Abstract
Background
Genome editing techniques, including ZFN, TALEN and CRISPR, have created a need to rapidly screen many F1 individuals to identify carriers of indels and determine the sequences of the mutations. Current techniques require multiple clones of the targeted region to be sequenced for each individual, which is inefficient when many individuals must be analyzed. Direct Sanger sequencing of a PCR amplified region surrounding the target site is efficient, but Sanger sequencing genomes heterozygous for an indel results in a string of “double peaks” due to the mismatched region.
Results
In order to facilitate indel identification, we developed an online tool called Poly Peak Parser (available at http://yost.genetics.utah.edu/software.php) that is able to separate chromatogram data containing ambiguous base calls into wild-type and mutant allele sequences. This tool allows the nature of the indel to be determined from a single sequencing run per individual performed directly on a PCR product spanning the targeted site, without cloning.
Conclusions
The method and algorithm described here facilitate rapid identification and sequence characterization of heterozygous mutant carriers generated by genome editing. Although designed for screening F1 individuals, this tool can also be used to identify heterozygous indels in many contexts.
Keywords: Indel Identification, Sanger Sequencing, Genome Editing
Introduction
Over the last few years, several “genome editing” technologies utilizing customizable DNA binding domains to mutate specific locations in the genome have been developed. These technologies, including Zinc-finger Nucleases (ZFN)(Bibikova et al., 2002; Gupta et al., 2012), Transcription activator-like effector nucleases (TALEN)(Joung and Sander, 2013; Miller et al., 2011; Zu et al., 2013) and clustered regularly interspaced short palindromic repeats (CRISPR)(Hwang et al., 2013), work by creating double-strand breaks in the DNA at a specific site in the genome, stimulating Non-Homologous End Joining (NHEJ) mechanisms that often edit the genome by removing and/or adding bases to the DNA during the repair process (reviewed in Gaj et al., 2013). The customizable nature of these enzymes has led to their adoption in a number of model organisms including, but not limited to, human cell lines, zebrafish, fly and mouse.
Although these genome-editing techniques have proven widely applicable and successful, the resulting mutant allele sequence is not predictable and a single treatment often results in several distinct mutations. These mutations can be passed through the germline of a single mutagenized individual to the F1 generation. Therefore, screening to identify germline carriers for a desired set of mutations must rely on Sanger sequencing of the targeted region in a large number of F1 offspring or clonal cell lines.
Current sequencing procedures often incorporate a prescreening step utilizing High-Resolution Melt Analysis (HRMA), and/or Sanger sequencing of PCR products generated in the region surrounding the targeted site to eliminate wild-type individuals from subsequent analysis (Parant et al., 2009; Dahlem et al., 2012). This prescreening approach identifies potential F1 heterozygous carriers, but does not reveal the sequence of the mutations. Researchers must then clone PCR fragments containing the target site, transform bacteria and sequence the inserts of several resulting clones. Statistically, at least 5 clones per individual F1 carrier must be sequenced to reduce the probability of missing a mutation to less than 5% and at least 7 clones must be sequenced per individual to reduce the false negative rate to less than 1%. Therefore, the number of clones necessary to analyze all F1 offspring quickly becomes very large, increasing the time and cost required.
Direct Sanger sequencing of PCR products encompassing the mutation target site is faster, but produces chromatogram traces containing two strong peaks for each position starting at the upstream end of the indel, making their interpretation difficult (Figure 1A). Several tools have been previously published that seek to segregate these double peaks into two separate sequences. However, they are either unavailable online (Dmitriev and Rakitov, 2008; Tenney et al., 2007), designed for clean insertions or deletions without alternative base insertions (Bhangale et al., 2006) or require numerous traces (Chen et al., 2007). Therefore, we developed an online tool, called Poly Peak Parser, to separate wild-type and mutant sequence calls from Sanger sequencing trace files, facilitating the identification and characterization of heterozygous mutants by direct Sanger sequencing of PCR products from F1 individuals. Poly Peak Parser can be run online (http://yost.genetics.utah.edu/software.php) or locally via the “sangerseqR” R package (http://www.bioconductor.org/packages/release/bioc/html/sangerseqR.html). Using a Poly Peak Parser-based workflow, heterozygous indels can be identified in less than 48 hours using an overnight DNA extraction protocol or 24 hours if DNA can be extracted and PCR amplified in the same day (Table 1). Conversely, the traditional cloning method takes up to 5 days using an overnight DNA extraction protocol. Reducing the amount of time the procedure takes not only helps the researcher be more efficient, but reduces the stress on animals, such as zebrafish, that must be kept in isolation during the genotyping process. This report focuses on using this technique for F1 genotyping, but it is applicable to any situation where heterozygous indels in diploid individuals or clonal colonies have been sequenced by Sanger sequencing.
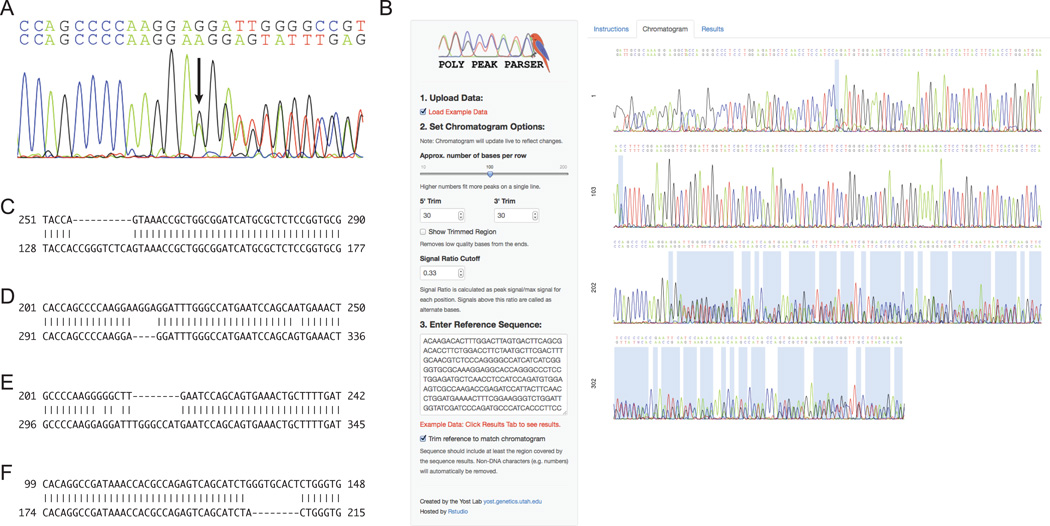
Figure 1. Overview of Poly Peak Parser.
(A) Example of a Sanger Sequencing chromatogram when a heterozygous indel (arrow) is present. (B) Poly Peak Parser screenshot. User input includes a Sanger sequencing file (abif or scf) containing a seed region of homozygous peaks followed by double peaks and a reference (wild-type) sequence for the region. The user can also optionally alter the peak ratio cutoff for calling heterozygous vs. homozygous positions (default = 0.33) and the parameters for trimming the Sanger sequencing results from the 5’ end (default = 30) and 3’ end (default = 30). Shaded regions in the chromatogram show double peak calls. (C-F) Partial view of alignments from Poly Peak Parser output showing the following apparent mutation types: a 10 bp deletion (C), a 4 bp insertion (D), a 14 bp deletion coupled with a 6 bp insertion (E), and a 1 bp deletion coupled with an 9 bp insertion (F). The mutant allele (query) is the top line and the user-provided WT sequence (subject) is the bottom line in each case.
Table 1. Comparison of a typical cloning-based and the Poly Peak Parser workflows.
The Poly Peak Parser-based workflow can be completed in less than 48 hours and requires only one sequencing reaction per individual. Typical cloning based workflows require 5 days and 5–7 sequencing reactions per individual.
| Cloning Workflow | Poly Peak Parser Workflow | |
|---|---|---|
| Day 1 | Extract DNA | Extract DNA |
| Day 2 | PCR amplification/cloning | PCR amplification/Send for sequencing |
| Day 3 | Pick colonies (5–7X per individual) | Analyze results |
| Day 4 | Mini-prep DNA/Send for sequencing (5–7X per individual) | |
| Day 5 | Analyze results |
Method Overview
1. Prescreening (optional)
The germline transmission rate of indels created by genomic editing is highly variable. In our experience using genome editing in zebrafish, germline transmission has ranged from a few percent to almost 100%. If the transmission rate to the F1 is low, or unknown, it may be beneficial to prescreen the F1 offspring to find mutant allele carriers by high resolution melt analysis (HRMA, Parant et al., 2009) or, if the equipment and reagents for HRMA are not available, restriction fragment length polymorphism (RFLP) can be used. HRMA is preferred because it does not restrict target site selection and is less prone to false-positives arising from poor DNA digestion.
2. PCR primer design, amplification and Sanger Sequencing
In order to identify and characterize mutant carriers, the targeted region must be amplified. Several considerations should be taken into account when designing the PCR reaction. First, the PCR product must contain at least 40–50 bases upstream of the target site (relative to the sequencing primer). Second, the primers should be far enough away from the target site to ensure that one or both primer sites are not removed as a result of a deletion. Failing to do so will cause only the wild-type allele to be amplified. Third, the length of the PCR product should be appropriate for Sanger sequencing. Sanger Sequencing runs are typically 600–1000 bp long, so PCR products should not be much longer than this. On the opposite end, they should not be so short that sequencing runs yield few bases of high quality sequencing. Finally, care should be taken to prevent multiple priming or other problems that can negatively impact sequencing results. In order to meet these requirements, we recommend that primers be designed to amplify a 400–800 bp region centered on the target site or suspected mutation. These primers should be carefully checked for potential secondary target sites in the genome using Primer Blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) or a similar tool.
Sanger Sequencing is performed directly using the PCR product and standard protocols. Either one of the primers used in the PCR amplification step can simply be used as a sequencing primer, or an internal sequencing primer can be designed. The resulting chromatogram file is then used for data analysis (Figure 1A). The read should begin with a region of single peaks, followed by a region of double peaks at least 20 basepairs from the beginning of the sequencing results. Substantial noise, background or triple peaks in the chromatogram will interfere with subsequent base calling and prevent appropriate alignment of the results to the reference sequence.
3. Data analysis
If the individual being genotyped is heterozygous for an insertion or deletion, the sequencing peaks for the mutant allele will shift. The result is a series of double peaks beginning at the mutation site (See Figure 1A). Therefore, the Sanger sequencing results from mutant carriers must be parsed to separate the sequences into wild-type and mutant alleles. We have created a web-based tool, called Poly Peak Parser, to automate this parsing. Poly Peak Parser requires only two inputs: (1) The Sanger sequencing results file (.ab1 and .scf formats are supported) from the previous step containing the double peaks to be separated by the program, and (2) a text string of the wild-type sequence. Example data for both requirements is included on the website. The ends of the reference sequence provided do not need to perfectly match the beginning and/or end of the sequencing results, but should ideally encompass the entire sequenced region. It should also be noted that Poly Peak Parser tolerates occasional snps well, but is sensitive to indels in the double peak region that result in both alleles not matching the reference sequence. Therefore, care should be taken to ensure that the reference sequence matches the genotyped animals as closely as possible. This can generally be achieved by selecting a uniform population for genomic targeting. Also, in the absence of a good reference annotation, sequence (potentially generated from the same sequencing primer) from a wild-type sibling or parent can be used to generate the reference sequence.
Both the Sanger sequencing file and the wild-type sequence are entered into a web form and submitted to a server side script (Figure 1B). The script trims the sequence (default 30 bases; user adjustable) to remove the poor base calls typically found at the beginning and end of the sequencing runs. It then extracts base calls for positions where the signal for only one base is greater than a user-defined ratio (default 0.33), i.e. the single peak regions. This sequence is aligned to the wild-type reference to determine the position of the sequencing results relative to the reference sequence. Next, primary and secondary base calls are made for double peak positions and compared to the reference sequence to build the mutant allele sequence. Finally, the resulting mutant sequence is aligned to the reference and both the mutant sequence and the alignment are returned to the user, allowing easy visualization of the indel.
Results and Discussion
Genome editing techniques rely on the removal or non-templated insertion of DNA bases as part of the Non-homologous End Joining process. Therefore, they can generate four classes of mutations: a clean insertion, a clean deletion, a deletion coupled with a smaller insertion or an insertion coupled with a smaller deletion. In order to test the ability of Poly Peak Parser to identify mutants containing simple or complex indels, we ran the software on PCR-based DNA sequencing of four mutations generated using ZFNs or TALENs in our lab and previously identified by sequencing multiple clones (Figure 1C-F). These mutants included the following apparent indels: a 10-bp deletion (Figure 1C), a 4-bp insertion (Figure 1D), a 14-bp deletion combined with a 6-bp insertion (Figure 1E), and a 9-bp insertion coupled with a 1-bp deletion (Figure 1F), collectively representing all four mutant classes created by genome editing. PCR products containing the target site were generated and parsed using Poly Peak Parser. Complete Poly Peak Parser results for these genes can be found in the supplemental materials. In each case, Poly Peak Parser was able to correctly identify the sequence of the mutant allele; despite the challenges of poor base calls at the ends of the sequencing results and the presence of snps within the PCR product due to the high polymorphism rate in zebrafish.
There are specific situations where Poly Peak Parser may not perform well. For example, regions with multiple repeats may not align well with the reference sequence, making the base calls less certain. However, because Poly Peak Parser seeks for the longest match between the reference sequence and sequencing results, this is only a concern if the repeat region encompasses the entire single-peak region used to align the sequencing results to the user provided reference sequence. Carefully selecting targets to avoid these regions or ensuring that the sequenced region contains non-repetitive bases upstream of the suspected indel can minimize this concern. Poly Peak Parser also requires that the reference sequence provided by the user matches one of the two alleles well. As mentioned earlier, it will tolerate snps, but the presence of indels in both alleles carried by the individual may cause it to fail. In these rare cases, it may be necessary to redesign the PCR to encompass larger segments of reference sequence or to clone and sequence the PCR products.
In conclusion, the Poly Peak Parser-based workflow for characterizing heterozygous indels fulfills an increasingly important need to characterize genome editing-derived indels and can be used in a number of other cases where Sanger sequencing has been performed on heterozygous individuals. The tool is available on the web (http://yost.genetics.utah.edu/software.php) and requires only a minimal amount of input data. Although demonstrated here for zebrafish mutants, the approach is species agnostic and should be applicable to any diploid species, including humans. Using this tool will save substantial amounts of time and resources by eliminating the need to create and sequence multiple clones for each heterozygous individual. In addition, the underlying algorithms have been submitted to the Bioconductor Repository for R packages (http://www.bioconductor.org/packages/release/bioc/html/sangerseqR.html) as the “sangerseqR” package, creating a common sanger sequencing object class that can be generated from abif and scf files, as well as allowing chromatogram plotting and poly peak parsing in R for the first time. The package also contains a full copy of Poly Peak Parser that can be run locally in a web browser.
Experimental Procedures
HRMA Screening
HRMA screening was performed as described previously (Parant et al., 2009). Briefly, tail clips of individual F1 zebrafish from each of the lines used in our confirmation analyses were taken and digested in ELB buffer (10 mM TRIS pH 8.3, 50 mM KCl, 0.3% Tween 20, 0.3% NP40, 1 mg/ml Proteinase K) followed by PCR of the targeted region using the following primers and conditions: Tbx2a line B11 (Figure 1C) CTTGGACATCACCACCAGGC and GTGGAATTGATCCCAAAGCTCC, Tm=60C; 3ost5 line F96 (Figure 1D) and 3ost5 line M21 (Figure 1E) CAGCTGACGGTGGAAAAGACT and AATCTGATACGAGCCTCTCTGC, Tm=57C; Left2 line F19 (Figure 1F) GCACCACAGGCCGATAAAC and CATACCGTGAGTCTAGGAGTG, Tm=51C. PCR reactions used standard buffers with the addition of 1X LCGreen Plus intercalating dye and were run in a Lightcycler 480 or standard thermocycler with the following program: 95C for 1 minute; 50 cycles of 95C for 12 seconds, 57C for 12 seconds and 72C for 12 seconds; 95C for 8 seconds; 45C for 10 seconds and 95C for 5 minutes. Melt curves were then generated by measuring the decrease in LCGreen fluorescence as temperature increased using either a Roche LightCycler 480 system or an Idaho Technologies (now Biofire) LightScanner with the built in software and default settings.
PCR Amplification
PCR reactions for the genomic editing target regions used the following primers and annealing temperatures: Tbx2a line B11 (Figure 1C) TGGCAAGAGAGCGACAGTCAG and AGAGGCTTCGATGCTATGTCAG, Tm=60C; 3ost5 line F96 (Figure 1D) and 3ost5 line M21 (Figure 1E) TGGACCTTCTAATGCTTCGAC and GGCCTTGTATTTGGGGTTG, Tm=50C; Left2 line F19 (Figure 1F) TTACCGGAGAAATCAAGTACTCGGACAC and AGAGGCTTCGATGCTATGTCAG, Tm=59C. PCR reactions were amplified in a standard thermocycler using the following program: 94C for 2 minutes followed by 45 cycles of 94C for 30 seconds, annealing temperature for 30 seconds and 68C for 30 seconds. PCR reactions were then gel purified by running on a 1% polyacrylamide gel in 1X TAE buffer or directly used for subsequent steps.
Subcloning
PCR products were cloned using the TA Cloning Dual Promoter Kit (Invitrogen #K2060-01) and transformed into DH5alpha chemically competent E. coli cells. Colonies (6-8 for each gene) were selected the next day and grown in LB containing Kanamycin overnight in a shaking incubator set at 37C. Plasmids were purified the next day using the Qiaprep Miniprep kit (Qiagen # 27104). Eluates from the miniprep were quantified using a Nanodrop 2000c spectrophotometer (Thermo Scientific).
Sanger Sequencing and Data Analysis
Sanger sequencing of PCR products or cloned inserts was performed at the University of Utah DNA Sequencing Core using the 5’ PCR primer for each fragment. For sequencing the PCR products, 1 ul of the PCR reaction was diluted 1:10 and combined with the appropriate primer before sending. Plasmids were diluted to 100 ng/ul and 10 ul of the resulting solution mixed with the sequencing primer. Sequencing was performed using a proprietary chemistry based on Big Dye 3.1 on an Applied Biosystems 3730xl DNA Analyzer. Base calling was performed using Applied Biosystems Sequencing Analysis Software version 5.3.1 with default settings.
Supplementary Material
Acknowledgements
Funding: NHLBI Bench-to-Bassinet Consortium (benchtobassinet.com) grant to HJY (U01HL0981), a core facilities support grant to New England Research Institute (U01HL098188) and an NRSA fellowship (1F32HL115881) to JTH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
References
- Bhangale TR, Stephens M, Nickerson DA. Automating resequencing-based detection of insertion-deletion polymorphisms. Nat. Genet. 2006;38:1457–1462. doi: 10.1038/ng1925. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, McLellan MD, Ding L, Wendl MC, Kasai Y, Wilson RK, Mardis ER. PolyScan: an automatic indel and SNP detection approach to the analysis of human resequencing data. Genome Res. 2007;17:659–666. doi: 10.1101/gr.6151507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev Da, Rakitov Ra. Decoding of superimposed traces produced by direct sequencing of heterozygous indels. PLoS Comput. Biol. 2008;4:e1000113. doi: 10.1371/journal.pcbi.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Christensen RG, Rayla AL, Lakshmanan A, Stormo GD, Wolfe SA. An optimized two-finger archive for ZFN-mediated gene targeting. Nat. Methods. 2012;9:588–590. doi: 10.1038/nmeth.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh J-RJ, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Parant JM, George SA, Pryor R, Wittwer CT, Yost HJ. A rapid and efficient method of genotyping zebrafish mutants. Dev. Dyn. 2009;238:3168–3174. doi: 10.1002/dvdy.22143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney AE, Wu JQ, Langton L, Klueh P, Quatrano R, Brent MR. A tale of two templates: automatically resolving double traces has many applications, including efficient PCR-based elucidation of alternative splices. Genome Res. 2007;17:212–218. doi: 10.1101/gr.5661407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Y, Tong X, Wang Z, Liu D, Pan R, Li Z, Hu Y, Luo Z, Huang P, Wu Q, Zhu Z, Zhang B, Lin S. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods. 2013;10:329–331. doi: 10.1038/nmeth.2374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.