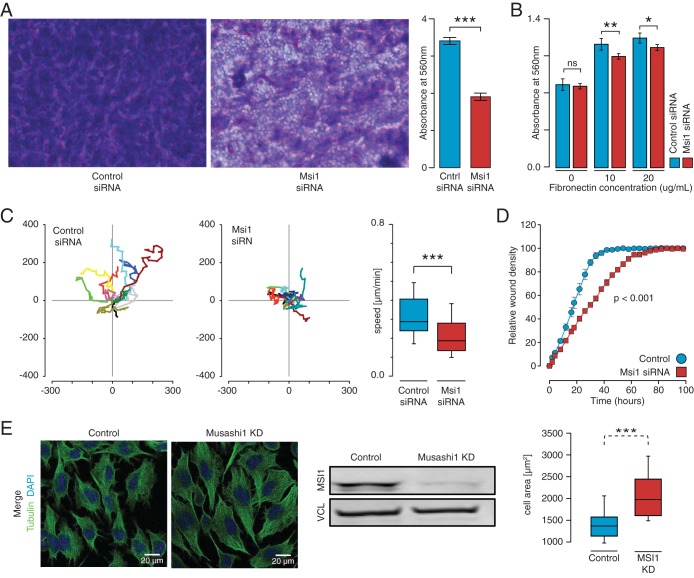
MSI1's involvement in cell proliferation, apoptosis, and the cell cycle has been previously described in other systems (47–51), and it was here corroborated in glioblastoma cells (see Fig. S6 in the supplemental material). In addition, our analysis uncovered a novel association of MSI1 with adhesion pathways/functions and set out to investigate the effect of modulating MSI1 on the relevant phenotypes. Interesting, MSI1 knockdown impaired cell adhesion and contributed to changes in cell morphology (Fig. 5B and E; see also Fig. S7B and D in the supplemental material). To interrogate the involvement of MSI1 in migration and invasion, we first performed a cell invasion transwell assay. A reduction in MSI1 expression promoted a decrease in the number of cells that were able to invade the basement membrane compared to the control siRNA transfection (Fig. 5A; see also Fig. S7A in the supplemental material). Subsequently, an in vitro scratch assay demonstrated that MSI1 silencing disrupts the ability to close a wound in cell culture, indicating that MSI1 promotes a cell migratory phenotype (Fig. 5D; see also Fig. S7C in the supplemental material). Corroborating these results, we conducted single-cell migration analysis and determined that MSI1 knockdown affected migration speed (Fig. 5C). Altogether, these results indicate that MSI1 contributes to GBM adhesion, migration, and invasion.
FIG 5.
MSI1 impact on cell adhesion, morphology, migration, and invasion in U251 glioblastoma cells. (A) A colorimetric cell invasion assay was used to determine the effects of MSI1 on the invasiveness of U251 cells through basement membrane. Cells were resuspended in serum-free media, and serum-containing medium was used as a chemoattractant. A basement membrane was used as a barrier between both chambers. After 24 h of incubation, cells were stained, extracted, and quantified. A denser staining (in purple) was observed in the control siRNA cells compared to the MSI1 siRNA cells at ×40 magnification. Measuring the extracted cell solution demonstrated that MSI1 silencing impairs in vitro cell invasion. The data were analyzed with a Student t test and are presented as means ± the standard errors of the mean. (B) A fibronectin cell adhesion assay was used to determine the effects of MSI1 on cell adhesion in glioblastoma cells. MSI1 silencing reduced the capacity of U251 cells to adhere to the fibronectin substrate. The data were analyzed with Student t test and are presented as means ± the standard errors of the mean. *, P < 0.05; **, P < 0.01. (C) U251 cells were transfected with indicated siRNAs and seeded on a collagen-coated 24-well plate before single-cell tracking using time-lapse microscopy (1 frame/5 min for 10 h). Representative cell tracks for 10 cells are shown; in each case, control and MSI1 knockdown are shown in the left and middle panels. The mean speed of the cells was determined by the manual tracking plugin for ImageJ in three independent analyses. The data for control and MSI1 knockdown are shown as box plots in the right panel (n > 40 cells per condition). Migration of MSI1 knockdown cells was significantly slower compared to the control cells, whereas directionality was not affected (data not shown). Representative movies are provided as supplemental material. Bar, 50 μm. Error bars indicate the standard deviations of at least three independent experiments. ***, P < 0.001. (D) An in vitro scratch assay was used to further assess the effects of MSI1 on the ability for U251 cells to migrate. An IncuCyte automated microscopy system was used to evaluate scratch wound closure in real time. The data show the relative wound density. Silencing of MSI1 resulted in an impairment of wound closure in vitro. The data were analyzed with analysis of variance and are presented as means ± the standard errors of the mean. (E) Impact of MSI1 silencing on cell morphology. U251 cells were transfected with control and MSI1 siRNAs, fixed, and processed for indirect immunostaining. Representative images are shown. The cell area of transfected cells (n = 25) is shown as a box plot (right panels). Bar, 25 μm. Error bars indicate the standard deviations of at least three independent experiments. ***, P < 0.001.
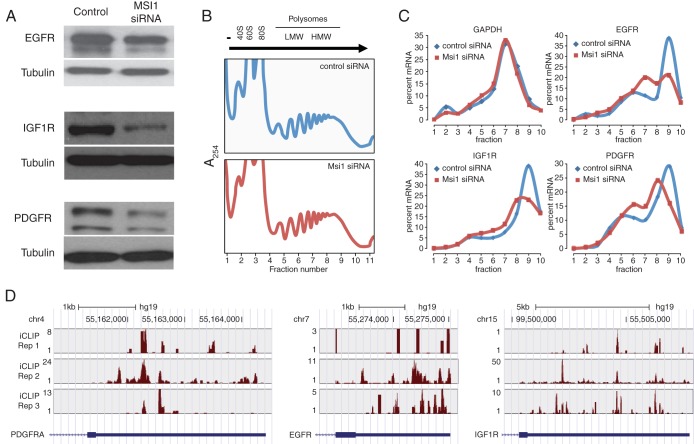
To further examine the impact of MSI1 on cell adhesion and related processes, we investigated whether MSI1 contributes to the translation of the genes for platelet-derived growth factor receptor alpha (PDGFRα), EGFR, and IGF1R. These three genes contain MSI1 binding sites in their 3′ UTRs and are known to be critical to glioblastoma development thanks to their adhesion related functions. Western blot experiments showed a reduction of PDGFRα, EGFR, and IGF1R levels upon MSI1 knockdown (Fig. 6A). Our RNA-Seq data did not show a statistically significant difference in mRNA levels for these three genes between control and MSI1 knockdown conditions (P > 0.05 [edgeR]). To confirm that observed changes in protein levels are driven by MSI1 impact on the translation of these three genes, we fractionated cytoplasmic components of control and MSI1 knockdown cells through sucrose gradients (Fig. 6B) and investigated the distribution of their mRNAs along the gradient using qRT-PCR. As shown in Fig. 6C, a decrease in MSI1 levels reduced the presence of these three mRNA species in the heavy polysome fractions. GAPDH mRNA, which serves as a negative control, did not experience any shift in its profile. These results suggest that MSI1 acts as a translational activator of these three mRNAs.
FIG 6.
MSI1 controls translation of key targets in adhesion pathways. (A) A Western blot of MSI1 targets (IGF1R, EGFR, and PDGFRα) demonstrates changes in protein levels upon MSI1 silencing. Tubulin was used as an endogenous, loading control. (B) Aspect of the polysomal gradient of control and MSI11 knockdown samples. (C) Analysis of MSI1 targets involved in adhesion pathways. Polysomal gradients of iCLIP-identified MSI1 targets (IGF1R, EGFR, and PDGFRα mRNAs) and GAPDH mRNA (control) in control and MSI1 knockdown cells. RNA isolated from different fractions was analyzed by qRT-PCR. The leftward shift in polysome distribution curves after MSI1 knockdown suggests that MSI1 promotes their translation. (D) UCSC genome browser plots showing iCLIP read coverage in the 3′ UTRs of the PDGFRA, EGFR, and IGF1R genes; iCLIP coverage was minimal in other regions of these genes.
DISCUSSION
Several studies have described high MSI1 expression in different tumor types and subsequent association with poor prognosis. However, only a few recent studies have reported on the biological consequence of MSI1 in cancer. In colon cancer, silencing of MSI1 induced apoptosis, mitosis, G2/M arrest, and tumor regression (51). In breast cancer, silencing of MSI1 resulted in decreased tumor mammosphere formation, decreased proliferation, and reduced breast cancer xenograft growth (50). It was also shown that tenascin C (TNC) promotes the fitness of metastasis-initiating breast cancer cells by enhancing the expression of LGR5 and MSI1 (52). In lung cancer, MSI1 silencing reduced spheroid colony formation with inhibition of the Wnt and Notch pathways (49). Silencing of MSI1 in Daoy medulloblastoma cells decreased proliferation, tumor growth, and neurosphere formation and induced differentiation and apoptosis (27, 47). MSI1's impact on tumorigenesis has frequently been explained based on regulation of the Notch inhibitor Numb and cyclin-dependent kinase inhibitor p21. Our results strongly suggest that this view is incomplete. In fact, neither Numb nor p21 appeared as direct targets of MSI1 in the CLIP analysis presented here and in our previous ribonucleoprotein immunoprecipitation followed by microarray analysis (RIP-chip) experiments (27, 53). The same is true for Msi2 (54). We propose that the observed effects on the Notch pathway do not occur via Numb and that changes in p21 expression upon MSI1 knockdown are indirect. Our results suggest that rather than working via a small group of target genes, MSI1 functions within an RNA operon (55) by coordinating the posttranscriptional regulation of several mRNAs that encode proteins that are functionally related.
Musashi1 target site.
The high resolution of the iCLIP assay provided us with the opportunity to closely examine the MSI1 binding site. After normalizing for region length, iCLIP sites are mostly enriched in untranslated regions, primarily 3′ UTRs, providing further evidence of MSI1's role in translational control. The binding site favors single-stranded conformation and contains the UAG triplet as its core. Interestingly, we observed differences in the enrichment of trimers around sites in different contexts. The strongest enrichment for the UAG core was observed in 3′ UTR iCLIP sites, followed by those in introns. However, sites in coding regions did not show UAG as the strongest motif. Moreover, triplets enriched in the coding sequence showed weaker signal than their counterparts in other contexts. In contrast to the sequence motif, the secondary structure preference showed remarkably little variation between contexts. In light of this, it seems likely that if MSI1 does bind to coding regions (as our iCLIP data suggest), it is targeted by a weaker sequence motif. More generally, we propose that the primary sequence is the major modulator of site strength, while the secondary structure serves as a refinement feature. We then investigated how often the two RRMs act together to recognize target mRNAs. Recent in vitro work has suggested GUAG and UAG as cores of the primary sequence motif recognized by each MSI1 RRM (43). Around MSI1 iCLIP sites in 3′ UTRs but not in other locations in the RNA, NUAG (where N is any nucleotide other than G) and GUAG tetramers appear significantly more often in pairs than in flanking regions, establishing that a proportion of MSI1 binding locations preferentially contain binding sequences for both RRMs. This difference in target site recognition (3′ UTRs versus other regions) might lead to differences in the quality and magnitude of the functional outcome. However, this remains to be investigated. The detected intronic iCLIP sites may be due to retained introns or unannotated transcripts, although the large number of sites argues against this. However, an interesting possibility emerged from the nuclear presence of MSI1 in glioblastoma tumors and cell lines; we identified as nuclear protein partners of MSI1 the chromatin remodeling factors INO80C and SMARCD1. We are testing a model whereby MSI1 uses its binding sites in nascent RNA to bridge its interaction with these proteins to regulate chromatin function (P. R. de Araujo, unpublished data).
Musashi1 involvement in cell adhesion, invasion, and migration.
Gene ontology and pathway analyses revealed that a large number of MSI1 targets are associated with adhesion and related functions. These associations translated into a direct impact on cell morphology, migration, cell adhesion and invasion and revealed novel functions of MSI1 (Fig. 3 to 6; see also Fig. S6 and S7 in the supplemental material). Unlike other tumor types, glioblastoma and other central nervous system tumors are unique in that cell migration and invasion are the main mechanisms of spread, rather than metastasis. Invasion into adjacent normal tissue is dependent on many different factors, including the interaction between the cell and the extracellular matrix and intrinsic changes to the cell itself to allow for active cell movement (reviewed in reference 56). In order to accomplish this, the cell body must change its morphology/polarization and adhere itself to the surrounding extracellular matrix (57). For this reason, proteins involved in cell adhesion play extremely important roles in the pathogenesis of cancer (58).
Among glioma-relevant targets in adhesion pathways, we could first highlight the mediators of cell motility beta- and gamma-actin. Inhibition of actin polymerization with molecules such as vanadate and phenylarsine oxide reduced cell spreading, migration, and invasion of glioma cells (50). The actin-associated protein actinin-4 was also identified as a target of MSI1. Actinin-4 is highly expressed in high-grade astrocytoma cells, and its knockdown resulted in reduced cell motility and cell adhesion (59, 60).
Three different integrins were identified as MSI1 targets: integrin-α1, integrin-αV, and integrin-β8. The integrin family is composed of 24 heterodimeric transmembrane cell surface receptors that connect the cytoskeleton of a cell with that of the extracellular matrix, thus allowing for cellular adhesion as well as signal transduction. Alterations of integrin expression, particularly overexpression (61, 62), are thought to contribute to the malignant character of glioblastoma by affecting cell adhesion, cell migration, angiogenesis (63), and radioresistance (64). Therapeutic targeting of integrins with compounds such as cilengitide (65), a synthetic pentapeptide (Arg-Gly-Asp), are proving effective at minimizing the cytotoxic effects of ionizing radiation (66). Integrin-α1 has been shown to be involved in focal adhesion formation through binding of other intracellular proteins (67), as well as acting as receptors for transmitting the signal required for cell migration and invasion (68). Integrin-αV, usually found in association with integrin-β3, has been widely implicated in cell motility and cell adhesion in glioblastoma. Inhibition of integrin-αV using monoclonal antibodies was able to inhibit the migration of astrocytoma cells (69). Integrin-αV may also be involved in brain metastasis since immunohistochemistry of brain metastases shows its increased expression (70).
Other adhesion-related MSI1 targets that deserve mention include PIK3R1, tenascin C, IGF1R, EGFR, and PDGFRα. p85α, encoded by PIK3R1, is a regulatory protein involved in the phosphatidylinositol 3-kinase (PI3K) complex. PIK3R1 knockdown in glioblastoma cell lines reduced proliferation and decreased migration velocity (71). Although very rare in common cancers (72) somatic mutations of PIK3R1 found in GBM can reduce the regulatory effect of p85α, resulting in an increased kinase activity and enhanced PI3K pathway activation (73). Tenascin C (TNC) was also identified as a target of MSI1, and its association with glioblastoma is well documented. The degree of malignancy in glioblastoma is positively correlated with TNC expression (74), most likely through mediating cell adhesion and cell motility. TNC mediates cell adhesion through its interaction with the FNIII domain of the receptor protein tyrosine phosphatase beta (71, 75). Notably, in breast cancer, MSI1 functions as a downstream effector of TNC in metastases (76, 77). Therefore, an interesting feedback loop involving MSI1 and TNC might exist. IGF1R is a tyrosine kinase that activates PI3K/Akt and RAS-RAF-MAPK signaling pathways (78). In glioblastoma, the stimulation of IGF1R by IGF1 or IGF2 leads to STAT3 activation, increasing the transcription factor hypoxia-inducible factor 1 (HIF1) protein levels and activity. The activation of HIF1 induces the expression of VEGF and IGF2; the first promotes angiogenesis, and the second is responsible for keeping the autocrine loop active (79). Interestingly, both HIF1 and VEGF were identified as MSI1 targets. PDGFRα encodes a cell surface tyrosine kinase receptor for members of the platelet-derived growth factor family; it can function in an autocrine and paracrine manner that results in cell proliferation, increased angiogenesis, and altered cell adhesion via deregulated integrin signaling (80). EGFR is the cell surface receptor for members of the epidermal growth factor family; it has been well characterized as an important signal transduction molecular in glioblastoma and other cancers. EGFR can also mediate cell-extracellular matrix interactions by interacting with CD44 (another target of MSI1), resulting in cell invasiveness (81). EGFR receives signal from focal adhesion kinase (FAK), mediating cell migration (82).
MSI1 could also influence adhesion related phenotypes via splicing. The majority of affected genes have clear roles in tumor invasiveness, adhesion, and growth, similar to what was found in the UTR analysis. One of the best examples is ZAK/MLTK; MSI1 promotes usage of the longer isoform (MLTKα; see Fig. 2), which, in contrast to the shorter isoform, is involved in the disruption of actin stress fibers and cell shrinkage (76), potentially implicating MLTK in tumor invasiveness.
In summary, our results indicate that MSI1 plays a significant role in GBM by coordinating the expression of an RNA operon associated with adhesion pathways, influencing therefore morphology, adhesion, migration, and invasion. The changes triggered by MSI1 expression will result in a boost in tumor spread and relapse, agreeing with the fact that high MSI1 expression is an indicator of poor prognosis (83).
Supplementary Material
ACKNOWLEDGMENTS
The results published here are in whole or part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/).
L.O.F.P. and A.D.S. conceived the study, designed the experiments, and coordinated data analysis and the writing. P.J.U. conducted the iCLIP and RNA-Seq data analysis, performed most of the subsequent computational and bioinformatics analyses, and coordinated the writing. D.T.V. performed the majority of the biological assays and was a major contributor to the writing process. P.R.D.A. conducted cell adhesion and migration studies and alternative splicing and performed several Western analyses. R.P. and C.K. designed and conducted experiments to determine the impact of MSI1 on cell migration and morphology. S.C.B. performed all iCLIP experiments and helped with biological assays. E.B.-S. helped with iCLIP analysis and subsequent computational analyses and performed secondary structure studies. M.Q. conducted Western analyses and several biological assays. J.U. helped with the implementation of the iCLIP experiments and the analysis platform. M.G., J.L.M., and K.A. contributed to MSI1 target analysis; R.D.S.A. helped with bioinformatics analysis. H.I.N. performed pathway analyses. B.R.C. performed bioinformatic analyses. All authors have read and approved the submission.
J.L.M., K.A., and M.G. were supported by the National Institute on Aging Intramural Research Program, National Institutes of Health. This study was supported by the Max and Minnie Tomerlin Voelcker Fund, Voices Against Brain Cancer, IIMS-CTRC (UTHSCSA), NIH R01HG006015, and 1R21CA175875-01A1. P.R.D.A. is supported by CPRIT RP140105 (Texas).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00410-15.
REFERENCES
- 1.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, Colman H, Chakravarti A, Pugh S, Won M, Jeraj R, Brown PD, Jaeckle KA, Schiff D, Stieber VW, Brachman DG, Werner-Wasik M, Tremont-Lukats IW, Sulman EP, Aldape KD, Curran WJ Jr, Mehta MP. 2014. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med 370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, Carpentier AF, Hoang-Xuan K, Kavan P, Cernea D, Brandes AA, Hilton M, Abrey L, Cloughesy T. 2014. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med 370:709–722. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N. 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN, Cancer Genome Atlas Research . 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galante PA, Sandhu D, de Sousa Abreu R, Gradassi M, Slager N, Vogel C, de Souza SJ, Penalva LO. 2009. A comprehensive in silico expression analysis of RNA binding proteins in normal and tumor tissue: identification of potential players in tumor formation. RNA Biol 6:426–433. doi: 10.4161/rna.6.4.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin R, Zhou J, Chen C, Xu T, Yan Y, Ma Y, Zheng Z, Shen Y, Lu Y, Fu D, Chen J. 2014. LIN28 is involved in glioma carcinogenesis and predicts outcomes of glioblastoma multiforme patients. PLoS One 9:e86446. doi: 10.1371/journal.pone.0086446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han W, Xin Z, Zhao Z, Bao W, Lin X, Yin B, Zhao J, Yuan J, Qiang B, Peng X. 2013. RNA-binding protein PCBP2 modulates glioma growth by regulating FHL3. J Clin Invest 123:2103–2118. doi: 10.1172/JCI61820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filippova N, Yang X, Wang Y, Gillespie GY, Langford C, King PH, Wheeler C, Nabors LB. 2011. The RNA-binding protein HuR promotes glioma growth and treatment resistance. Mol Cancer Res 9:648–659. doi: 10.1158/1541-7786.MCR-10-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolognani F, Gallani AI, Sokol L, Baskin DS, Meisner-Kober N. 2012. mRNA stability alterations mediated by HuR are necessary to sustain the fast growth of glioma cells. J Neurooncol 106:531–542. doi: 10.1007/s11060-011-0707-1. [DOI] [PubMed] [Google Scholar]
- 11.Filippova N, Yang X, King P, Nabors LB. 2012. Phosphoregulation of the RNA-binding protein Hu antigen R (HuR) by Cdk5 affects centrosome function. J Biol Chem 287:32277–32287. doi: 10.1074/jbc.M112.353912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen AJ, Paik JH, Zhang H, Shukla SA, Mortensen R, Hu J, Ying H, Hu B, Hurt J, Farny N, Dong C, Xiao Y, Wang YA, Silver PA, Chin L, Vasudevan S, Depinho RA. 2012. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes Dev 26:1459–1472. doi: 10.1101/gad.189001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toda M, Iizuka Y, Yu W, Imai T, Ikeda E, Yoshida K, Kawase T, Kawakami Y, Okano H, Uyemura K. 2001. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia 34:1–7. doi: 10.1002/glia.1034. [DOI] [PubMed] [Google Scholar]
- 14.Kanemura Y, Mori K, Sakakibara S, Fujikawa H, Hayashi H, Nakano A, Matsumoto T, Tamura K, Imai T, Ohnishi T, Fushiki S, Nakamura Y, Yamasaki M, Okano H, Arita N. 2001. Musashi1, an evolutionarily conserved neural RNA-binding protein, is a versatile marker of human glioma cells in determining their cellular origin, malignancy, and proliferative activity. Differentiation 68:141–152. doi: 10.1046/j.1432-0436.2001.680208.x. [DOI] [PubMed] [Google Scholar]
- 15.Strojnik T, Rosland GV, Sakariassen PO, Kavalar R, Lah T. 2007. Neural stem cell markers, nestin and musashi proteins, in the progression of human glioma: correlation of nestin with prognosis of patient survival. Surg Neurol 68:133–144. doi: 10.1016/j.surneu.2006.10.050. [DOI] [PubMed] [Google Scholar]
- 16.Vo DT, Qiao M, Smith AD, Burns SC, Brenner AJ, Penalva LO. 2011. The oncogenic RNA-binding protein Musashi1 is regulated by tumor suppressor miRNAs. RNA Biol 8:817–828. doi: 10.4161/rna.8.5.16041. [DOI] [PubMed] [Google Scholar]
- 17.Vo DT, Abdelmohsen K, Martindale JL, Qiao M, Tominaga K, Burton TL, Gelfond JA, Brenner AJ, Patel V, Trageser D, Scheffler B, Gorospe M, Penalva LO. 2012. The oncogenic RNA-binding protein Musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells. Mol Cancer Res 10:143–155. doi: 10.1158/1541-7786.MCR-11-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glazer RI, Vo DT, Penalva LO. 2012. Musashi1: an RBP with versatile functions in normal and cancer stem cells. Front Biosci (Landmark ed) 17:54–64. doi: 10.2741/3915. [DOI] [PubMed] [Google Scholar]
- 19.Muto J, Imai T, Ogawa D, Nishimoto Y, Okada Y, Mabuchi Y, Kawase T, Iwanami A, Mischel PS, Saya H, Yoshida K, Matsuzaki Y, Okano H. 2012. RNA-binding protein Musashi1 modulates glioma cell growth through the posttranscriptional regulation of Notch and PI3 kinase/Akt signaling pathways. PLoS One 7:e33431. doi: 10.1371/journal.pone.0033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagadec C, Vlashi E, Frohnen P, Alhiyari Y, Chan M, Pajonk F. 2014. The RNA-binding protein Musashi-1 regulates proteasome subunit expression in breast cancer- and glioma-initiating cells. Stem Cells 32:135–144. doi: 10.1002/stem.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahlrot RH, Hansen S, Herrstedt J, Schroder HD, Hjelmborg J, Kristensen BW. 2013. Prognostic value of Musashi-1 in gliomas. J Neurooncol 115:453–461. doi: 10.1007/s11060-013-1246-8. [DOI] [PubMed] [Google Scholar]
- 22.Sakakibara S, Imai T, Hamaguchi K, Okabe M, Aruga J, Nakajima K, Yasutomi D, Nagata T, Kurihara Y, Uesugi S, Miyata T, Ogawa M, Mikoshiba K, Okano H. 1996. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev Biol 176:230–242. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- 23.Sakakibara S, Okano H. 1997. Expression of neural RNA-binding proteins in the postnatal CNS: implications of their roles in neuronal and glial cell development. J Neurosci 17:8300–8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Good P, Yoda A, Sakakibara S, Yamamoto A, Imai T, Sawa H, Ikeuchi T, Tsuji S, Satoh H, Okano H. 1998. The human Musashi homolog 1 (MSI1) gene encoding the homologue of Musashi/Nrp-1, a neural RNA-binding protein putatively expressed in CNS stem cells and neural progenitor cells. Genomics 52:382–384. doi: 10.1006/geno.1998.5456. [DOI] [PubMed] [Google Scholar]
- 25.Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, Sawamoto K, Ogawa Y, Toyama Y, Miyata T, Okano H. 2000. Musashi1: an evolutionally conserved marker for CNS progenitor cells, including neural stem cells. Dev Neurosci 22:139–153. doi: 10.1159/000017435. [DOI] [PubMed] [Google Scholar]
- 26.Okano H, Imai T, Okabe M. 2002. Musashi: a translational regulator of cell fate. J Cell Sci 115:1355–1359. [DOI] [PubMed] [Google Scholar]
- 27.Vo DT, Subramaniam D, Remke M, Burton TL, Uren PJ, Gelfond JA, de Sousa Abreu R, Burns SC, Qiao M, Suresh U, Korshunov A, Dubuc AM, Northcott PA, Smith AD, Pfister SM, Taylor MD, Janga SC, Anant S, Vogel C, Penalva LO. 2012. The RNA-binding protein Musashi1 affects medulloblastoma growth via a network of cancer-related genes and is an indicator of poor prognosis. Am J Pathol 181:1762–1772. doi: 10.1016/j.ajpath.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruitt KD, Tatusova T, Brown GR, Maglott DR. 2012. NCBI reference sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res 40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler David. 2002. The human genome browser at UCSC. Genome Res 12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, Raney BJ, Pohl A, Malladi VS, Li CH, Lee BT, Learned K, Kirkup V, Hsu F, Heitner S, Harte RA, Haeussler M, Guruvadoo L, Goldman M, Giardine BM, Fujita PA, Dreszer TR, Diekhans M, Cline MS, Clawson H, Barber GP, Haussler D, Kent WJ. 2013. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith AD, Chung W-Y, Hodges E, Kendall J, Hannon G, Hicks J, Xuan Z, Zhang MQ. 2009. Updates to the RMAP short-read mapping software. Bioinformatics 25:2841–2842. doi: 10.1093/bioinformatics/btp533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uren PJ, Bahrami-Samani E, Burns SC, Qiao M, Karginov FV, Hodges E, Hannon GJ, Sanford JR, Penalva LO, Smith AD. 2012. Site identification in high-throughput RNA-protein interaction data. Bioinformatics 28:3013–3020. doi: 10.1093/bioinformatics/bts569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCaskill J. 1990. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 36.Huang DW, Sherman BT, Lempicki RA. 2008. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 37.Razick S, Magklaras G, Donaldson I. 2008. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics 9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing. 2009. The Sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S, Pyl PT, Huber W. 2015. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love M, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biology 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bahrami-Samani E, Vo DT, de Araujo PR, Vogel C, Smith AD, Penalva LOF, Uren PJ. 2015. Computational challenges, tools, and resources for analyzing co- and posttranscriptional events in high throughput. Wiley Interdiscip Rev RNA 6:291–310. doi: 10.1002/wrna.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohyama T, Nagata T, Tsuda K, Kobayashi N, Imai T, Okano H, Yamazaki T, Katahira M. 2012. Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions. Nucleic Acids Res 40:3218–3231. doi: 10.1093/nar/gkr1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. 2010. Genome-wide measurement of RNA secondary structure in yeast. Nature 467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cragle C, MacNicol AM. 2014. Musashi protein-directed translational activation of target mRNAs is mediated by the poly(A) polymerase, germ line development defective-2. J Biol Chem 289:14239–14251. doi: 10.1074/jbc.M114.548271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutledge CE, Lau H-T, Mangan H, Hardy LL, Sunnotel O, Guo F, MacNicol AM, Walsh CP, Lees-Murdock DJ. 2014. Efficient translation of Dnmt1 requires cytoplasmic polyadenylation and Musashi binding elements. PLoS One 9:e88385. doi: 10.1371/journal.pone.0088385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez-Diaz PC, Burton TL, Burns SC, Hung JY, Penalva LO. 2008. Musashi1 modulates cell proliferation genes in the medulloblastoma cell line Daoy. BMC Cancer 8:280. doi: 10.1186/1471-2407-8-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rezza A, Skah S, Roche C, Nadjar J, Samarut J, Plateroti M. 2010. The overexpression of the putative gut stem cell marker Musashi-1 induces tumorigenesis through Wnt and Notch activation. J Cell Sci 123:3256–3265. doi: 10.1242/jcs.065284. [DOI] [PubMed] [Google Scholar]
- 49.Wang XY, Yu H, Linnoila RI, Li L, Li D, Mo B, Okano H, Penalva LO, Glazer RI. 2013. Musashi1 as a potential therapeutic target and diagnostic marker for lung cancer. Oncotarget 4:739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang XY, Penalva LO, Yuan H, Linnoila RI, Lu J, Okano H, Glazer RI. 2010. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol Cancer 9:221. doi: 10.1186/1476-4598-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, Bishnupuri KS, Natarajan G, Anant S, Houchen CW. 2008. Knockdown of RNA binding protein Musashi-1 leads to tumor regression in vivo. Gastroenterology 134:1448–1458. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 52.Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massague J. 2011. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med 17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Sousa Abreu R, Sanchez-Diaz PC, Vogel C, Burns SC, Ko D, Burton TL, Vo DT, Chennasamudaram S, Le S-Y, Shapiro BA, Penalva LOF. 2009. Genomic analyses of Musashi1 downstream targets show a strong association with cancer-related processes. J Biol Chem 284:12125–12135. doi: 10.1074/jbc.M809605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park SM, Deering RP, Lu Y, Tivnan P, Lianoglou S, Al-Shahrour F, Ebert BL, Hacohen N, Leslie C, Daley GQ, Lengner CJ, Kharas MG. 2014. Musashi-2 controls cell fate, lineage bias, and TGF-β signaling in HSCs. J Exp Med 211:71–87. doi: 10.1084/jem.20130736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keene JD. 2007. RNA regulons: coordination of posttranscriptional events. Nat Rev Genet 8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 56.Kuersten S, Radek A, Vogel C, Penalva LOF. 2013. Translation regulation gets its ‘omics’ moment. Wiley Interdiscip Rev RNA 4:617–630. doi: 10.1002/wrna.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gotoh I, Adachi M, Nishida E. 2001. Identification and characterization of a novel MAP kinase kinase kinase, MLTK. J Biol Chem 276:4276–4286. doi: 10.1074/jbc.M008595200. [DOI] [PubMed] [Google Scholar]
- 58.Dziegiel P, Owczarek T, Plazuk E, Gomulkiewicz A, Majchrzak M, Podhorska-Okolow M, Driouch K, Lidereau R, Ugorski M. 2010. Ceramide galactosyltransferase (UGT8) is a molecular marker of breast cancer malignancy and lung metastases. Br J Cancer 103:524–531. doi: 10.1038/sj.bjc.6605750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Onodera T, Sakai T, Hsu JC, Matsumoto K, Chiorini JA, Yamada KM. 2010. Btbd7 regulates epithelial cell dynamics and branching morphogenesis. Science 329:562–565. doi: 10.1126/science.1191880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tao YM, Huang JL, Zeng S, Zhang S, Fan XG, Wang ZM, Yang HX, Yuan XH, Wang P, Wu F, Luo J, Zeng DY, Shen H. 2013. BTB/POZ domain-containing protein 7: epithelial-mesenchymal transition promoter and prognostic biomarker of hepatocellular carcinoma. Hepatology 57:2326–2337. doi: 10.1002/hep.26268. [DOI] [PubMed] [Google Scholar]
- 61.Canettieri G, Coni S, Della Guardia M, Nocerino V, Antonucci L, Di Magno L, Screaton R, Screpanti I, Giannini G, Gulino A. 2009. The coactivator CRTC1 promotes cell proliferation and transformation via AP-1. Proc Natl Acad Sci U S A 106:1445–1450. doi: 10.1073/pnas.0808749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Caunt M, Mak J, Liang WC, Stawicki S, Pan Q, Tong RK, Kowalski J, Ho C, Reslan HB, Ross J, Berry L, Kasman I, Zlot C, Cheng Z, Le Couter J, Filvaroff EH, Plowman G, Peale F, French D, Carano R, Koch AW, Wu Y, Watts RJ, Tessier-Lavigne M, Bagri A. 2008. Blocking neuropilin-2 function inhibits tumor cell metastasis. Cancer Cell 13:331–342. doi: 10.1016/j.ccr.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 63.Zhang H, Hu J, Recce M, Tian B. 2005. PolyA_DB: a database for mammalian mRNA polyadenylation. Nucleic Acids Res 33:D116–D120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheng Y, Miura RM, Tian B. 2006. Prediction of mRNA polyadenylation sites by support vector machine. Bioinformatics 22:2320–2325. doi: 10.1093/bioinformatics/btl394. [DOI] [PubMed] [Google Scholar]
- 65.Chaudhary A, Hilton MB, Seaman S, Haines DC, Stevenson S, Lemotte PK, Tschantz WR, Zhang XM, Saha S, Fleming T, St Croix B. 2012. TEM8/ANTXR1 blockade inhibits pathological angiogenesis and potentiates tumoricidal responses against multiple cancer types. Cancer Cell 21:212–226. doi: 10.1016/j.ccr.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Galland F, Lacroix L, Saulnier P, Dessen P, Meduri G, Bernier M, Gaillard S, Guibourdenche J, Fournier T, Evain-Brion D, Bidart JM, Chanson P. 2010. Differential gene expression profiles of invasive and non-invasive non-functioning pituitary adenomas based on microarray analysis. Endocrinol Relat Cancer 17:361–371. doi: 10.1677/ERC-10-0018. [DOI] [PubMed] [Google Scholar]
- 67.Courtois G. 2008. Tumor suppressor CYLD: negative regulation of NF-κB signaling and more. Cell Mol Life Sci 65:1123–1132. doi: 10.1007/s00018-007-7465-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. 2006. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-κB signaling. Cell 125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 69.Demuth T, Berens ME. 2004. Molecular mechanisms of glioma cell migration and invasion. J Neurooncol 70:217–228. doi: 10.1007/s11060-004-2751-6. [DOI] [PubMed] [Google Scholar]
- 70.Gladson CL, Cheresh DA. 1991. Glioblastoma expression of vitronectin and the αvβ3 integrin: adhesion mechanism for transformed glial cells. J Clin Invest 88:1924–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gingras MC, Roussel E, Bruner JM, Branch CD, Moser RP. 1995. Comparison of cell adhesion molecule expression between glioblastoma multiforme and autologous normal brain tissue. J Neuroimmunol 57:143–153. doi: 10.1016/0165-5728(94)00178-Q. [DOI] [PubMed] [Google Scholar]
- 72.Mizoguchi M, Nutt CL, Mohapatra G, Louis DN. 2004. Genetic alterations of phosphoinositide 3-kinase subunit genes in human glioblastomas. Brain Pathol 14:372–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chintala SK, Kyritsis AP, Mohan PM, Mohanam S, Sawaya R, Gokslan Z, Yung WK, Steck P, Uhm JH, Aggarwal BB, Rao JS. 1999. Altered actin cytoskeleton and inhibition of matrix metalloproteinase expression by vanadate and phenylarsine oxide, inhibitors of phosphotyrosine phosphatases: modulation of migration and invasion of human malignant glioma cells. Mol Carcinog 26:274–285. doi:. [DOI] [PubMed] [Google Scholar]
- 74.Quick Q, Skalli O. 2010. Alpha-actinin 1 and alpha-actinin 4: contrasting roles in the survival, motility, and RhoA signaling of astrocytoma cells. Exp Cell Res 316:1137–1147. doi: 10.1016/j.yexcr.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 75.Paulus W, Baur I, Schuppan D, Roggendorf W. 1993. Characterization of integrin receptors in normal and neoplastic human brain. Am J Pathol 143:154–163. [PMC free article] [PubMed] [Google Scholar]
- 76.Friedlander DR, Zagzag D, Shiff B, Cohen H, Allen JC, Kelly PJ, Grumet M. 1996. Migration of brain tumor cells on extracellular matrix proteins in vitro correlates with tumor type and grade and involves αV and β1 integrins. Cancer Res 56:1939–1947. [PubMed] [Google Scholar]
- 77.Vitolo D, Paradiso P, Uccini S, Ruco LP, Baroni CD. 1996. Expression of adhesion molecules and extracellular matrix proteins in glioblastomas: relation to angiogenesis and spread. Histopathology 28:521–528. doi: 10.1046/j.1365-2559.1996.d01-471.x. [DOI] [PubMed] [Google Scholar]
- 78.Skuli N, Monferran S, Delmas C, Favre G, Bonnet J, Toulas C, Cohen-Jonathan Moyal E. 2009. αvβ3/αvβ5 Integrins-FAK-RhoB: a novel pathway for hypoxia regulation in glioblastoma. Cancer Res 69:3308–3316. doi: 10.1158/0008-5472.CAN-08-2158. [DOI] [PubMed] [Google Scholar]
- 79.Burke PA, DeNardo SJ, Miers LA, Lamborn KR, Matzku S, DeNardo GL. 2002. Cilengitide targeting of alpha(v) beta(3) integrin receptor synergizes with radioimmunotherapy to increase efficacy and apoptosis in breast cancer xenografts. Cancer Res 62:4263–4272. [PubMed] [Google Scholar]
- 80.Mikkelsen T, Brodie C, Finniss S, Berens ME, Rennert JL, Nelson K, Lemke N, Brown SL, Hahn D, Neuteboom B, Goodman SL. 2009. Radiation sensitization of glioblastoma by cilengitide has unanticipated schedule dependency. Int J Cancer 124:2719–2727. doi: 10.1002/ijc.24240. [DOI] [PubMed] [Google Scholar]
- 81.Loster K, Vossmeyer D, Hofmann W, Reutter W, Danker K. 2001. α1 Integrin cytoplasmic domain is involved in focal adhesion formation via association with intracellular proteins. Biochem J 356:233–240. doi: 10.1042/0264-6021:3560233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tucker GC. 2006. Integrins: molecular targets in cancer therapy. Curr Oncol Rep 8:96–103. doi: 10.1007/s11912-006-0043-3. [DOI] [PubMed] [Google Scholar]
- 83.Vogetseder A, Thies S, Ingold B, Roth P, Weller M, Schraml P, Goodman SL, Moch H. 2013. αv-Integrin isoform expression in primary human tumors and brain metastases. Int J Cancer 133:2362–2371. doi: 10.1002/ijc.28267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.