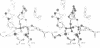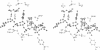Abstract
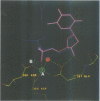
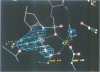
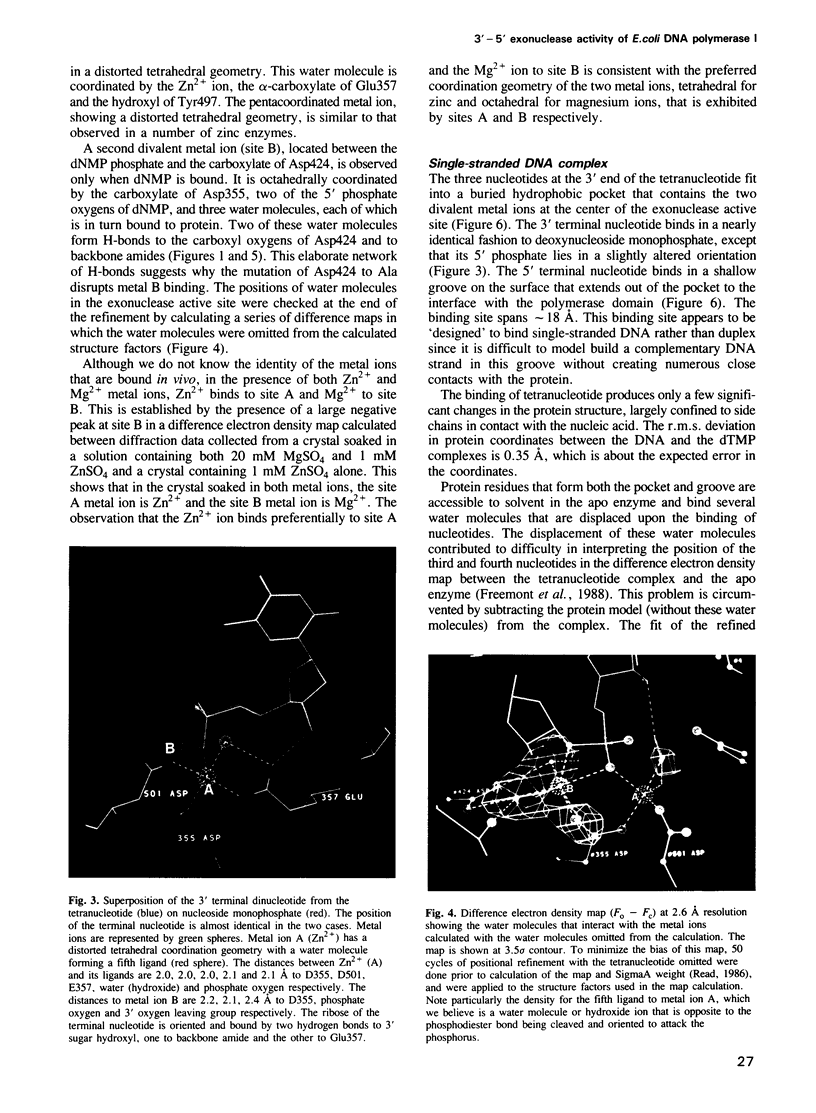
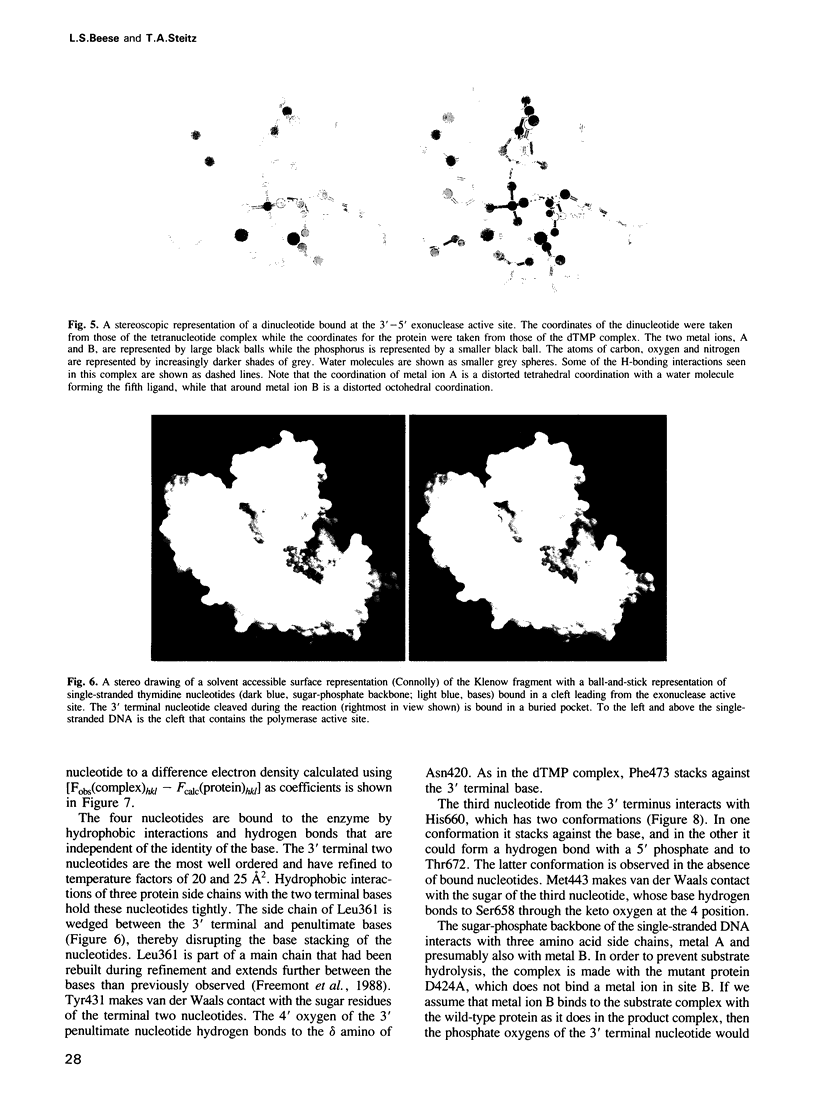
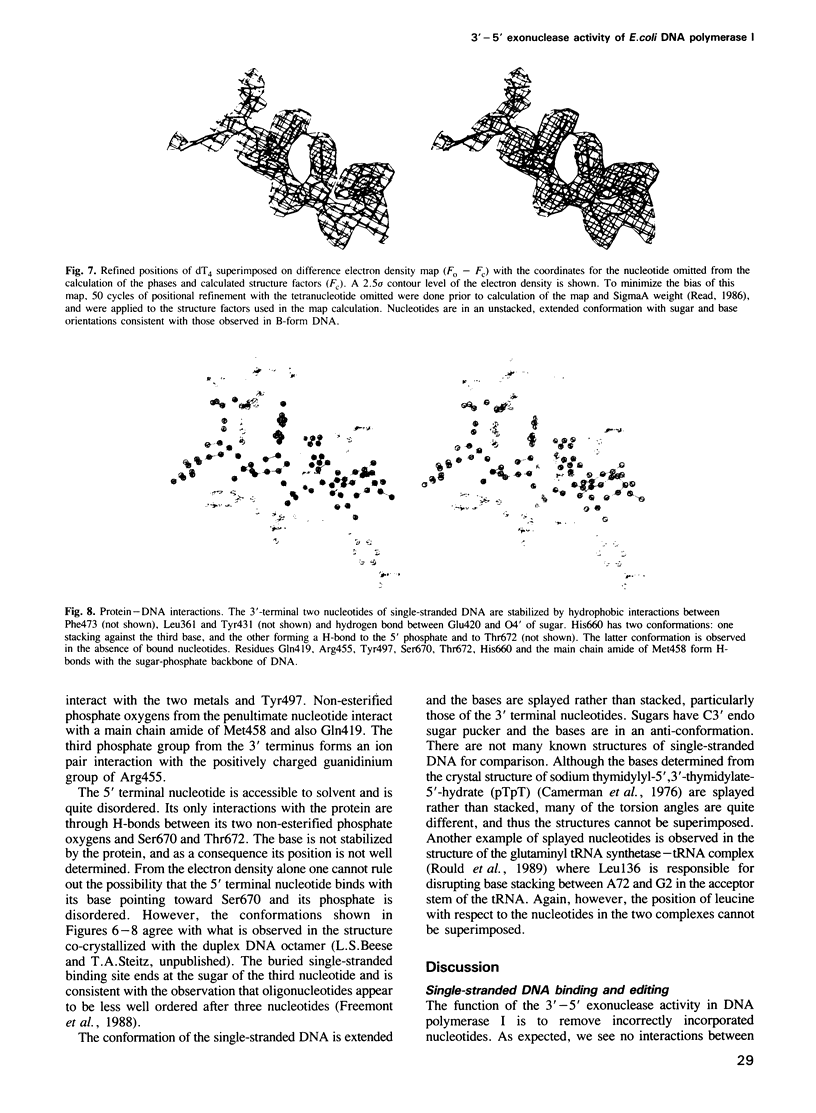
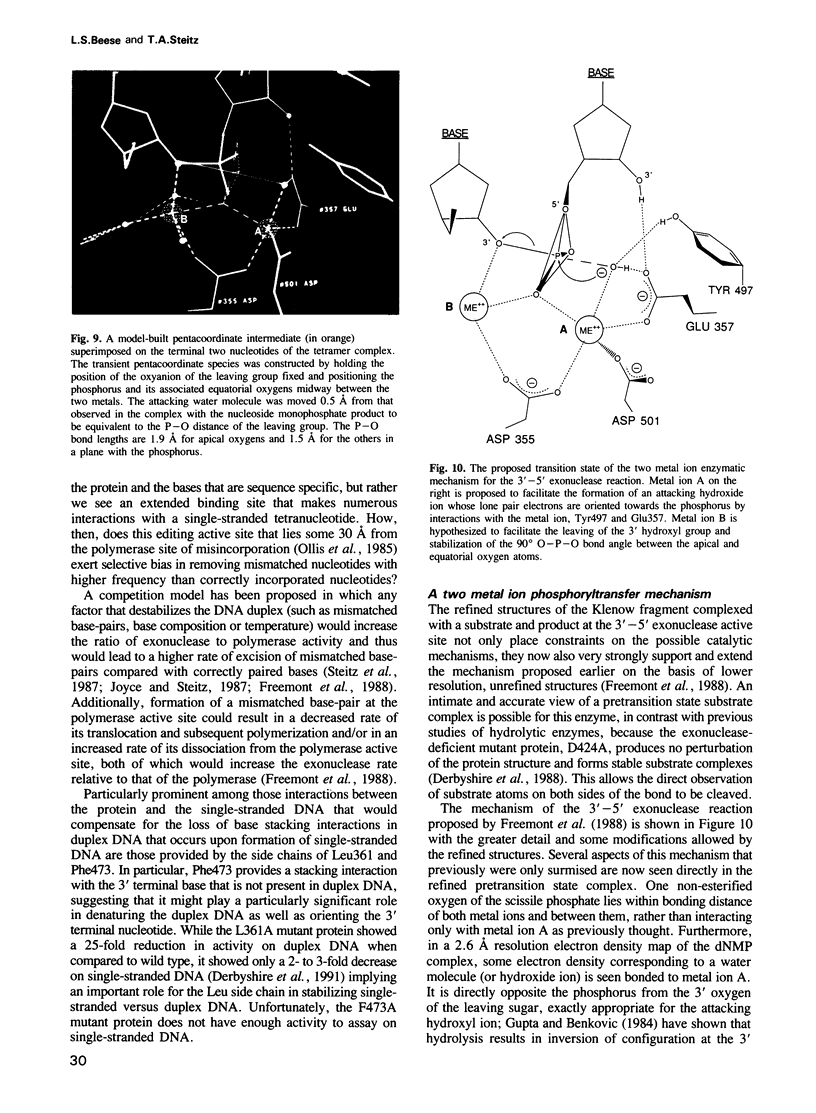
The refined crystal structures of the large proteolytic fragment (Klenow fragment) of Escherichia coli DNA polymerase I and its complexes with a deoxynucleoside monophosphate product and a single-stranded DNA substrate offer a detailed picture of an editing 3'-5' exonuclease active site. The structures of these complexes have been refined to R-factors of 0.18 and 0.19 at 2.6 and 3.1 A resolution respectively. The complex with a thymidine tetranucleotide complex shows numerous hydrophobic and hydrogen-bonding interactions between the protein and an extended tetranucleotide that account for the ability of this enzyme to denature four nucleotides at the 3' end of duplex DNA. The structures of these complexes provide details that support and extend a proposed two metal ion mechanism for the 3'-5' editing exonuclease reaction that may be general for a large family of phosphoryltransfer enzymes. A nucleophilic attack on the phosphorous atom of the terminal nucleotide is postulated to be carried out by a hydroxide ion that is activated by one divalent metal, while the expected pentacoordinate transition state and the leaving oxyanion are stabilized by a second divalent metal ion that is 3.9 A from the first. Virtually all aspects of the pretransition state substrate complex are directly seen in the structures, and only very small changes in the positions of phosphate atoms are required to form the transition state.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkinson M. R., Deutscher M. P., Kornberg A., Russell A. F., Moffatt J. G. Enzymatic synthesis of deoxyribonucleic acid. XXXIV. Termination of chain growth by a 2',3'-dideoxyribonucleotide. Biochemistry. 1969 Dec;8(12):4897–4904. doi: 10.1021/bi00840a037. [DOI] [PubMed] [Google Scholar]
- Bernad A., Blanco L., Lázaro J. M., Martín G., Salas M. A conserved 3'----5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989 Oct 6;59(1):219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Atkinson M. R., Setlow P., Kornberg A. An active fragment of DNA polymerase produced by proteolytic cleavage. Biochem Biophys Res Commun. 1969 Dec 4;37(6):982–989. doi: 10.1016/0006-291x(69)90228-9. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3' leads to 5' exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972 Jan 10;247(1):241–248. [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Camerman N., Fawcett J. K., Cameran A. Molecular structure of a deoxyribose-dinucleotide, sodium thymidylyl-(5' yields to 3')-thymidylate-(5') hydrate (pTpT), and a possible structural model for polythymidylate. J Mol Biol. 1976 Nov 15;107(4):601–621. doi: 10.1016/s0022-2836(76)80086-1. [DOI] [PubMed] [Google Scholar]
- Catalano C. E., Benkovic S. J. Inactivation of DNA polymerase I (Klenow fragment) by adenosine 2',3'-epoxide 5'-triphosphate: evidence for the formation of a tight-binding inhibitor. Biochemistry. 1989 May 16;28(10):4374–4382. doi: 10.1021/bi00436a038. [DOI] [PubMed] [Google Scholar]
- Cowart M., Gibson K. J., Allen D. J., Benkovic S. J. DNA substrate structural requirements for the exonuclease and polymerase activities of procaryotic and phage DNA polymerases. Biochemistry. 1989 Mar 7;28(5):1975–1983. doi: 10.1021/bi00431a004. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., Steitz T. A. Genetic and crystallographic studies of the 3',5'-exonucleolytic site of DNA polymerase I. Science. 1988 Apr 8;240(4849):199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Grindley N. D., Joyce C. M. The 3'-5' exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J. 1991 Jan;10(1):17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W., Tsui W. C. Kinetic basis of spontaneous mutation. Misinsertion frequencies, proofreading specificities and cost of proofreading by DNA polymerases of Escherichia coli. J Mol Biol. 1982 Mar 25;156(1):37–51. doi: 10.1016/0022-2836(82)90457-0. [DOI] [PubMed] [Google Scholar]
- Freemont P. S., Friedman J. M., Beese L. S., Sanderson M. R., Steitz T. A. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont P. S., Ollis D. L., Steitz T. A., Joyce C. M. A domain of the Klenow fragment of Escherichia coli DNA polymerase I has polymerase but no exonuclease activity. Proteins. 1986 Sep;1(1):66–73. doi: 10.1002/prot.340010111. [DOI] [PubMed] [Google Scholar]
- Gupta A. P., Benkovic S. J. Stereochemical course of the 3'----5'-exonuclease activity of DNA polymerase I. Biochemistry. 1984 Nov 20;23(24):5874–5881. doi: 10.1021/bi00319a029. [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A. Stereochemically restrained refinement of macromolecular structures. Methods Enzymol. 1985;115:252–270. doi: 10.1016/0076-6879(85)15021-4. [DOI] [PubMed] [Google Scholar]
- Holmes M. A., Matthews B. W. Structure of thermolysin refined at 1.6 A resolution. J Mol Biol. 1982 Oct 5;160(4):623–639. doi: 10.1016/0022-2836(82)90319-9. [DOI] [PubMed] [Google Scholar]
- Jones T. A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- Joyce C. M. How DNA travels between the separate polymerase and 3'-5'-exonuclease sites of DNA polymerase I (Klenow fragment). J Biol Chem. 1989 Jun 25;264(18):10858–10866. [PubMed] [Google Scholar]
- Kim H., Lipscomb W. N. Crystal structure of the complex of carboxypeptidase A with a strongly bound phosphonate in a new crystalline form: comparison with structures of other complexes. Biochemistry. 1990 Jun 12;29(23):5546–5555. doi: 10.1021/bi00475a019. [DOI] [PubMed] [Google Scholar]
- LEHMAN I. R., RICHARDSON C. C. THE DEOXYRIBONUCLEASES OF ESCHERICHIA COLI. IV. AN EXONUCLEASE ACTIVITY PRESENT IN PURIFIED PREPARATIONS OF DEOXYRIBONUCLEIC ACID POLYMERASE. J Biol Chem. 1964 Jan;239:233–241. [PubMed] [Google Scholar]
- Leavitt M. C., Ito J. T5 DNA polymerase: structural--functional relationships to other DNA polymerases. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4465–4469. doi: 10.1073/pnas.86.12.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljas A., Kannan K. K., Bergstén P. C., Waara I., Fridborg K., Strandberg B., Carlbom U., Järup L., Lövgren S., Petef M. Crystal structure of human carbonic anhydrase C. Nat New Biol. 1972 Feb 2;235(57):131–137. doi: 10.1038/newbio235131a0. [DOI] [PubMed] [Google Scholar]
- Lindskog S., Coleman J. E. The catalytic mechanism of carbonic anhydrase. Proc Natl Acad Sci U S A. 1973 Sep;70(9):2505–2508. doi: 10.1073/pnas.70.9.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. II. Purification of the alpha subunit, devoid of nuclease activities. J Biol Chem. 1985 Oct 25;260(24):12987–12992. [PubMed] [Google Scholar]
- Ollis D. L., Brick P., Hamlin R., Xuong N. G., Steitz T. A. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. 1985 Feb 28-Mar 6Nature. 313(6005):762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Scheuermann R. H., Echols H. A separate editing exonuclease for DNA replication: the epsilon subunit of Escherichia coli DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7747–7751. doi: 10.1073/pnas.81.24.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer E. K., Rush J., Fung C., Reha-Krantz L. J., Karam J. D., Konigsberg W. H. Primary structure of T4 DNA polymerase. Evolutionary relatedness to eucaryotic and other procaryotic DNA polymerases. J Biol Chem. 1988 Jun 5;263(16):7478–7486. [PubMed] [Google Scholar]
- Steitz T. A., Beese L., Freemont P. S., Friedman J. M., Sanderson M. R. Structural studies of Klenow fragment: an enzyme with two active sites. Cold Spring Harb Symp Quant Biol. 1987;52:465–471. doi: 10.1101/sqb.1987.052.01.053. [DOI] [PubMed] [Google Scholar]
- Steitz T. A., Freemont P. S., Ollis D. L., Joyce C. M., Grindley J. M. Functional implications of the Klenow fragment structure. Biochem Soc Trans. 1986 Apr;14(2):205–207. doi: 10.1042/bst0140205. [DOI] [PubMed] [Google Scholar]
- Weis W. I., Brünger A. T., Skehel J. J., Wiley D. C. Refinement of the influenza virus hemagglutinin by simulated annealing. J Mol Biol. 1990 Apr 20;212(4):737–761. doi: 10.1016/0022-2836(90)90234-D. [DOI] [PubMed] [Google Scholar]