Abstract
During spontaneous cell polarization of Dictyostelium discoideum cells, phosphatidylinositol (3,4,5)-triphoshpate (PIP3) and PTEN (phosphatase tensin homolog) have been identified as key signaling molecules which govern the process of polarization in a self-organized manner. Recent experiments have quantified the spatio-temporal dynamics of these signaling components. Surprisingly, it was found that membrane-bound PTEN can be either in a high or low state, that PIP3 waves were initiated in areas lacking PTEN through an excitable mechanism, and that PIP3 was degraded even though the PTEN concentration remained low. Here we develop a reaction-diffusion model that aims to explain these experimental findings. Our model contains bistable dynamics for PTEN, excitable dynamics for PIP3, and postulates the existence of two species of PTEN with different dephosphorylation rates. We show that our model is able to produce results that are in good qualitative agreement with the experiments, suggesting that our reaction-diffusion model underlies the self-organized spatio-temporal patterns observed in experiments.
Keywords: chemotaxis, polarization, modeling
1. Introduction
Motile eukaryotic cells are often polarized and display a distinct cell morphology in which the cytoskeleton is organized to form a protruding front and a retracting tail. This polarity can occur spontaneously [1] or can be induced by external directional cues, including the presence of chemoattractant gradients [2–4]. Many of the signaling molecules that are involved in polarity formation and the subsequent motility have been identified through careful and quantitative experiments [5]. In the polarized state, these components are often localized to either the back or the front of the cell, making it possible to distinguish these parts of the cell through the use of fluorescent markers. These experiments have also revealed that many components are shared among different organisms. Furthermore, experiments in which cells are perturbed in carefully controlled fashion, by changing the external chemoattractant concentration, are now being used to determine the wiring of the intracellular pathways that underly cell motility and polarity [6, 7]. These experimental studies have been used in theoretical work to couple signaling to cell motility, resulting in reaction-diffusion models for directional sensing and migration [8–16].
The mechanisms responsible for the establishment of cell polarity are not well understood. It is clear, however, that this establishment must be a dynamic process since cells are typically able to reverse their polarity in response to changing directional cues [17]. Recent experiments have shown that reaction-diffusion waves of specific signaling components might be involved in polarity formation. These self-organized waves were observed in neutrophils where actin-based waves of Hem-1, a constituent of the SCAR/WAVE complex regulating Arp2/3 activity, were found to be present on the cell surface closest to the substrate [18]. The waves move toward the front of the cells where they are involved in leading edge protrusions [19, 20]. Self-organized actin-based waves were also found in fibroblasts, fibrosarcoma cells and in mouse melanoma cells [21, 22].
Perhaps the most studied cell-type exhibiting self-organized waves of signaling components is the amoeboid Dictyostelium discoideum (D. discoideum). In these cells, actin waves are observed on the substrate-attached surface of randomly moving cells [21, 23–26]. Furthermore, the actin dynamics was found to be correlated with waves of the upstream signaling component PIP3 (phosphatidylinositol (3,4,5)tris-phosphate). PIP3 is synthesized by the kinase PI3K from PIP2 (phosphatidylinositol (4,5) bis-phosphate) and degraded back to PIP2 by PTEN (phosphatase tensin homolog) [27–29]. The latter is also known as a tumor suppressor in various eukaryotic cell lines as it controls the PIP3-dependent F-actin polymerization by down regulating PIP3 [30]. PIP3 is enriched at the front of migrating cells [31] where it increases the polymerization of actin [32] and its pathway has been linked to chemotaxis [33, 34].
A recent study by Gerisch and coworkers carefully quantified the dynamics of F-actin, PIP3 and PTEN waves in D. discoideum cells [35]. They observed that substrate-attached cell membranes in the resting state exhibit a very low concentration of PIP3 and a high concentration of PTEN. This high PTEN concentration is occasionally and spontaneously reduced in small regions of the membrane, resulting in spatial domains with a markedly lower PTEN concentration. These domains, termed ‘holes’ by Gerisch et al, were often transient, resulting in a rapid return to the high, uniform PTEN state of the membrane. Occasionally, however, the PIP3 concentration within these holes increased, resulting in a propagating wave of both PIP3 and PTEN that covered the entire substrate-attached cell body. The PIP3 dynamics was found to be consistent with excitable dynamics: a PIP3 wave was only triggered if the concentration reached a threshold, after which the PIP3 signal was strongly amplified, followed by a slow degradation. Importantly, PTEN declined without any PIP3 increase while, following the PIP3 wave creation, PIP3 decreased and returned to its basal level without an increase in PTEN. Thus, the PIP3 and PTEN dynamics cannot be explained by simple cross-inhibition between the two components. Additionally, the experiments showed that once the PIP3 level was back to its basal level, a PTEN wave reentered from the cell perimeter, resulting in a fast return to the original resting state characterized by low PIP3 and high PTEN levels. This lateral wave was observed to be initiated at a location that was closest to the site of nucleation of the PIP3 wave. Thus, the PTEN concentration switches between a low state, which allows the formation of a PIP3 wave, and a high state. A representative sample of the experimental data is shown in the left panel of figure 1(A), which shows seven snapshots of the PTEN and PIP3 dynamics visualized by fluorescent markers.
Figure 1.
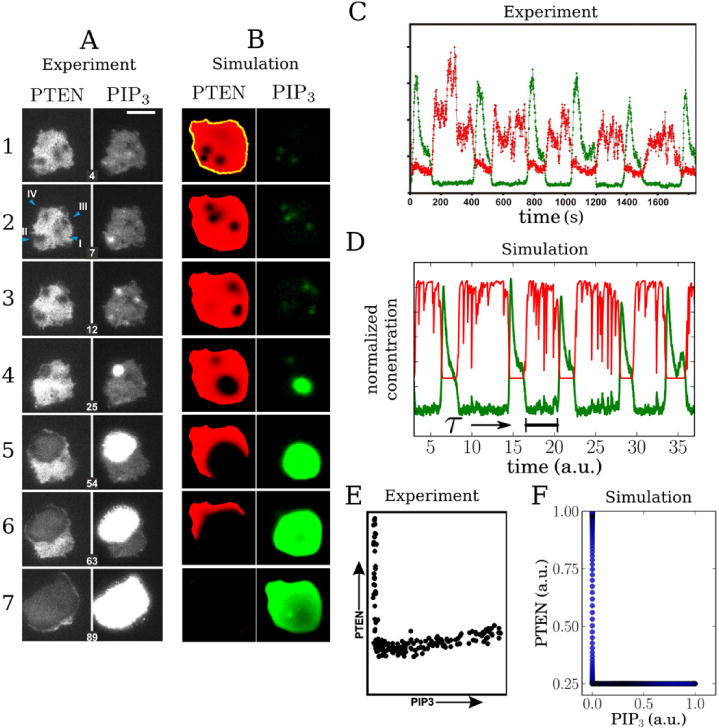
Comparison of experimental results obtained by Gerisch et al [35] (A, C, and E) and captured by our simulations (B, D, and F). (A) Polarization in Dictyostelium discoideum cells labeled with PTEN and PIP3 fluorescent markers and measured using TIRF microscopy. An incomplete polarization cycle is shown (without PTEN reentry). The first three panels from top to bottom for each marker exhibit transient hole formations of PTEN with transient clustering of PIP3. The 4th row shows the nucleation of a PIP3 wave inside a PTEN hole. The remaining panels show the propagation of the PIP3 and PTEN wave. Time (in min) is indicated by the number in each panel set. (B) Simulated polarization cycle of PIP3 (green) and PTEN (red) following the model presented in this paper and using the parameter values of supplementary table 1. The cell morphology is indicated by the yellow dashed line and was taken from an experiment. (C) Local time traces of the normalized PTEN (red) and PIP3 (green) concentration as observed in the experiments [35]. These traces were computed by averaging a 3×3 pixel array at an arbitrary position on the membrane. (D) Corresponding local time traces as determined in our simulations with PTEN in red and PIP3 in green. The quantity τ represents the average minimum time delay between the return of PIP3 to basal levels and a novel excitation. (E) Experimental trajectory of the fluorescence intensity in the PIP3/PTEN phase plane obtained at the location of a hole. (F) Corresponding phase trajectory obtained in our simulation.
The experiments of Gerisch and coworkers were performed in D. discoideum cells that are recovering from the actin depolymerization drug Latrunculin A. Similar wave behavior was also reported in non-treated cells during growth and the early starvation stage [20] while excitable PIP3 enriched domains have been found in Latrunculin treated cells [36]. We should also note that excitable behavior appears not to be limited to PIP3: recent experiments in D. discoideum have revealed that other components also exhibit excitability [37]. Thus, excitable dynamics and self-organizing waves appear to be present under general conditions and it has been suggested that they play a role in polarity formation and might be related to membrane pearling [38].
The new experimental results raise several important questions that can not be addressed with current models in which PIP3 is degraded by PTEN through a simple cross-inhibition scheme [14, 20].
What is the mechanism for the switch-like behavior of the membrane-bound PTEN concentration?
What causes the slow degradation of PIP3 within the hole where the PTEN concentration remains low?
Why does PTEN always reenter from a location at the cell perimeter that is closest to the original PIP3 wave nucleation point?
The aim of this paper is to address these questions using a mathematical reaction-diffusion model that can provide further insight into the role of self-organized wave in cell polarity and movement.
2. Results and discussion
2.1. Model
The reaction scheme of our model is shown in figure 2 and focuses on the interplay between membrane-bound PTEN and PIP3. We do not include a detailed description of downstream actin events, as carried out in earlier numerical work [39], since the questions we aim to address are independent of the dynamics of the actin waves. Furthermore, as the PIP2 concentration on the membrane significantly exceeds the PIP3 concentration [40], the depletion of PIP2 cannot be a limiting factor for the PIP3 dephosphorylation. Thus, it is unlikely that PIP2 plays a role in the localization of PIP3 and we do not consider the PIP2 concentration in this model. Further evidence for this simplification is given by the experiments of Gerisch et al [41], where the direct labeling of PIP2 during a self-organized wave only shows a weak change in concentration.
Figure 2.
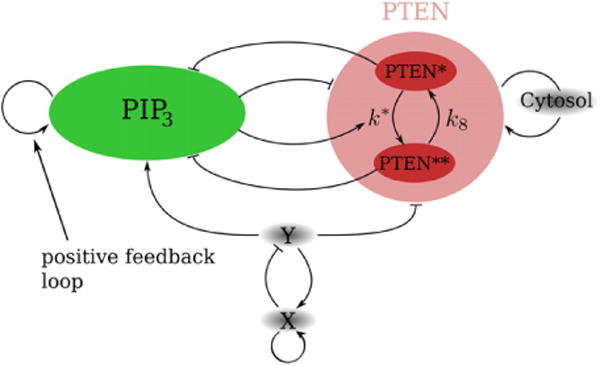
Schematic representation of the reaction-diffusion model consisting of membrane-bound and diffusible PIP3 and PTEN. PTEN is assumed to exist in two different forms, PTEN* and PTEN**, with different dephosphorylation rates. Furthermore, PIP3 and PTEN exhibit a positive feedback loop, resulting in excitable and bistable dynamics, respectively. The substances X and Y prime the formation of holes in the PTEN layer through a stochastic, excitable reaction-diffusion mechanism. Finally, PTEN can detach from the membrane in a PIP3-dependent fashion and can quickly reattach from a constant cytosolic PTEN pool using a positive feedback loop, which is activated by PTEN diffusing back from the non-attached membrane.
In our model, we represent the cell membrane as a 2D sheet on which the model components PTEN and PIP3 can diffuse. PIP3 is degraded by PTEN, as further detailed below, while PTEN detaches from the membrane in a PIP3-dependent fashion as was proposed before [25] and can be reattached from a constant and uniform PTEN cytosolic pool. The boundary conditions for this sheet are implemented using a stationary phase-field method, allowing us to examine complex boundaries [42]. The results presented here are derived from the non-dimensional form of the reaction-diffusion equations which are detailed in the supplementary data, along with their parameter values. We will now address the essential ingredients of the model that enable us to answer the three questions posed above.
2.1.1. What is the mechanism for the switch-like behavior of the membrane-bound PTEN concentration?
The experimental data suggest that PTEN exhibits two stable states, one with a low concentration on the membrane (corresponding to holes) and one with a much higher membrane-bound concentration (before the formation of holes and corresponding to the resting state). To capture this observation, we assume a coupling between PTEN and PIP3 such that the stable fixed points of the PTEN equation are qualitatively different for different PIP3 concentrations. Specifically, we assume that for low values of PIP3 the PTEN dynamics is described by a bistable equation. This nonlinear equation includes a positive feedback, consistent with experimental findings [43], and a nonlinear interaction between PIP3 and PTEN (supplementary data). The phase diagram of the PTEN equation is shown in figure 3(A) where the blue line corresponds to the kinetics for PIP3 = 0, demonstrating that PTEN can be either in a low (P0) or high (P2) state. The intermediate value P1 constitutes a threshold, indicated by the dashed gray line, such that initial values smaller than this threshold will flow to P0 while higher values will be attracted to the P2 state. For higher values of PIP3, however, we assume that the PTEN equation only admits the single low (P0) solution (red and green lines in figure 3(A)).
Figure 3.
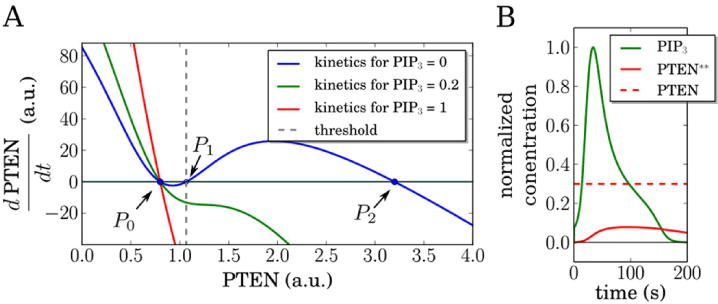
Dynamics of the proposed model. (A) Local kinetics (neglecting diffusion and noise) of PTEN for different constant PIP3 values. If the PIP3 concentration is low (blue curve), PTEN will show bistability corresponding two stable fixed points (P0 = 0.8 and P2 = 3.2) and an unstable one (P1 = 1.1). For higher values of PIP3 only the lower stable fixed point P0 remains. Remaining parameter values as in supplementary table 1. (B) Local normalized concentration profiles (neglecting diffusion and noise) of PIP3 and PTEN** for constant concentration of PTEN. PTEN* can be calculated from PTEN* = PTEN − PTEN**. Parameter values in (A) and (B) are given in supplementary table 1. PIP3 is normalized to its maximum value while PTEN and PTEN* are normalized by P0.
The transition between the high and low state corresponds to crossing the threshold and results in the creation of a hole. In the experiments, these holes are formed by the amplification of negative fluctuations in the PTEN concentration. Since the experiments show that PTEN holes are formed when PIP3 levels are low, they cannot be due to cross-inhibiting activity of PIP3. The precise biological mechanism for the negative fluctuations are unclear but might involve an autocatalytic Ser/Thr-kinase that phosphorylates PTEN [35]. Another possibility is that spontaneous polymerization of small F-actin spots results in detachment of PTEN molecules from the membrane. The latter scenario is supported by the experimental observation that the concentration of F-actin is elevated inside PTEN holes [35] and that the PIP3 nucleation rate is significantly decreased by inhibition of F-actin [20]. Since the purpose of our model is to be as simple as possible while still capturing the most salient features of the biological system, we take a generic noisy excitable system [44] to be responsible for the transient and finite-size hole formation. A detailed formulation of this system as well as its coupling to the PIP3/PTEN dynamics is given in the supplementary data.
2.1.2 What causes the dephosphorylation of PIP3 even though the PTEN concentration is low?
As Gerisch and coworkers have shown, the dynamics of PIP3 and PTEN are clearly coupled since the excitable PIP3 waves are only initiated in PTEN holes. These experiments also revealed, however, that a simple cross-inhibiting scheme where PTEN degrades PIP3 through first-order kinetics, as proposed in existing models [20, 25], is not sufficient to explain the experimental observations. Instead, a mechanism is required that degrades PIP3 while the overall detectable PTEN level stays low or even declines. This mechanism might involve PIP3 degrading enzymes other than PTEN with production rates that are controlled by PTEN levels or the inhibition of PI3K through an unknown component. Rather than introducing a new variable, we propose that the overall PTEN concentration detected in the experiments is composed of two PTEN species hereinafter named PTEN* and PTEN**, with PTEN** exhibiting a much higher dephosphorylation rate than PTEN*. We assume that PTEN* molecules can undergo a state transition to PTEN** while bound to PIP3, resulting in fast PIP3 degradation. The fact that membrane-bound PTEN molecules can undergo state transitions was recently shown using single molecule tracking techniques [45]. Once the PIP3 dephosphorylation to PIP2 is completed, PTEN** can either be converted back to PTEN* or can bind with a higher binding affinity than PTEN* to another PIP3 molecule (supplementary data). We should note that our model is the simplest implementation of multiple PTEN species and that it is possible to formulate a more complicated model with more than two PTEN states.
The excitability of PIP3 is incorporated into the model through a nonlinear positive feedback and a threshold of PIP3 activation, as is standard for excitable systems (see supplementary figure 1 for the PIP3–PTEN** phase space). We include a stochastic term which leads to PIP3 levels that occasionally rise above this threshold value. This, in turn, results in large bursts of PIP3 activity, followed by a gradual decline due to PTEN**-mediated dephosphorylation. To illustrate our reaction scheme we plot in figure 3(B) the time trace of the PIP3 concentration in a simplified version of the model in which the components do not diffuse and in which the PTEN concentration is constant (dashed line). Following the noise-induced excitation, PIP3 increases rapidly, reaches a peak, and subsequently decays through the action of PTEN** which has a much higher degradation rate than PTEN*. The time course of PTEN** is also shown in the figure; it exhibits low values before the PIP3 rise and increases following the PIP3 excitation. The details of the ensuing PIP3 decay dynamics agree well with the experiments as is further detailed in supplementary data and supplementary figure 2. Simulations revealed that the ratio of degradation rates of PTEN** and PTEN* cannot be too large nor too small to account for the experimental PIP3 profiles (see supplementary figure 2). A qualitative comparison between the profiles resulted in a a good matching profile for a ratio of 90.
2.1.3. Why does PTEN always reenter from a location at the cell perimeter that is closest to the original PIP3 wave nucleation point?
Experiments show that once a PIP3 wave is initiated inside a PTEN hole it sweeps over the entire substrate-attached surface of the cell [35]. The global nature of this wave can be recapitulated in our model through the inclusion of a PIP3-dependent detachment rate of membrane-bound PTEN. The initiation of a PIP3 wave then results in higher PIP3 concentrations and increased detachment of membrane-bound PTEN, allowing a hole to expand. Following this global event, experiments also show that a wave of high PTEN enters the cell-substrate domain from the boundary at a location that is closest to the initial PIP3 wave nucleation [35]. Following additional experimental results [41], we assume that PTEN is always in the high state on the cell membrane that is not attached to the substrate, even when a PIP3 wave sweeps over the substrate-attached membrane (see figure 4(C)). In other words, at the edge of the surface-attached membrane, and thus at the boundary of our simulation domain, the concentration of PTEN is high. Therefore, we propose that the non-attached membrane functions as a reservoir for membrane-bound PTEN from which it can diffuse back to the surface attached membrane. Incorporating this into a boundary condition, which also includes a positive feedback [43] as detailed in supplementary data, we find that the P0 to P2 transition is always initiated at the edge of the domain. Furthermore, since the PIP3 concentration is lowest at the boundary closest to the original wave nucleation site, the lateral PTEN reentry wave will be initiated from a location close to the original PIP3 nucleation site.
Figure 4.
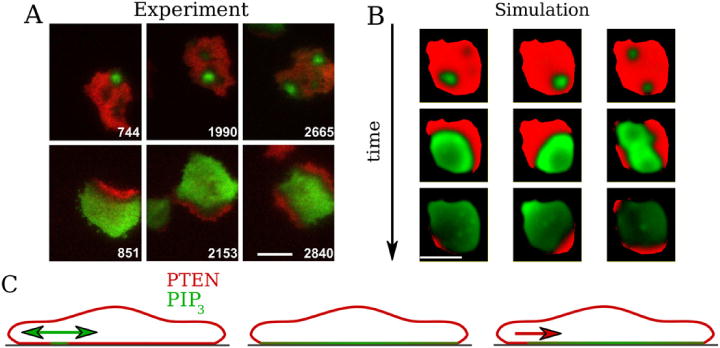
(A) Merged fluorescent images of PIP3 (green) and PTEN (red) measured by Gerisch et al [35], illustrating the coordinated reentry of PTEN. The upper panels show the PIP3 nucleation spots and the corresponding lower panels represent the PTEN reentry sides at a later time. PTEN always reenters from the location on the cell perimeter which is closest to the PIP3 nucleation spot. Time is seconds and scale bar represents 10 μm. (B) Snapshots of PTEN reentry (PIP3 (green) and PTEN (red)) according to our simulations (bar 10 μm). (C) Schematic sketch of PTEN/PIP3 distributions on the membrane postulated in our model and based on experimental results. It illustrates (from left to right) hole formation, PIP3 wave propagation and coordinated reentry of PTEN. The membrane not attached to the surface continuously exhibits high levels of PTEN even when the surface-attached PTEN membrane level is low and, thus, acts as a reservoir for membrane-bound PTEN.
2.2. Full model results
In figure 1(B) we have plotted snapshots of our simulation results, using a typical cell morphology, for seven different times. For comparison, the computational domain in this simulation was taken to be identical to the cell-substrate area of an experimental cell [35]. The left panel of the figure shows the PTEN concentration in red while the right panel shows the PIP3 concentration in green. As in the experiments, the PTEN coverage of the membrane is not uniform but displays holes in which the PTEN concentration is markedly lower (corresponding to the black regions in the figure). These holes are created in a stochastic fashion through an excitable system (supplementary data). Once the PIP3 concentration within such a hole crosses a threshold for activation, a PIP3 wave ensues, shown as a green expanding region. The agreement with the experimental picture, shown in figure 1(A), is remarkably good. A complete sequence of events in our model is presented in supplementary movie 1.
A further comparison between experiments and simulations is presented in figure 1(D) where we have plotted the ‘local’ time series of PIP3 and PTEN in the simulations. These concentrations were determined by taking the spatial average of a small, square membrane region (3 × 3 grid points) within our simulation domain. The corresponding experimental results are shown in figure 1 and were determined by taking the spatial average of 122 pixels in the middle of the surface area [35]. As is the case for the corresponding experimental results, the numerical PTEN concentration (red line) is fluctuating around its high stable level. Occasionally, and due to noise, PTEN within part or the entire averaging domain crosses its threshold value and drops to the low stable state. The local average PIP3 level, shown as a green line, shows fluctuations and can cross an excitation threshold once the PTEN level drops to its low value. Consistent with the experimental data, this only happens sporadically. When it happens, however, it quickly reaches a maximum value, spreads to all points within the averaging domain, and subsequently decays while the PTEN level remains low (supplementary movie 1). By matching the minimum delay τ between two activation cycles in the simulations with the experimentally obtained results, estimated to be 180 s, we can determine the temporal scaling of our model equations (see supplementary data).
The local concentrations of the model components can also be examined in the PIP3/PTEN phase plane. The simulated phase trajectories for the PIP3 and PTEN levels on the membrane-attached surface are shown in figure 1(F). For clarity, this trajectory is shown for the deterministic version of our model. Figure 1(F) was obtained using initial conditions that correspond to the conditions inside a hole. Specifically, a hole was initiated by setting the PTEN value to P0 on a 10 × 10 square grid. After several time steps, a wave was initiated by assigning PIP3 on the 10 × 10 grid to a value above the threshold. Following this wave excitation the PIP3 level increases and subsequently decreases while the PTEN level remains low. Finally, the PTEN level rises again when the PIP3 level is low resulting in an angular trajectory. This trajectory is consistent with the experimentally obtained phase trajectory shown in figure 1(E).
A final comparison between our simulation results and experimental data is shown in figures 4(A) and (B) and illustrates the lateral reentry of PTEN. In (B) we have plotted snapshots of the PTEN and PIP3 distributions for three different simulations. The upper row corresponds to a time point right after the initiation of a PIP3 wave inside a hole. The three columns in (A) show the PTEN (red) and PIP3 (green) fluorescent intensity during PIP3 wave initiation (upper row) and during lateral PTEN wave reentry (bottom row). Since the formation of a hole is a stochastic event, some simulations and experiments displayed two holes, along with two PIP3 waves (right panels in (A) and (B)). The middle row of figure 4(B) shows a time frame where the PIP3 wave covers almost the entire cell membrane. Note the spatial gradient in PIP3 level with a marked decrease at the initiation site. The bottom row shows the onset of the lateral PTEN wave reentry and shows that this always occurs at a site close to the location of the initial PTEN hole, consistent with experimental observations.
3. Conclusions
In this study we have developed a reaction-diffusion model for the dynamics of PTEN and PIP3 at the substrate-attached membrane in D. discoideum cells. Similar self-organized waves are present in a wide variety of cell types and are thought to be involved in cell polarity and cell movement and have been proposed to underly phagocytosis [46]. Crucial in the dynamics of these waves is their excitability, a feature shared with many other biological systems. Excitability results in a threshold, refractoriness, and a recovery phase and can, combined with noise, trigger waves that cover the entire domain.
Our model assumes that the initiation of both the PTEN holes and the PIP3 wave inside these holes are stochastic and excitable events and contains three crucial mechanisms. First, our model includes a mechanism that is able to degrade PIP3 even though the PTEN level remains low. Such a mechanism is required since experiments have shown that the PTEN level remains constant and low throughout the PIP3 degradation phase (figure 1(C)). In our model, we assume that PTEN can exist in two different forms with unequal degradation capabilities. By assuming appropriate conversion kinetics between the two forms the model is able to degrade PIP3 while keeping the overall level of PTEN low (figures 1(D) and 3(B)). Interestingly, we find that the ratio of degradation rates needs to be large (supplementary figure 2). This suggests that alternative mechanisms, for example different PIP3 degrading enzymes or shutting off of PI3K, require components that have large degradation rates. Second, our model captures the experimentally observed abrupt transition from high to low PTEN levels (and vice versa) by incorporating bistable dynamics for PTEN. This bistable dynamics, coupled to PTEN fluctuations, naturally leads to rapid conversion from a high to a low PTEN state during the hole formation and from low to high PTEN values during the lateral reentry wave. Third, the location of these PTEN reentry waves are determined by the experimentally motivated boundary condition of high PTEN levels.
Together, these three mechanisms result in PTEN and PIP3 dynamics that is schematically shown in figure 4(C). Stochastic fluctuations can lead to regions of depressed PTEN levels on the cell surface closest to the substrate. Within these regions, PIP3 waves can be initiated through an excitatory mechanism, resulting in PTEN and PIP3 waves that sweep over the entire substrate-attached membrane. The membrane returns to its initial state through the lateral entry of a PTEN wave, initiated by the high values of PTEN concentration on the membrane that is not attached to the substrate. These three mechanisms result in PIP3/PTEN dynamics that agrees well with the experimental findings (figures 1(B), (D), (F)) and 4(A)). Our model does not yet address the coupling of actin dynamics to PTEN and PIP3 waves. Future work should include this coupling and should investigate how the waves determine cell morphology.
Supplementary Material
Acknowledgments
We would like to thank A Bae, G Gerisch and W F Loomis for helpful suggestions and fruitful discussions. This work was supported by the National Institutes of Health (P01 GM078586) and the SFB 937 Collective behavior of soft and biological matter.
Footnotes
Online supplementary data available from stacks.iop.org/pb/11/046002/mmedia
References
- 1.Altschuler SJ, Angenent SB, Wang Y, Wu LF. On the spontaneous emergence of cell polarity. Nature. 2008;454:886–9. doi: 10.1038/nature07119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 3.Parent CA, Devreotes PN. A cell’s sense of direction. Science. 1999;284:765–70. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 4.Rappel WJ, Loomis WF. Eukaryotic chemotaxis. Wiley Interdiscip Rev Syst Biol Med. 2009;1:141–9. doi: 10.1002/wsbm.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swaney KF, Huang CH, Devreotes PN. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–89. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang CJ, Bergmann A, Lin B, Kim K, Levchenko A. Diverse sensitivity thresholds in dynamic signaling responses by social amoebae. Sci Signal. 2012;5:ra17. doi: 10.1126/scisignal.2002449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeda K, Shao D, Adler M, Charest PG, Loomis WF, Levine H, Groisman A, Rappel WJ, Firtel RA. Incoherent feedforward control governs adaptation of activated ras in a eukaryotic chemotaxis pathway. Sci Signal. 2012;5:ra2. doi: 10.1126/scisignal.2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meinhardt H. Orientation of chemotactic cells and growth cones: models and mechanisms. J Cell Sci. 1999;112:2867. doi: 10.1242/jcs.112.17.2867. [DOI] [PubMed] [Google Scholar]
- 9.Narang A, Subramanian KK, Lauffenburger DA. A mathematical model for chemoattractant gradient sensing based on receptor-regulated membrane phospholipid signaling dynamics. Ann Biomed Eng. 2001;29:677–91. doi: 10.1114/1.1385805. [DOI] [PubMed] [Google Scholar]
- 10.Gamba A, de Candia A, di Talia S, Coniglio A, Bussolino F, Serini G. Diffusion-limited phase separation in eukaryotic chemotaxis. Proc Natl Acad Sci. 2005;102:16927–32. doi: 10.1073/pnas.0503974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine H, Kessler DA, Rappel WJ. Directional sensing in eukaryotic chemotaxis: a balanced inactivation model. Proc Natl Acad Sci USA. 2006;103:9761–6. doi: 10.1073/pnas.0601302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beta C, Amselem G, Bodenschatz E. A bistable mechanism for directional sensing. New J Phys. 2008;10:083015. [Google Scholar]
- 13.Xiong Y, Huang CH, Iglesias PA, Devreotes PN. Cells navigate with a local-excitation, global-inhibition-biased excitable network. Proc Natl Acad Sci USA. 2010;107:17079–86. doi: 10.1073/pnas.1011271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibata T, Nishikawa M, Matsuoka S, Ueda M. Modeling the self-organized phosphatidylinositol lipid signaling system in chemotactic cells using quantitative image analysis. J Cell Sci. 2012;125:5138–50. doi: 10.1242/jcs.108373. [DOI] [PubMed] [Google Scholar]
- 15.Shi C, Huang CH, Devreotes PN, Iglesias PA. Interaction of motility, directional sensing, and polarity modules recreates the behaviors of chemotaxing cells. PLoS Comput Biol. 2013;9:e1003122. doi: 10.1371/journal.pcbi.1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibata T, Nishikawa M, Matsuoka S, Ueda M. Intracellular encoding of spatiotemporal guidance cues in a self-organizing signaling system for chemotaxis in. Dictyostelium cells Biophys J. 2013;105:2199–209. doi: 10.1016/j.bpj.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meier B, Zielinski A, Weber C, Arcizet D, Youssef S, Franosch T, Radler JO, Heinrich D. Chemotactic cell trapping in controlled alternating gradient fields. Proc Natl Acad Sci USA. 2011;108:11417–22. doi: 10.1073/pnas.1014853108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW. An actin-based wave generator organizes cell motility. PLoS Biol. 2007;5:e221. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroth-Diez B, Gerwig S, Ecke M, Hegerl R, Diez S, Gerisch G. Propagating waves separate two states of actin organization in living cells. HFSP J. 2009;3:412–27. doi: 10.2976/1.3239407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taniguchi D, Ishihara S, Oonuki T, Honda-Kitahara M, Kaneko K, Sawai S. Phase geometries of two-dimensional excitable waves govern self-organized morphodynamics of amoeboid cells. Proc Natl Acad Sci USA. 2013;110:5016–21. doi: 10.1073/pnas.1218025110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vicker MG. Eukaryotic cell locomotion depends on the propagation of self-organized reaction-diffusion waves and oscillations of actin filament assembly. Exp Cell Res. 2002;275:54–66. doi: 10.1006/excr.2001.5466. [DOI] [PubMed] [Google Scholar]
- 22.Kunida K, Matsuda M, Aoki K. FRET imaging and statistical signal processing reveal positive and negative feedback loops regulating the morphology of randomly migrating HT-1080. cells J Cell Sci. 2012;125:2381–92. doi: 10.1242/jcs.096859. [DOI] [PubMed] [Google Scholar]
- 23.Bretschneider T, Diez S, Anderson K, Heuser J, Clarke M, Muller-Taubenberger A, Kohler J, Gerisch G. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr Biol. 2004;14:1–10. doi: 10.1016/j.cub.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Asano Y, Nagasaki A, Uyeda TQ. Correlated waves of actin filaments and PIP3 in Dictyostelium cells. Cell Motil Cytoskeleton. 2008;65:923–34. doi: 10.1002/cm.20314. [DOI] [PubMed] [Google Scholar]
- 25.Arai Y, Shibata T, Matsuoka S, Sato MJ, Yanagida T, Ueda M. Self-organization of the phosphatidylinositol lipids signaling system for random cell migration. Proc Natl Acad Sci USA. 2010;107:12399–404. doi: 10.1073/pnas.0908278107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brzeska H, Pridham K, Chery G, Titus MA, Korn ED. The association of myosin IB with actin waves in Dictyostelium requires both the plasma membrane-binding site and actin-binding region in the myosin tail. PLoS ONE. 2014;9:e94306. doi: 10.1371/journal.pone.0094306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by pi 3-kinase and pten mediates chemotaxis. Cell. 2002;109:611–23. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 28.Iijima M, Devreotes P. Tumor suppressor pten mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 29.Tang M, Iijima M, Kamimura Y, Cen L, Long Y, Devreotes P. Disruption of PKB signaling restores polarity to cells lacking tumor suppressor. PTEN Mol Biol Cell. 2011;22:437–47. doi: 10.1091/mbc.E10-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vazquez F, Devreotes P. Regulation of PTEN function as a PIP3 gatekeeper through membrane interaction. Cell Cycle. 2006;5:1523–7. doi: 10.4161/cc.5.14.3005. [DOI] [PubMed] [Google Scholar]
- 31.Parent CA. Making all the right moves: chemotaxis in neutrophils and Dictyostelium. Curr Opin Cell Biol. 2004;16:4–13. doi: 10.1016/j.ceb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki AT, et al. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol. 2007;178:185–91. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider IC, Haugh JM. Mechanisms of gradient sensing and chemotaxis: conserved pathways, diverse regulation. Cell Cycle. 2006;5:1130–4. doi: 10.4161/cc.5.11.2770. [DOI] [PubMed] [Google Scholar]
- 34.Stephens L, Milne L, Hawkins P. Moving towards a better understanding of chemotaxis. Curr Biol. 2008;18:R485–494. doi: 10.1016/j.cub.2008.04.048. [DOI] [PubMed] [Google Scholar]
- 35.Gerisch G, Schroth-Diez B, Muller-Taubenberger A, Ecke M. PIP3 waves and PTEN dynamics in the emergence of cell polarity. Biophys J. 2012;103:1170–8. doi: 10.1016/j.bpj.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishikawa M, Horning M, Ueda M, Shibata T. Excitable signal transduction induces both spontaneous and directional cell asymmetries in the phosphatidylinositol lipid signaling system for eukaryotic chemotaxis. Biophys J. 2014;106:723–34. doi: 10.1016/j.bpj.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang CH, Tang M, Shi C, Iglesias PA, Devreotes PN. An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration. Nat Cell Biol. 2013;15:1307–16. doi: 10.1038/ncb2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinrich D, Ecke M, Jasnin M, Engel U, Gerisch G. Reversible membrane Pearling in live cells upon destruction of the actin cortex. Biophys J. 2014;106:1079–91. doi: 10.1016/j.bpj.2013.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khamviwath V, Hu J, Othmer HG. A continuum model of actin waves. Dictyostelium discoideum PLoS ONE. 2013;8:e64272. doi: 10.1371/journal.pone.0064272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leslie NR, Downes CP. PTEN: The down side of PI 3-kinase signalling. Cell Signal. 2002;14:285–95. doi: 10.1016/s0898-6568(01)00234-0. [DOI] [PubMed] [Google Scholar]
- 41.Gerisch G, Ecke M, Wischnewski D, Schroth-Diez B. Different modes of state transitions determine pattern in the phosphatidylinositide-actin system. BMC Cell Biol. 2011;12:42. doi: 10.1186/1471-2121-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kockelkoren J, Levine H, Rappel WJ. Computational approach for modeling intra- and extracellular dynamics. Phys Rev E. 2003;68:037702. doi: 10.1103/PhysRevE.68.037702. [DOI] [PubMed] [Google Scholar]
- 43.Iijima M, Huang YE, Luo HR, Vazquez F, Devreotes PN. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J Biol Chem. 2004;279:16606–13. doi: 10.1074/jbc.M312098200. [DOI] [PubMed] [Google Scholar]
- 44.Hecht I, Kessler DA, Levine H. Transient localized patterns in noise-driven reaction-diffusion systems. Phys Rev Lett. 2010;104:158301. doi: 10.1103/PhysRevLett.104.158301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuoka S, Shibata T, Ueda M. Asymmetric PTEN distribution regulated by spatial heterogeneity in membrane-binding state transitions. PLoS Comput Biol. 2013;9:e1002862. doi: 10.1371/journal.pcbi.1002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerisch G, Ecke M, Schroth-Diez B, Gerwig S, Engel U, Maddera L, Clarke M. Self-organizing actin waves as planar phagocytic cup structures. Cell Adh Migr. 2009;3:373–82. doi: 10.4161/cam.3.4.9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


