Abstract
Although early studies suggested that little compartmentalization exists within the nucleus, more recent studies on metazoan systems have identified a still increasing number of specific subnuclear compartments. Some of these compartments are dynamic structures; indeed, protein and RNA-protein components can cycle between different domains. This is particularly evident for RNA processing components. In plants, lack of tools has hampered studies on nuclear compartmentalization and dynamics of RNA processing components. Here, we show that transient expression of fluorescent protein fusions of U1 and U2 small nuclear ribonucleoprotein particle (snRNP)-specific proteins U1-70K, U2B″, and U2A ′, nucleolar proteins Nop10 and PRH75, and serine-arginine-rich proteins in plant protoplasts results in their correct localization. Furthermore, snRNP-specific proteins also were correctly assembled into mature snRNPs. This system allowed a systematic analysis of the cellular localization of Arabidopsis serine-arginine-rich proteins, which, like their animal counterparts, localize to speckles but not to nucleoli and Cajal bodies. Finally, markers for three different nuclear compartments, namely, nucleoli, Cajal bodies, and speckles, have been established and were shown to be applicable for colocalization studies in living plant protoplasts. Thus, transient expression of proteins tagged with four different fluorescent proteins is a suitable system for studying the nuclear organization of spliceosomal proteins in living plant cells and should therefore allow studies of their dynamics as well.
INTRODUCTION
Removal of introns from eukaryotic pre-mRNAs takes place in the spliceosome, the most complex molecular machine characterized thus far. Spliceosome assembles from five small nuclear ribonucleoprotein particles (snRNPs) and numerous additional proteins. Each snRNP consists of RNA and seven so-called Sm proteins (LSm in U6 snRNP), SmB, SmD1, SmD2, SmD3, SmE, SmF, and SmG, which are common to U1, U2, U4, and U5 snRNPs. In addition, each snRNP contains a set of snRNP-specific proteins (Krämer, 1996; Burge et al., 1999; Kambach et al., 1999; Will and Lührmann, 2001). After synthesis, snRNAs (except U6 snRNA) are exported to the cytoplasm where they assemble with seven Sm core proteins and acquire a trimethylguanosine (m3G) cap structure at their 5′ end. Such preassembled snRNPs are imported back to the nucleus where they finally associate with snRNP-specific proteins (Kambach et al., 1999; Will and Lührmann, 2001). The majority of proteins involved in splicing, including snRNP-specific proteins, enter the nucleus separately by an active transport mechanism. On nuclear entry spliceosomal components do not distribute randomly, rather they localize into distinct subnuclear compartments. The best studied and the most prominent nuclear compartment is the nucleolus; however, splicing factors usually do not localize into it. Exceptions are snRNPs, which transiently pass through the nucleolus on their nuclear maturation pathway (Sleeman and Lamond 1999a). Splicing factors accumulate in the nucleus in an irregular network called splicing factor compartment or speckles (Misteli and Spector, 1998; Sleeman and Lamond, 1999b; Lamond and Spector, 2003). At the ultrastructural level speckles can be divided into perichromatin fibrils, which contain nascent transcripts and are usually at the periphery of splicing factor compartments, and interchromatin granule clusters (IGCs). Active genes are not found in the IGCs, which led to the proposal that this nuclear compartment serves as storage/assembly factory for spliceosomal components. Consistent with this idea are observations that 1) splicing factors move from speckles to their periphery (perichromatin fibrils) upon activation of transcription, 2) transcription active sites do not colocalize with IGCs but are rather found on their periphery, and 3) IGCs increase in size and in splicing factor content upon inhibition of splicing or transcription (Dundr and Misteli, 2001; Lamond and Spector, 2003). Thus, speckles are dynamic structures, and both their protein and RNA components cycle continuously between speckles and other nuclear compartments (Misteli and Spector, 1998; Carmo-Fonseca, 2002a; Lamond and Spector, 2003). In mammalian cells speckles can be easily visualized by fluorescence microscopy by using antibodies against various splicing factors. The most widely used markers for speckles are members of serine-arginine-rich (SR) phosphoproteins, which are involved in splice site selection in both constitutive and alternative splicing. Localization of SR proteins in nuclear speckles depends on their phosphorylation status, and phosphorylation/dephosphorylation is considered as an important regulatory mechanism for recruiting splicing factors from speckles to transcriptionally active sites (Misteli and Spector, 1998; Carmo-Fonseca, 2002a; Lamond and Spector, 2003).
In addition to speckles, spliceosomal snRNPs also accumulate in a sphere nuclear compartment called Cajal body (Carmo-Fonseca et al., 1991a,b; Wu et al., 1991; Matera and Ward, 1993; Visa et al., 1993). Cajal bodies are found in the nuclei of plant and animal cells, and the number and size of Cajal bodies per cell are subject to change, depending on cell type and cell cycle (Gall, 2000; Ogg and Lamond, 2002). Cajal bodies are very often located at the nucleolar periphery and even within nucleoli. This feature is particularly prominent in plant cells where virtually all Cajal bodies localize to nucleolar periphery (Beven et al., 1995; Boudonck et al., 1998). The most intriguing feature of Cajal bodies is their movement; however, their function is still unclear (Boudonck et al., 1999; Platani et al., 2000; Snaar et al., 2000). Recent evidence suggests that Cajal bodies might be involved in maturation and transport of snRNPs and small nucleolar ribonucleoprotein particles within the nucleus (Sleeman and Lamond, 1999a; Carmo-Fonseca, 2002b; Ogg and Lamond, 2002). Moreover, some other functions, in particular their involvement in the assembly of the transcriptional machinery have been proposed (Gall, 2000, 2001).
Little is known about the organization of spliceosomal machinery in plant cell nuclei (Simpson and Filipowicz, 1996). One reason is that this issue was not yet systematically addressed, presumably because of the lack of good quality antibodies against plant splicing factors. By using monoclonal antibody against the human U2 snRNP-specific protein U2B″, and later, green fluorescent protein (GFP) fusion of potato U2B″, it has been shown that U2 snRNP accumulates in Cajal bodies and in a diffuse nucleoplasmic pool in bean, tobacco, Arabidopsis, and onion cells (Beven et al., 1995; Boudonck et al., 1998; Cui and Moreno Diaz de la Espina, 2003). In addition, in situ hybridization with U2 snRNA-specific probes revealed the same staining pattern, indicating that U2B″ is a good marker for Cajal bodies in plant cells (Beven et al., 1995; Boudonck et al., 1998). Similarly, by using U6 snRNA-specific probes it has been shown that U6 snRNP also accumulates in Cajal bodies as well as in a diffuse nucleoplasmic pool (Beven et al., 1995). In contrast, U1 snRNP was rarely found in Cajal bodies (Beven et al., 1995). More recently, cellular localization of some SR proteins have been analyzed in transgenic Arabidopsis plants expressing fluorescent protein (FP)-tagged proteins (Ali et al., 2003; Docquier et al., 2004; Fang et al., 2004). In addition, RSZ33, a plant-specific SR protein, also was shown to be a nuclear protein localized in a speckled pattern when expressed in tobacco protoplasts (Lopato et al., 2002).
In this work, we set out to develop plasmids that can be used in transient transformation assays in plant protoplasts. Several proteins with known or unknown subnuclear localization were fused to GFP, red fluorescent protein (RFP), monomeric RFP (mRFP) (Campbell et al., 2002), yellow fluorescent protein (YFP), and cyan fluorescent protein (CFP) and analyzed for their localization in transiently transformed tobacco and Arabidopsis protoplasts by using confocal laser scanning microscopy. We show that transient expression of spliceosomal proteins in plant protoplasts results in their correct localization, and in case of snRNP-specific proteins, in correct assembly into mature snRNPs, making this system suitable for studying nuclear organization of the spliceosomal machinery in living plant cells.
MATERIALS AND METHODS
Construction of Plasmids Expressing RFP, mRFP, YFP, and CFP
pDEDH-GFP has been described by Lambermon et al. (2000). pDEDH-HA has been described by Genschik et al. (1997). pDEDH-RFP, pDEDH-mRFP, pDEDH-YFP, and pDEDH-CFP were constructed by cloning of RFP (dsRED; BD Biosciences Clontech, Palo Alto, CA), mRFP (Campbell et al., 2002), YFP (BD Biosciences Clontech), and CFP (BD Biosciences Clontech) open reading frames into pDEDH/Nco (Goodall and Filipowicz, 1989; Lambermon et al., 2000). RFP and mRFP were polymerase chain reaction (PCR) amplified with the following primers: RFP5′ CTGCAGGGATCCATGGTGCGCTCCTCCAAGAACGTC, RFP3′ CTGGACATGCATCTACAGGAACAGGTGGTGGCG, mRFP5′ GTCAGCGGATCCATGGCCTCCTCCGAGGACGTCATC,andmRFP3′GACTGCATGCATTTCGGCGCCGGTGGAGTGGC, which introduce BamHI and NsiI restriction sites (in bold) in front of start and after stop codons, respectively. YFP and CFP were PCR amplified with the following primers: YFP5′ CGTATGGATCCATGGTGAGCAAGGGCGAGGAGC, YFP3′ GTCATCTGCAGTTACTTGTACAGCTCGTCCATG, CFP5′ CGTACGGATCCGTGAGCAAGGGCGAGGAGC, and CFP3′ ATGCGACTGCAGTTAGCTCTTGTACAGCTCGTCCA, which introduce BamHI and PstI restriction sites in front of start and after stop codons, respectively. Cut PCR products were ligated into BamHI and PstI opened pDEDH/Nco.
Construction of Plasmids for the Expression of Fluorescent Protein Fusion Proteins
To clone full-length SCL28, SCL30, SCL30a, SCL33, atSC35 (Lopato et al., 2002), SRp30, SRp34 (Lopato et al., 1999b), RSZp21, RSZp22 (Lopato et al., 1999a), RSZ33 (Lopato et al., 2002), RSp31, RSp40 (Lopato et al., 1996), U1-70K (Golovkin and Reddy, 1996), U2B″ (at2g30260), U2A′ (at1g09760), Nop10 (at2g20490), SmB (at4g20440), and PRH75 (Lorkovic et al., 1997) into plant expression vectors pDEDH-GFP and pDEDH-RFP, the coding regions were PCR amplified by using the following primers: SCL28 5′ primer, TGCGCCCGGGAATAAACCATGGCTAGAGCGAGAAGCCGGA, 3′ primer, TATTGCGGATCCACGACTTAAGGATCGAGAAC; SCL30 5′ primer, TGCGGGTCGACAATAAACCATGAGGAGATACAGTCCGCCTTA,3′primer,TATTGCAGATCTTGGAGATACCTCCACAGACC; SCL30a 5′ primer, TGCGGGTCGACAATAAACCATGAGAGGAAGGAGCTACAC, 3′ primer, TATTGCAGATCTCTGCTTGGAGAACGGTCTC; SCL33 5′ primer, TGCGGGTCGACAATAAACCATGAGGGGAAGGAGCTACACTCC,3′primer,TATTGCGGATCCCTGGCTTGGTGAACGGTCTT; atSC35 5′ primer, TGCGGGTCGACAATAAACCATGTCGCACTTCGGAAGGTCAGG,3′primer,TATTGCGGATCCTTCCGCAGCATAAGGAGATTG; SRp30 5′ primer, TGCGGCCCGGGAATAAACCATGGGTAGCCGATGGAATCGTACG, 3′ primer, TATTGCGGATCCACCAGATATCACAGGTGAAAC; SRp34 5′ primer, TGCAGGTCGACAATAAACCATGAGCAGTCGTTCGAGTAGA, 3′ primer, TATTGCGGATCCCCTCGATGGACTCCTAGTGTG; RSp31 5′ primer, TGCGCCCGGGAATAAACCATGAGGCCAGTGTTCGTCGGCA; 3′ primer, TATTGCGGATCCAGGTCTTCCTCTTGGGACTG; RSp40 5′ primer, TGCGGGTCGACAATAAACCATGAAGCCAGTCTTCTGTGGGAA, 3′ primer, TATTGCGGATCCCTCGTCAGCTGGTGGCGAAC; RSZp21 5′ primer, TGCGGCCCGGGAATAAACCATGGCGAGGGTTTATGTCGGGA, 3′ primer, TATTGCGGATCCCACCCCATTGGCATATGGCG; RSZp22 5′ primer, TGCGGGTCGACAATAAACCATGTCACGTGTGTACGTCGGA, 3′ primer, TATTGCGGATCCGCTCCTGCTTCTGCGTCTT; RSZ33 5′ primer, TGCGGGTCGACAATAAACCATGCCTCGCTATGATGATCGCTA; 3′ primer, TATTGCGGATCCAGGAGACTCACTTCCTCTAG; U1-70K 5′ primer, TGCAGGTCGACAATAAACCATGGGAGACTCCGGCGATCCT, 3′ primer, TATGGC-AGATCTACGAACATACTCTCGCGATTC; U2B″ 5′ primer, GTCGAGTCGACAATAAACCATGTTAACGGCAGATATACCA, 3′ primer, TATTGCGGATCCTTTCTTGGCGAAAGAGATG; U2A' 5′ primer, ACTAGGTCGACAATAAACCATGGTGAAGCTCACGGCTGA, 3′ primer, AGCATGGATCCTTCCTCCATGGGAGCAGAG; SmB 5′ primer, ACTAGGTCGAC- AATAAACCATGTCGATGTCGAAGAG, 3′ primer, AGCATGGATCCCTGCTGCTGATTATGTG; Nop10 5′ primer, ACTAGGTCGACAATAAACCATGTATCTTCAGTGCT, 3′ primer, AGCATGGATCCGTACTGGAGAGGTGC; PRH75 5′ primer, TGCGGGTCGACAATAAACCATGCCTTCCCTAATGTTATCTGATAAG, 3′ primer, TTATTGCGGATCCATATCTCTGGCCTCTACCA. The 5′ primers for SCL28, SRp30, RSZp21, RSp31 introduced XmaI restriction sites; 5′ primers of SCL30, SCL30a, SCL33, atSC35, SRp34, RSZp22, RSZ33, RSp40, U2B″, U2A′, SmB, Nop10, and PRH75 introduced SalI restriction sites (in bold), followed by the plant consensus translation initiation sequence (in italics). The 3′ primers for SCL30 and SCL30a, and U1-70K introduced BglII restriction sites (in bold) in place of the stop codon, whereas all other 3′ primers introduced BamHI restriction sites. The PCR products were cut with the respective restriction enzymes and ligated into correspondingly opened pDEDH-GFP, pDEDH-RFP, pDEDH-mRFP, pDEDH-YFP, and pDEDH-CFP, resulting in C-terminally tagged GFP, RFP, mRFP, YFP, and CFP fusion constructs, respectively. The same PCR products were used for construction of hemagglutinin (HA)-tagged proteins in pDEDH-HA.
Preparation and Transient Transformation of Tobacco and Arabidopsis Protoplasts
Nicotiana tabacum SR1 mesophyll protoplasts were isolated and transformed with 20 μg of plasmid DNA per 1 million of protoplasts by the polyethylene glycol method as described by Koop et al. (1996). Arabidopsis cell suspension protoplasts were prepared and transformed as described in Meskiene et al. (2003). Protoplasts were analyzed 24 h after transformation by confocal laser scanning microscopy.
Preparation of Whole Cell Extracts from Transiently Transformed Protoplasts and Immunoprecipitation
Twenty-four hours after transformation protoplasts were collected by centrifugation (15 min, 70 × g), frozen in liquid nitrogen, and resuspended in protoplast extraction buffer (PEB400; 50 mM HEPES-KOH, pH 7.9, 400 mM KCl, 2.5 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 0.1% Triton X-100), supplemented with EDTA-free protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). One hundred microliters of PEB400 was used per 1 million protoplasts. The suspension was shortly vortexed and then sonicated three times for 6 s. An additional 100 μl of PEB400 was added to the extract, and the suspension was incubated on ice for 20 min with occasional mixing. After 15-min centrifugation in an Eppendorf centrifuge at maximum speed at 4°C, the supernatant was mixed with PEB without KCl to adjust KCl concentration to 250 mM (PEB250). Protein extracts prepared from transformed protoplasts were incubated with anti-GFP mAb (Roche Diagnostics) for 1 hour on ice and then mixed with protein A-Sepharose (Amersham Biosciences AB, Uppsala, Sweden) and incubated on a rotary shaker additional 3 h at 4°C. After three washings with 1 ml of PEB200 and one washing with 1 ml of PEB250 the beads were resuspended in 100 μl of TE buffer containing 0.5% SDS and extracted with phenol/chloroform. RNA was precipitated by addition of 40 μg of glycogen and 2.5 volumes of ethanol, labeled by [32P]pCp ligation and analyzed on 8% denaturing PAA gels. Proteins were precipitated from the organic phase by addition of 7 volumes of acetone, resuspended in 30 μl of loading buffer, and analyzed by SDS-PAGE and Western blotting. In cases where only precipitated proteins were analyzed (immunoprecipitations with anti-m3G and anti-PRH75 antibodies), immunoprecipitates were directly resuspended in 30 μl of SDS-PAGE loading buffer and analyzed by Western blotting. In all cases, control immunoprecipitation was performed with protein A-Sepharose beads alone. Protein extract prepared from 2 million transformed cells was used for one immunoprecipitation. Immunoprecipitations with anti-m3G (Lührmann et al., 1982) and anti-PRH75 antibodies were performed as described above, except that the antibodies were first bound to protein A-Sepharose and then mixed with protein extract.
Confocal Microscopy
Images were obtained with a TCS-SP confocal microscope (Leica Microsystems, Heidelberg, Germany). GFP and RFP were excited with an ArKr laser at 476 and 568 nm, respectively. GFP was detected at 510 to 550 nm, and RFP and mRFP were detected at 630 to 680 nm. YFP was excited by two photon equipment at 820 nm. Images were exported to Adobe Photoshop software and prepared for presentation.
SDS-PAGE and Western Blotting Analysis
Proteins were separated by SDS-PAGE on 12% gels and were electroblotted on a polyvinylidene difluoride membrane (Millipore, Billerica, MA). Blots were developed by using enhanced chemiluminescence system (Amersham Biosciences, UK, Little Chalfont, Buckinghamshire, United Kingdom). Antibodies were diluted as follows: mouse monoclonal anti-GFP 7.1 and 13.1 (Roche Diagnostics), 1:1000; rat monoclonal anti-HA 3F10 (Roche Diagnostics), 1:1000; rabbit polyclonal anti-PRH75, 1:500; rabbit polyclonal anti-RZ1, 1:500; goat anti-rat IgG horseradish peroxidase-conjugated (Sigma-Aldrich, St. Louis, MO), 1:10,000; goat anti-mouse IgG horseradish peroxidase-conjugated (Bio-Rad, Hercules, CA), 1:10,000; and goat anti-rabbit IgG horseradish peroxidase-conjugated (Sigma-Aldrich), 1:20,000.
RESULTS
The lack of specific antibodies against plant spliceosomal proteins made it impossible to analyze the subnuclear organization of the splicing machinery in plant cells by indirect immunofluorescence. However, correct localization of GFP-tagged proteins in plant cells, among them also some splicing factors (Boudonck et al., 1999; Lopato et al., 2002; Ali et al., 2003; Docquier et al., 2004; Fang et al., 2004), has been well documented. For example, a GFP fusion of potato U2 snRNP-specific protein U2B″ expressed in stably transformed tobacco BY-2 cells and in Arabidopsis plants was shown to localize into a pattern that includes diffuse nucleoplasmic staining and strong accumulation in Cajal bodies (Boudonck et al., 1999). In addition, analyses of transgenic Arabidopsis plants expressing SR proteins SR45, RSp31, SCL33/SR33, SRp34/SR1, and SRp30 revealed speckled nuclear localization patterns (Ali et al., 2003; Docquier et al., 2004; Fang et al., 2004). However, generation of stable transgenic plants or cell suspension lines is time-consuming and does not allow simultaneous analysis of many proteins. In particular, colocalization studies that require at least two different fluorescent markers are very laborious when using transgenic plants. Therefore, we constructed plasmids for expression of RFP, mRFP, YFP, and CFP proteins, which together with the previously described pDEDH-GFP (Lambermon et al., 2000) allow construction of C-terminal FP fusions of the protein of interest and analysis of fusion proteins in transient assays in plant protoplasts. All plasmids, along with pDEDH-GFP (Lambermon et al., 2000), were transformed into tobacco mesophyll and Arabidopsis cell suspension protoplasts and 24 h after transformation protoplasts were analyzed for the expression of fluorescent proteins by confocal microscopy. All four fluorescent proteins are successfully expressed in both cell types, showing localization in both cytoplasm and the nucleus (Supplemental Figure 1), as reported previously for GFP (Lambermon et al., 2000). In addition, expression of GFP, CFP, and YFP proteins also was analyzed by Western blotting, which indicated similar expression levels (our unpublished data), as expected from the fact that all proteins are expressed from the same promoter, with the same 5′ leader sequence and the same 3′ untranslated region.
Transient Expression of U2 snRNP-specific Proteins U2B″ and U2A ′ : Markers for Cajal Bodies
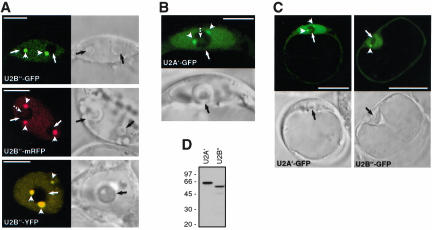
To find out whether transient expression of FP-tagged spliceosomal proteins can be used for studying nuclear organization of the spliceosomal machinery in living plant protoplasts, FP fusions of Arabidopsis U2 snRNP-specific proteins U2B″ and U2A′ were constructed. Both, U2B″-GFP (Figure 1A) and U2A′-GFP (Figure 1B) fusion proteins as well as U2B″-mRFP, U2B″-YFP and U2B″-CFP (Figure 1A; our unpublished data) were found in the nucleus within a diffuse nucleoplasmic pool and in Cajal bodies, which in most cases localized next to the nucleoli (seen as black areas). Virtually all cells transformed showed the same localization pattern. In a small percentage of transformed cells, irrespective of the FP tag used, both U2B″ and U2A′ also were found in the central nucleolar vacuole (Figure 1A, see U2B″-mRFP, and Figure 1B, broken arrows). The same nuclear patterns were observed with GFP-tagged SmB protein, a core component of U1, U2, U4, and U5 snRNPs (our unpublished data; but see third rows in Figure 4, A and E). The observed nuclear patterns of U2B″ and U2A′ resemble that described previously by means of 1) indirect immunofluorescence with U2B″-specific antibodies (Beven et al., 1995; Boudonck et al., 1998); 2) RNA fluorescence in situ hybridization with U2 snRNA probe (Beven et al., 1995); and 3) expression of U2B″-GFP fusion protein in transgenic Arabidopsis plants and tobacco BY-2 cells (Boudonck et al., 1999). However, we have to mention here that the majority of transformed cells also showed localization of U2B″-GFP, U2B″-mRFP, U2B″-YFP, U2B″-CFP, and U2A′-GFP fusion proteins in the cytoplasm (Figure 1C; shown are only cells expressing GFP fusion proteins), even 48 h after transformation (also see DISCUSSION). Western analysis of cell lysates prepared from transformed cells showed that U2B″-GFP and U2A′-GFP fusion proteins are expressed at similar levels without detectable degradation products (Figure 1D).
Figure 1.
Analysis of FP-tagged U2 snRNP-specific proteins U2B″ and U2A′ in transiently transformed Arabidopsis protoplasts. (A) Localization of U2B″-GFP, U2B″-mRFP, and U2B″-YFP fusion proteins. Arrows and arrowheads point to nucleoli and Cajal bodies, respectively. Broken arrow points to the central nucleolar vacuole. (B) Localization of U2A′-GFP fusion protein. Arrows and arrowheads point to nucleoli and Cajal bodies, respectively. (C) U2A′-GFP and U2B″-GFP fusion proteins localize to nucleus and cytoplasm. Arrowheads point to Cajal bodies and arrows to nucleoli. Bars, 7 μm (A and B) and 25 μm (C). In A-C, single confocal sections with corresponding DIC images are shown. (D) Immunodetection of U2A′ and U2B″ GFP fusion proteins in protein extract from transformed protoplasts. Total protein extracts were analyzed by SDS-PAGE and Western blotting with anti-GFP antibody. Molecular mass standards in kilodaltons are indicated on the left.
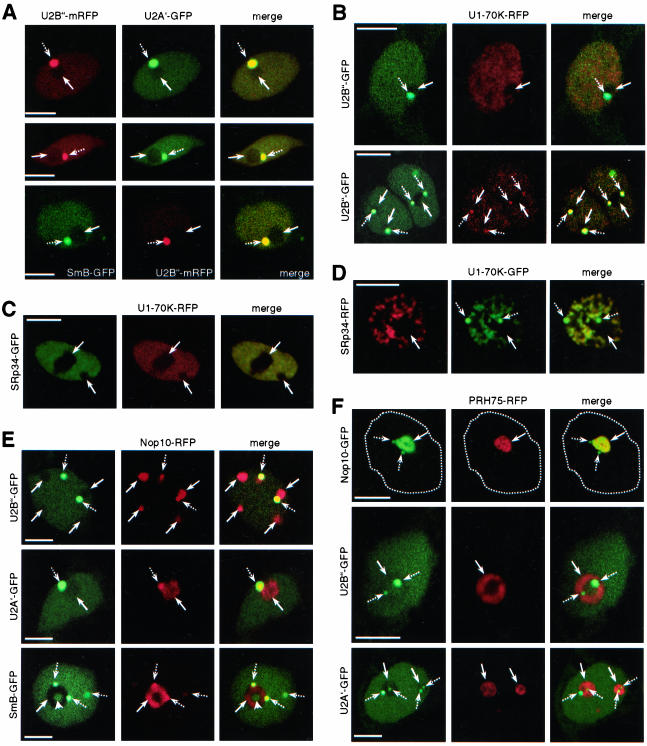
Figure 4.
Colocalization studies with established markers for Cajal bodies and nucleoli. (A) Colocalization of U2B″-mRFP and U2A′-GFP (two top rows), and SmB-GFP and U2B″-mRFP (third row). (B) Colocalization of U2B″-GFP and U1-70K-RFP. (C) Colocalization of U1-70K-RFP and SRp34-GFP in Arabidopsis cell. (D) Colocalization of U1-70K-RFP and SRp34-GFP in tobacco cell. (E) Colocalization of U2B″-GFP, U2A′-GFP, and SmB-GFP with Nop10-RFP. (F) Colocalization of PRH75-RFP with Nop10-GFP, U2B″-GFP, and U2A′-GFP. Broken line in the first row delineates the nucleus. In A-F, arrows and broken arrows point to nucleoli and Cajal bodies, respectively. Arrowhead in SmB row in E points to central nucleolar vacuole. Merged images show superimposition of GFP and RFP (mRFP) signals, which look yellow in case of colocalization. All images shown are single confocal sections. Bars, 4 μm (A-F).
Transient Expression of U1 snRNP-specific Protein 70K
To substantiate results obtained with U2 snRNP-specific proteins, analysis of Arabidopsis U1 snRNP-specific protein 70K (Golovkin and Reddy, 1996) fused to GFP or RFP was performed. Transient expression of RFP- (Figure 2, A and B) or GFP (Figure 2C)-tagged Arabidopsis U1-70K protein in tobacco (Figure 2, A and B; and Figure 2C, left) or in Arabidopsis (Figure 2C, right) protoplasts resulted in accumulation of fusion proteins in the nucleus, in a characteristic network of speckles. Only a small portion of both cell types used showed localization of U1-70K in Cajal bodies (Figure 2C, broken arrow on the right), consistent with the fact that U1 snRNP does not accumulate in Cajal bodies in plant cells (Beven et al., 1995). In contrast to FP-tagged U2B″ and U2A′ proteins, RFP- and GFP-tagged U1-70K was found exclusively in the nucleus already at the time point when fluorescence can be first detected (usually 4-5 h after transformation).
Figure 2.
Analysis of GFP- and RFP-tagged U1 snRNP-specific protein 70K in transiently transformed tobacco and Arabidopsis protoplasts. (A) Localization of RFP-tagged U1-70K protein in tobacco protoplasts. Autofluorescence of chloroplasts was collected in blue channel. Arrowheads point to nuclei. (B) Nucleus of the cell shown in A at higher resolution. Arrows point to nucleolus. (C) Localization of GFP-tagged U1-70K protein in tobacco (left image) and Arabidopsis protoplasts (right image). Only nuclei are shown. Arrows point to nucleoli and broken arrow points to Cajal body. In A-C, single confocal sections are shown, with corresponding DIC images in A and B. Bars, 50 μm (A) and 4 μm (B and C).
Thus, results obtained with transient expression of FP-tagged U2 and U1 snRNP-specific proteins in protoplasts provide strong evidence that this system can be used to study the nuclear organization of the spliceosomal machinery in living plant cells.
Transient Expression of U1 and U2 snRNP-specific Proteins Results in Correct Assembly into Mature snRNPs
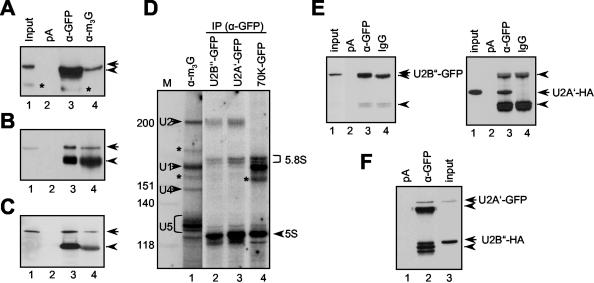
To further substantiate confocal microscopy results obtained with U1 and U2 snRNP-specific proteins, immunoprecipitations with protein extracts prepared from protoplasts transformed with plasmids expressing U1-70K-GFP, U2B″-GFP and U2A′-GFP fusion proteins were performed. All three proteins were efficiently precipitated with anti-GFP antibody (Figure 3, A-C, lane 3), and more importantly with antibody against the m3G cap at the 5′ end of U snRNAs (Figure 3, A-C, lane 4). None of the proteins was found in the pellet after incubation with protein A-Sepharose beads alone (Figure 3, A-C, lane 2). In addition, analysis of immunoprecipitates obtained with anti-GFP antibody for the presence of snRNAs revealed efficient coprecipitation of U1 snRNA with U1-70K-GFP (Figure 3D, lane 4) and U2 snRNA with U2B″-GFP and U2A′-GFP fusion proteins (Figure 3D, lanes 2 and 3), but not of other spliceosomal snRNAs. Finally, immunoprecipitations with protein extracts prepared from protoplasts coexpressing U2B″-GFP and U2A′-HA or U2A′-GFP and U2B″-HA fusion proteins resulted in efficient coprecipitation of the two proteins (Figure 3E, lane 3, and Figure 3F, lane 2), indicating correct assembly of transiently expressed proteins into mature snRNPs.
Figure 3.
Transiently expressed U1- and U2 snRNP-specific proteins assemble into mature snRNP. Immunoprecipitation of U2B″-GFP (A), U2A′-GFP (B), and U1-70K-GFP (C) fusion proteins. Lane 1, input protein extract. Lane 2, protein extracts incubated with protein A-Sepharose (pA). Lane 3, immunoprecipitations with anti-GFP antibody (α-GFP). Lane 4, immunoprecipitations with anti-m3G antibody (α-m3G). The blots are probed with anti-GFP antibody. Arrows in A-C point to U2B″, U2A′, and U1-70K-specific bands, and arrowheads point to immunoglobulin heavy chains. Bands marked with an asterisk in A are degradation products of U2B″-GFP. Only relevant portions of the blots are shown. (D) Analysis of anti-GFP immunoprecipitates for the presence of snRNAs. Lane 1, RNA immunoprecipitated with anti-m3G antibody. Lanes 2 and 3, RNA coprecipitated with U2B″-GFP and U2A′-GFP, respectively. Lane 4, RNA coprecipitated with U1-70K-GFP. Four spliceosomal snRNAs are indicated. Bands marked with asterisks are of unknown identity. Size markers in nucleotides are indicated on the left. 5S and 5.8S RNAs, which occur as background, are indicated on the right. (E) Coprecipitation of U2B″-GFP and U2A′-HA proteins. Protein extracts from protoplasts cotransformed with plasmids expressing U2B″-GFP and U2A′-HA were immunoprecipitated with anti-GFP antibody. Lane 1, input protein extract. Lane 2, protein extract incubated with protein A-Sepharose beads alone. Lane 3, immunoprecipitation with anti-GFP antibody. Lane 4, immunoprecipitation with unspecific antibody. The same blot was probed with anti-GFP (left) and after stripping anti-HA antibodies (right). (F) Coprecipitation of U2A′-GFP and U2B″-HA. Protein extracts from protoplasts cotransformed with plasmids expressing U2A′-GFP and U2B″-HA were immunoprecipitated with anti-GFP antibody. Lane 1, protein extract incubated with protein A-Sepharose beads alone. Lane 2, immunoprecipitation with anti-GFP antibody. Lane 3, input protein extract. Arrowheads in E and F point to immunoglobulin heavy and light chains.
Together, immunoprecipitation data and localization studies by confocal microscopy clearly demonstrated that transient expression of U1 and U2 snRNP-specific proteins in protoplasts results not only in correct localization but also in efficient incorporation into mature snRNPs. In addition, U2 snRNP-specific proteins U2B″ fused to GFP, mRFP, and YFP, and U2A′ fused to GFP proved to be good markers for Cajal bodies in transiently transformed plant protoplasts.
Colocalization of FP-Tagged Proteins in Transiently Transformed Plant Protoplasts
Having established a transient expression system in which FP-tagged proteins localize and assemble correctly, we asked next whether this system can be used for colocalization studies of two different proteins. Arabidopsis protoplasts were cotransformed with plasmid combinations indicated in Figure 4, and 24 h later analyzed by confocal laser scanning microscopy. In general, >50% of transformed cells showed expression of both FP-tagged proteins.
Because U2B″ and U2A′ are components of the same snRNP, we asked whether they colocalize in cotransformed cells. Arabidopsis protoplasts coexpressing U2B″-mRFP and U2A′-GFP showed perfect colocalization of proteins in Cajal bodies and nucleoplasm (Figure 4A, two top rows). This is consistent with the coprecipitation of these two proteins (Figure 3, E and F). In addition, coexpression of SmB-GFP and U2B″-mRFP resulted in colocalization of the two proteins in Cajal bodies and nucleoplasm (Figure 4A, third row), consistent with the fact that SmB is component of the U2 snRNP as well.
U1-70K was shown to localize in a speckled pattern in the nucleoplasm. In addition, in a small percentage of cells localization in Cajal bodies also was observed (Figure 2C). This localization pattern is different from those obtained with U2B″ and U2A′ proteins. Coexpression of RFP-tagged U1-70K with U2B″-GFP resulted in partial colocalization in the nucleoplasm but not in Cajal bodies (Figure 4B, first row). In cells where U1-70K localized to Cajal bodies, colocalization with U2B″ also was observed (Figure 4B, second row, broken arrows), although not to the extent observed in cells coexpressing U2B″ and U2A′ (compare second row in Figure 4B with A). In contrast, coexpression of U1-70K with SRp34/SR1, an Arabidopsis SR protein that localizes to speckles (see below), resulted in perfect colocalization of the two proteins in speckles (Figure 4, C and D) but not in Cajal bodies (Figure 4D, broken arrows), consistent with the fact that SR proteins do not localize to Cajal bodies (also see Figure 7).
Figure 7.
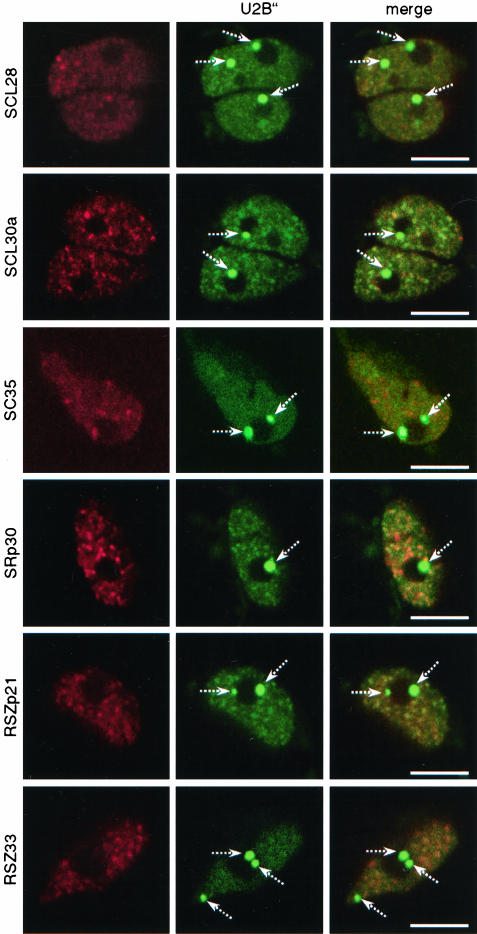
Colocalization of SR proteins with U2B″ in Arabidopsis protoplasts. Protoplasts were cotransformed with plasmids expressing RFP-tagged SR proteins and GFP-tagged U2B″. All images shown are single confocal sections. Broken arrows point to Cajal bodies. Bars, 5 μm.
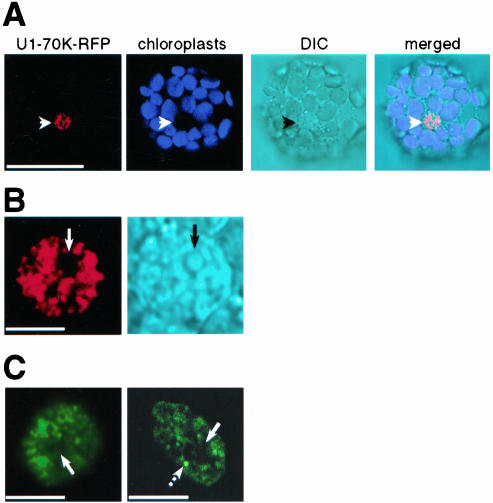
Analysis of Arabidopsis protoplasts coexpressing U2B″-GFP or U2A′-GFP and Nop10-RFP, an established marker for nucleoli and Cajal bodies (Supplemental Figure 2A) showed that Nop10 colocalizes with both proteins in Cajal bodies (Figure 4E, broken arrows) but not in the nucleoli (Figure 4E, arrows). Colocalization was not observed in nucleoplasm. These results are consistent with nuclear patterns obtained for each protein separately in which U2B″ and U2A′ localized to Cajal bodies and nucleoplasm (Figure 1, A and B), whereas Nop10 localized to nucleoli and Cajal bodies (Supplemental Figure 2A). Like with U2B″ and U2A′, coexpression of SmB-GFP with Nop10-RFP resulted in colocalization of the two proteins in Cajal bodies but not in nucleolus. In addition, portion of SmB present in the central nucleolar vacuole did not colocalize with Nop10 (Figure 4E, arrowhead in SmB-GFP row).
PRH75 is another established marker for nucleoli (Supplemental Figure 2, C-E). However, Nop10 also localized to Cajal bodies (Supplemental Figure 2A), whereas no evidence for localization of PRH75 in Cajal bodies was provided (Supplemental Figure 2E). In Arabidopsis cells coexpressing Nop10-GFP and PRH75-RFP, the two proteins colocalized in nucleoli but not in Cajal bodies (Figure 4F). This indicated that PRH75 is not associated with Cajal bodies. Consistent with this, in cells coexpressing either U2B″-GFP or U2A′-GFP and PRH75-RFP no colocalization was observed (Figure 4F), confirming that PRH75 indeed does not localize in Cajal bodies.
Together, colocalization studies presented in Figure 4 clearly demonstrate that coexpression of two proteins with different subnuclear localizations does not influence each others localization. Moreover, colocalization was observed only with protein combinations where this was expected, indicating that transient expression of two different FP-tagged proteins is suitable for colocalization studies in living plant cells.
FP-tagged Arabidopsis SR Proteins Localize into Speckles
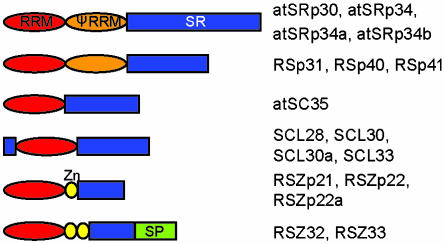
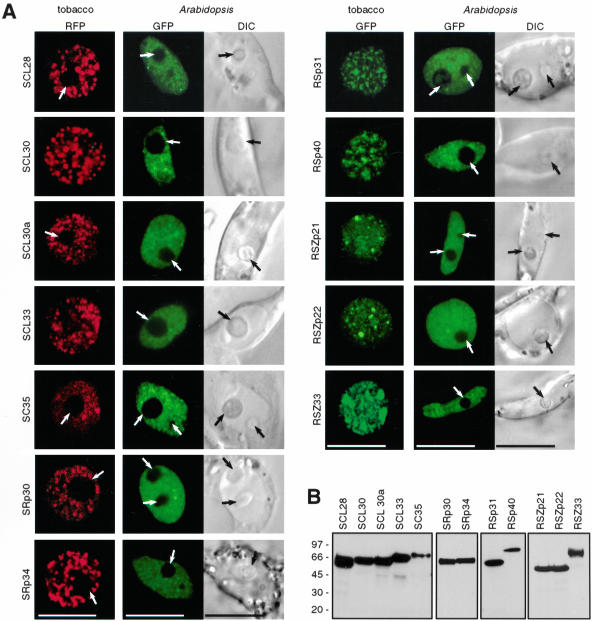
In mammalian cells, the majority of pre-mRNA splicing factors show speckled nuclear localization pattern (Sleeman and Lamond, 1999b; Dundr and Misteli, 2001; Lamond and Spector, 2003). SR proteins have often been used as markers for nuclear speckles, also called SC35 domain, because of localization of SR protein SC35 in them (Lamond and Spector, 2003). The Arabidopsis genome encodes for 18 SR proteins (Lorkovic and Barta, 2002) that can be grouped into subfamilies according to their domain organization (Figure 5). The established transient expression system allowed us to extend the localization studies to plant SR proteins. Plasmids expressing GFP and RFP fusions of 12 of 18 Arabidopsis SR proteins were constructed, and localization of fusion proteins was analyzed in tobacco and Arabidopsis protoplasts. In both cell types, RFP and GFP fusions of all SR proteins analyzed were found in the nucleus in a pattern resembling speckles in mammalian cells (Figure 6A). The appearance of speckles in tobacco cells varied from irregularly shaped to more regularly shaped dot-like structures (Figure 6A, compare for example SRp34 and SCL30). In addition, a diffuse nucleoplasmic staining also was observed. In contrast, speckles in Arabidopsis cells were less prominent and smaller, and more protein was found in a diffuse nucleoplasmic pool irrespective of the FP-tag used (Figure 6A and Supplemental Figure 3). Analysis of protein expression by Western blotting with anti-GFP and anti-RFP antibodies revealed that the stronger speckled appearance in tobacco cells cannot be accounted to aggregation of degraded proteins, because no major degradation products were detected (Figure 6B) (our unpublished data). All images shown in Figure 6A were taken with living cells 24 h after transformation; however, analysis of transformed cells as early as 4-5 h after transformation also revealed nuclear localization of all proteins analyzed, indicating that the import of SR proteins into nuclei is very rapid. In addition, it became clear that none of the analyzed SR proteins associates with nucleoli, seen as black areas inside the nucleus (Figure 6A, arrows; compare also fluorescence image with differential interference contrast [DIC] image in Arabidopsis cells). Because all images in Figure 6A are single confocal sections, nucleoli are not visible in all nuclei shown. However, cotransformation of representatives of each subgroup of Arabidopsis SR proteins fused to GFP with the established nucleolar marker Nop10 fused to RFP clearly demonstrated that none of the SR proteins associates with nucleoli (Supplemental Figure 4).
Figure 5.
Schematic representation of Arabidopsis SR proteins. RRM, RNA recognition motif, red ovals; PseudoRRM, orange ovals; SR domains, blue squares; Zinc knuckles (Zn), yellow ovals; and serine/proline (SP)-rich domain, green squares.
Figure 6.
Subnuclear localization of Arabidopsis SR proteins. (A) Localization of GFP or RFP-tagged SR proteins in transiently transformed tobacco and Arabidopsis protoplasts. DIC images are shown to visualize the nuclei in Arabidopsis cells. Arrows point to nucleoli. All images shown are single confocal sections. Bars, 10 μm. (B) Expression analysis of GFP-tagged SR proteins in tobacco protoplasts. Whole cell extracts were analyzed by Western blotting with anti-GFP antibody. Molecular mass standards in kilodaltons are indicated on the left.
Because some splicing factors, particularly snRNPs, have been found in Cajal bodies (Carmo-Fonseca et al., 1991a,b; Spector et al., 1991; Raska et al., 1991; Matera and Ward, 1993; Beven et al., 1995; this work), we asked next whether SR proteins localize into this nuclear compartment. Arabidopsis cell suspension protoplasts were cotransformed with plasmids expressing SR proteins fused to RFP and U2B″ or U2A′ proteins fused to GFP. Both U2B″ and U2A′ proteins localized into Cajal bodies and in a diffuse/speckled pattern in which partial colocalization with SR proteins was detected. None of the SR proteins localized to Cajal bodies, as evident from the lack of colocalization with U2B″ or U2A′ at these sites (Figure 7; our unpublished data). Thus, we conclude that in Arabidopsis, like in mammalian cells, SR proteins do not localize in Cajal bodies.
DISCUSSION
In mammalian cells, splicing factors are found in the nucleoplasm in a diffuse pool and in an irregular network called speckles (Sleeman and Lamond 1999b; Dundr and Misteli, 2001; Lamond and Spector, 2003). In addition, some splicing factors, particularly snRNPs, accumulate in Cajal bodies (Carmo-Fonseca et al., 1991a,b; Raska et al., 1991; Spector et al., 1991; Matera and Ward, 1993; Gall, 2000; Dundr and Misteli, 2001). The majority of knowledge about nuclear organization of spliceosomal machinery comes from experiments in which tagged proteins of interest were transiently expressed in HeLa cells, followed by indirect immunofluorescence detection. By the onset of fluorescent protein tags it became possible to analyze not only localization of the proteins but also their dynamics in the nucleus (Sleeman et al., 1998; Boudonck et al., 1999; Sleeman and Lamond, 1999a; Misteli, 2001; Carmo-Fonseca, 2002a; Lamond and Spector, 2003).
Here, we constructed plasmids that allow expression of RFP, mRFP, YFP, and CFP fusion proteins in plant protoplasts from the same transcription unit, resulting in comparable expression levels. Four different FP tags allowed localization and colocalization studies of plant splicing factors in living plant protoplasts and in combination with a plasmid designed for the expression of HA-tagged proteins (Genschik et al., 1997) coprecipitation of transiently expressed proteins also was possible. Advantages of this system are that there is no need for the fixation of the cells and that in contrast to generation of transgenic plants or cell suspension lines, a large number of samples can be analyzed in a very short time.
Plasmids expressing several Arabidopsis snRNP-specific proteins fused to FP tags developed in this study were constructed and tested for their functionality in transiently transformed tobacco and Arabidopsis protoplasts. Transiently expressed U2B″ fused to GFP, mRFP, or YFP in Arabidopsis cell suspension protoplasts was found in Cajal bodies and in a diffuse nucleoplasmic pool. Essentially the same localization pattern was observed for U2A′-GFP fusion protein. Both fusion proteins also were found to be efficiently incorporated into mature U2 snRNP, as determined by coprecipitation of the U2 snRNA (Figure 3), indicating that the observed nuclear patterns are functionally relevant. In addition, these results correlate well with previously reported expression of the GFP-tagged potato U2B″ in transgenic Arabidopsis plants and in a stable BY-2 cell line (Boudonck et al., 1998; Boudonck et al., 1999) as well as with patterns described in mammalian cells in which U2 snRNP also strongly accumulate in Cajal bodies (Carmo-Fonseca et al., 1991a,b; Matera and Ward, 1993). In contrast, the U1 snRNP-specific protein U1-70K fused to GFP or RFP localized in a speckled pattern, and it was rarely found in Cajal bodies, although it was efficiently incorporated into mature U1 snRNP (Figure 3). Subnuclear distribution of U1 snRNP differs from that of the other spliceosomal snRNPs in both mammalian and plant cells. In particular, by using U1 snRNA antisense probes and antibodies against U1-70K it has been shown that U1 snRNP in mammalian cells localizes in speckles (Carmo-Fonseca et al., 1991a; Carmo-Fonseca et al., 1991b; Wu et al., 1991; Matera and Ward, 1993; Visa et al., 1993). Localization in Cajal bodies also was observed; however, U1 snRNP does not accumulate in them (Matera and Ward, 1993). A similar situation has been described in pea root cells by using U1 snRNA antisense probe, with <1% of cells showing prominent labeling of Cajal bodies (Beven et al., 1995). Together, results obtained with snRNP-specific proteins clearly demonstrated that transient expression of FP-tagged spliceosomal proteins in protoplasts results in correct localization of fusion proteins and in their assembly into mature snRNPs. Therefore, this system allows studies of subnuclear organization of the spliceosomal machinery in living plant cells.
Using four different FP tags also allowed coexpression of two different proteins and colocalization studies without time consuming indirect immunofluorescence procedures. Indeed, by using cotransformation we were able to demonstrate that U2B″ and U2A′ proteins perfectly colocalize, consistent with the fact that they are components of the U2 snRNP. In addition, U2B″ and U2A′ colocalized with Nop10 in Cajal bodies but did not in nucleoli and nucleoplasm. Moreover, the two nucleolar markers Nop10 and PRH75 did colocalize in nucleoli but did not in Cajal bodies. Also, this allowed us to show that plant SR proteins do not localize to either nucleoli or Cajal bodies (Figure 7 and Supplemental Figure 4). Thus, coexpression of two proteins with different subnuclear localization did not affect each others localization, making this transient expression system suitable for colocalization studies as well.
In addition to chromatin (chromosomes), the nucleus contains numerous compartments, including nucleoli, Cajal bodies, gems, splicing speckles, and promyelocytic leukemia bodies (Dundr and Misteli, 2001; Spector, 2001). Compartmentalization of the nucleus is considered as an important feature for proper gene expression (Lamond and Earnshaw, 1998; Misteli and Spector, 1998; Misteli, 2001; Carmo-Fonseca, 2002a). Because little is known about compartmentalization of plant cell nuclei, one of the goals of this study was to establish FP-tagged markers for different nuclear compartments. The results with U2 snRNP-specific proteins U2B″ and U2A′ demonstrated that these two proteins can be used as markers for Cajal bodies in transiently transformed protoplasts. In addition to markers for Cajal bodies, Nop10 was established as marker for nucleoli. By using GFP and RFP fusions of PRH75, a previously reported nuclear RNA helicase, it was possible in combination with other approaches to unambiguously show that it is primarily a nucleolar protein (Supplemental Figure 2, C, D, and E). Moreover, analysis of nuclear localization of Arabidopsis SR proteins by using the same system revealed that they are good markers for speckles. In both tobacco and Arabidopsis cells, SR proteins localized into speckles (Figure 6); however, speckles were less prominent in Arabidopsis cells. Similarly, expression of U2B″-GFP in BY-2 cells and in Arabidopsis plants (Boudonck et al., 1998, 1999), fluorescence in situ hybridization experiments with U2, U1, and U6 snRNA probes in pea root cells (Beven et al., 1995), and indirect immunofluorescence on pea and onion root cells with monoclonal antibodies against U2B″ (Beven et al., 1995; Cui and Moreno Diaz de la Espina, 2003) also revealed a more diffuse than speckled nucleoplasmic pattern. Consistent with this, transiently expressed U2B″-GFP, U2B″-mRFP, U2B″-YFP, and U2A′-GFP proteins showed more uniform than speckled nucleoplasmic localization in Arabidopsis protoplasts. Similarly, BY-2 cells expressing FP-tagged SR1 (SRp34) and PK12, an SR protein specific kinase, also showed more diffuse than speckled localization patterns (Savaldi-Goldstein et al., 2003), indicating that this feature might be intrinsic to plant cells grown in suspension. The reason for this could be a difference in chromatin condensation in tobacco and Arabidopsis nuclei. In contrast to highly differentiated tobacco leaf mesophyll cells, Arabidopsis suspension cells grow and divide very fast, and therefore they likely have more decondensed or active chromatin. Thus, higher gene activity could result in the recruitment of more splicing factors to assembling spliceosome at nascent pre-mRNAs which, in turn, would result in smaller speckles and more uniformly distributed splicing factors. This would be in agreement with the widely accepted role of speckles as reservoirs that supply splicing factors for splicing of nascent pre-mRNAs at nearby genes (O'Keefe et al., 1994; Misteli et al., 1997; Lamond and Spector, 2003). Variations in the appearance of speckles have also been reported for different mammalian cells, different strains of HeLa cells and for the same strain of cells grown at different temperatures (Carmo-Fonseca et al., 1991b; Matera and Ward, 1993).
In all cells expressing U2 snRNP-specific proteins, a clear cytoplasmic staining was observed in addition to a predominant nuclear localization (Figure 1C). This does not seem to be due to overexpression of fusion proteins, as cells with very low expression levels also showed cytoplasmic localization even 48 h after transformation. One possible explanation of this phenomenon is that the import of U2B″ and U2A′ proteins into the nucleus is generally slow. Indeed, injection of U2B″ and U1A proteins into Xenopus oocytes resulted in their slower accumulation in the nuclei than reported for other karyophilic proteins (Kambach and Mattaj, 1992). It has been speculated that the import of U2B″ and U1A proteins depends on the number of free binding sites on their cognate snRNAs in the nucleus (Kambach and Mattaj, 1992; Kambach and Mattaj, 1994). It is also possible that FP tags impair nuclear import to some extent. However, nuclear accumulation of the U1 snRNP-specific protein 70K and of all plant SR proteins analyzed in this work was found to be very fast, as no cytoplasmic fluorescence was observed at any post-transformation time point. This indicates that overexpression of the fusion proteins may not be a limiting factor for efficient nuclear import. It is not clear why U2 snRNP localizes into diffuse nucleoplasmic pattern, whereas the U1 snRNP occurs in speckled pattern. Consistent with this observation, U2B″ and U1-70K did not show significant colocalization when coexpressed in Arabidopsis protoplasts (Figure 4B), whereas U1-70K colocalized with SRp34/SR1 in nuclear speckles (Figure 4, C and D). Further experimentation, which is beyond the scope of this work, is required to determine specific features of the import of snRNP-specific proteins into the nucleus and their assembly into mature snRNPs.
Supplementary Material
Acknowledgments
We thank Reinhard Lührmann for anti-m3G antibody, Roger Y. Tsien for mRFP cDNA, Elisabeth Waigmann for initial help with the confocal microscope, and Maria Kalyna for critical reading of the manuscript and helpful discussions. This work was supported by grants from the Österreichischer Fonds zur Förderung der wissenschaftlichen Forschung to A.B. (SFB-F017/10/11).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E04-01-0055. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-01-0055.
Abbreviations used: CFP, cyan fluorescent protein; FP, fluorescent protein; GFP, green fluorescent protein; mRFP, monomeric red fluorescent protein; RFP, red fluorescent protein; snRNP, small nuclear ribonucleoprotein; SR, serine-arginine; YFP, yellow fluorescent protein.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Ali, G.S., Golovkin, M., and Reddy, A.S. (2003). Nuclear localisation and in vivo dynamics of a plant-specific serine/arginine-rich protein. Plant J. 36, 883-893. [DOI] [PubMed] [Google Scholar]
- Beven, A.F., Simpson, G.G., Brown, J.W.S., and Shaw, P.J. (1995). The organisation of spliceosomal components in the nuclei of higher plants. J. Cell Sci. 108, 509-518. [DOI] [PubMed] [Google Scholar]
- Boudonck, K., Dolan, L., and Shaw, P.J. (1998). Coiled body numbers in the Arabidopsis root epidermis are regulated by cell type, developmental stage and cell cycle parameters. J. Cell Sci. 111, 3687-3694. [DOI] [PubMed] [Google Scholar]
- Boudonck, K., Dolan, L., and Shaw, P.J. (1999). The movement of Cajal bodies visualised in living plant cells by the green fluorescent protein. Mol. Biol. Cell 10, 2297-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. (1999). Splicing of precursors to mRNAs by the spliceosome. In: The RNA World, 2nd ed., ed. R.F. Gesteland, T.-R. Cech, and J.F. Atkins, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 525-560.
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca, M. (2002a). The contribution of nuclear compartmentalisation to gene regulation. Cell 108, 513-521. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca, M. (2002b). New clues to the function of the Cajal body. EMBO Rep. 3, 726-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca, M., Pepperkok, R., Sproat, B.S., Ansorge, W., Swanson, M.S., and Lamond, A.I. (1991a). In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO J. 10, 1863-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca, M., Tollervey, D., Barabino, S.M., Merdes, A., and Brunner, C. (1991b). Mammalian nuclei contain foci which are highly enriched in components of the pre-mRNA splicing machinery. EMBO J. 10, 195-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, P., and Moreno Diaz de la Espina, S. (2003). Sm and U2B″proteins redistribute to different nuclear domains in dormant and proliferating onion cells. Planta 217, 21-31. [DOI] [PubMed] [Google Scholar]
- Docquier, S., Tillemans, V., Deltour, R., and Motte, P. (2004). Nuclear bodies and compartmentalisation of pre-mRNA splicing factors in higher plants. Chromosoma 112, 255-266. [DOI] [PubMed] [Google Scholar]
- Dundr, M., and Misteli, T. (2001). Functional architecture of the cell nucleus. Biochem. J. 356, 297-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y., Hearn, S., and Spector, D.L. (2004). Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis. Mol. Biol. Cell 15, 2664-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall, J.G. (2000). Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16, 273-300. [DOI] [PubMed] [Google Scholar]
- Gall, J.G. (2001). A role for Cajal bodies in assembly of the nuclear transcription machinery. FEBS Lett. 498, 164-167. [DOI] [PubMed] [Google Scholar]
- Genschik, P., Hall, J., and Filipowicz, W. (1997). Cloning and characterization of the Arabidopsis cyclic phosphodiesterase which hydrolyzes ADP-ribose 1″,2″-cyclic phosphate and nucleoside 2′,3′-cyclic phosphates. J. Biol. Chem. 272, 13211-13219. [DOI] [PubMed] [Google Scholar]
- Golovkin, M., and Reddy, A.S. (1996). Structure and expression of a plant U1 snRNP 70K gene: alternative splicing of U1 snRNP 70K pre-mRNAs produces two different transcripts. Plant Cell 8, 1421-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall, G.J., and Filipowicz, W. (1989). The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell 58, 473-483. [DOI] [PubMed] [Google Scholar]
- Kambach, C., and Mattaj, I.W. (1992). Intracellular distribution of the U1A protein depends on active transport and nuclear binding to U1 snRNA. J. Cell Biol. 118, 11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach, C., and Mattaj, I.W. (1994). Nuclear transport of the U2 snRNP-specific U2B″protein is mediated by both direct and indirect signalling mechanism. J. Cell Sci. 107, 1807-1816. [DOI] [PubMed] [Google Scholar]
- Kambach, C., Walke, S., and Nagai, K. (1999). Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 9, 222-230. [DOI] [PubMed] [Google Scholar]
- Koop, H.U., Steinmuller, K., Wagner, H., Rossler, C., Eibl, C., and Sacher, L. (1996). Integration of foreign sequences into the tobacco plastome via polyethylene glycol-mediated protoplast transformation. Planta 199, 193-201. [DOI] [PubMed] [Google Scholar]
- Krämer, A. (1996). The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65, 367-409. [DOI] [PubMed] [Google Scholar]
- Lambermon, M.H., Simpson, G.G., Wieczorek Kirk, D.A., Hemmings-Mieszczak, M., Klahre, U., and Filipowicz, W. (2000). UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J. 19, 1638-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond, A.I., and Earnshaw, W.C. (1998). Structure and function of the nucleus. Science 280, 547-553. [DOI] [PubMed] [Google Scholar]
- Lamond, A.I., and Spector, D.L. (2003). Nuclear speckles: a model for nuclear organelles. Nature Rev. Mol. Cell. Biol. 4, 605-612. [DOI] [PubMed] [Google Scholar]
- Lopato, S., Forstner, C., Kalyna, M., Hilscher, J., Langhammer, U., Indrapichate, K., Lorkovic, Z.J., and Barta, A. (2002). Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors. J. Biol. Chem. 277, 39989-39998. [DOI] [PubMed] [Google Scholar]
- Lopato, S., Gattoni, R., Fabini, G., Stevenin, J., and Barta, A. (1999a). A novel family of plant splicing factors with a Zn knuckle motif: examination of RNA binding and splicing activities. Plant Mol. Biol. 39, 761-773. [DOI] [PubMed] [Google Scholar]
- Lopato, S., Kalyna, M., Dorner, S., Kobayashy, R., Krainer, A.R., and Barta, A. (1999b). atRSp30, one of two SF2(ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 13, 987-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato, S., Waigmann, E., and Barta, A. (1996). Characterisation of a novel arginine/serine-rich splicing factor in Arabidopsis. Plant Cell 8, 2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic, Z.J., and Barta, A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30, 623-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic, Z.J., Herrmann, R.G., and Oelmüller, R. (1997). PRH75, a new nucleus-localised member of the DEAD-box protein family from higher plants. Mol. Cell. Biol. 17, 2257-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lührmann, R., Appel, B., Bringmann, P., Rinke, J., Rothe, S., and Bald, R. (1982). Isolation and characterisation of a rabbit anti-m3 2,2,7 G antibodies. Nucleic Acids Res. 10, 7103-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera, A.G., and Ward, D.C. (1993). Nucleoplasmic organisation of small nuclear ribonucleoproteins in cultured human cells. J. Cell Biol. 121, 715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskiene, I., Baudouin, E., Schweighofer, A., Liwosz, A., Jonak, C., Rodriguez, P.L., Jelinek, H., and Hirt, H. (2003). Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. J. Biol. Chem. 278, 18945-18952. [DOI] [PubMed] [Google Scholar]
- Misteli, T. (2001). Protein dynamics: implications for nuclear architecture and gene expression. Science 291, 843-847. [DOI] [PubMed] [Google Scholar]
- Misteli, T., Caseras, J.F., and Spector, D.L. (1997). The dynamics of a pre-mRNA splicing factor in living cells. Nature 387, 523-527. [DOI] [PubMed] [Google Scholar]
- Misteli, T., and Spector, D.L. (1998). The cellular organisation of gene expression. Curr. Opin. Cell Biol. 10, 323-331. [DOI] [PubMed] [Google Scholar]
- Ogg, S.C., and Lamond, A.I. (2002). Cajal bodies and coilin-moving towards function. J. Cell Biol. 159, 17-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe, R.T., Mayeda, A., Sadowski, C.L., Krainer, A.R., and Spector, D.L. (1994). Disruption of pre-mRNA splicing in vivo results in reorganisation of splicing factors. J. Cell Biol. 124, 249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani, M., Goldberg, I., Swedlow, J.R., and Lamond, A.I. (2000). In vivo analysis of Cajal body movement, separation, and joining in live human cells. J. Cell Biol. 151, 1561-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska, I., Andrade, L.E.C., Ochs, R.L., Chan, E.K.L., Chang, C.-M., Roos, G., and Tan, E.M. (1991). Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp. Cell Res. 195, 27-37. [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein, S., Aviv, D., Davydov, O., and Fluhr, R. (2003). Alternative splicing modulation by LAMMER kinase impinges on developmental and transcriptome expression. Plant Cell 15, 926-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, G.G., and Filipowicz, W. (1996). Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol. Biol. 32, 1-41. [DOI] [PubMed] [Google Scholar]
- Sleeman, J.E., and Lamond, A.I. (1999a). Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 9, 1065-1074. [DOI] [PubMed] [Google Scholar]
- Sleeman, J.E., and Lamond, A.I. (1999b). Nuclear organization of pre-mRNA splicing factors. Curr. Opin. Cell Biol. 11, 372-377. [DOI] [PubMed] [Google Scholar]
- Sleeman, J., Lyon, C.E., Platani, M., Kreivi, J.-P., and Lamond, A.I. (1998). Dynamic interactions between splicing snRNPs, Coiled bodies and nucleoli revealed using snRNP proteins fusions to the green fluorescent protein. Exp. Cell Res. 243, 290-304. [DOI] [PubMed] [Google Scholar]
- Snaar, S., Wiesmeijer, K., Jochemsen, A.G., Tanke, H.J., and Dirks, R.W. (2000). Mutational analysis of fibrillarin and its mobility in living human cells. J. Cell Biol. 151, 653-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector, D.L. (2001). Nuclear bodies. J. Cell Sci. 114, 2891-2893. [DOI] [PubMed] [Google Scholar]
- Spector, D.L., Fu, X.-D., and Maniatis, T. (1991). Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 10, 3467-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa, N., Puvion-Dutilleul, F., Bachellerie, J.P., and Puvion, E. (1993). Intranuclear distribution of U1 and U2 snRNA visualized by high-resolution in situ hybridisation-revelation of a novel compartment containing U1 but not U2 snRNA in HeLa cells. Eur. J. Cell Biol. 60, 308-321. [PubMed] [Google Scholar]
- Will, C.L., and Lührmann, R. (2001). Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13, 290-310. [DOI] [PubMed] [Google Scholar]
- Wu, Z., Murphy, C., Callan, H.G., and Gall, J.G. (1991). Small nuclear ribonucleoproteins and heterologous nuclear ribonucleoproteins in the amphibian germinal vesicle -loops, sheres and snurposomes. J. Cell Biol. 113, 465-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.