Abstract
Background
The aim of this study was to evaluate, the existence of a signature of differentially expressed microRNAs (miRNAs) during osteogenic differentiation of bone marrow MSCs from OA and healthy donors and to describe their possible implication in joint regeneration through modulation of molecular mechanisms involved in homeostatic control in OA pathophysiology.
Methods
Following phenotypic assessment of BM-MSCs obtained from OA diagnosed patients (n = 10) and non-OA (n = 10), total small RNA was isolated after osteogenic induction for 1, 10 and 21 days, miRNA profiles were generated using a commercial expression array of 754 well-characterized miRNAs. MiRNAs, with consistent differential expression were selected for further validation by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis.
Results
A total of 246 miRNAs were differentially expressed (fold change ≥ ± 2, P ≤0.05) between OA and non-OA BM-MSC samples; these miRNAs showed variable interactions depending on the cell and differentiation status. Two miRNAs, hsa-miR-210 and hsa-miR-335-5p out of 21 used for validation showed a significant downregulated expression during induced osteogenesis. In particular hsa-miR-335-5p, a critical regulator in bone homeostasis, was further studied. hsa-miR-335-5p downregulation in OA-MSCs, as well as their host coding gene, MEST, were also assessed.
Conclusions
To our knowledge, this study represents the most comprehensive assessment to date of miRNA expression profiling in BM-MSCs from OA patients and their role during osteogenic differentiation. We describe the existence of a correlation between miR-335-5p expression and OA indicating the putative role of this miRNA in OA features. These findings, may contribute to our understanding of the molecular mechanisms involved in MSCs mediated homeostatic control in OA pathophysiology that could be applicable in future therapeutic approaches.
Electronic supplementary material
The online version of this article (doi:10.1186/s12891-015-0652-9) contains supplementary material, which is available to authorized users.
Background
Osteoarthritis (OA) is a common musculoskeletal disorder with high socioeconomic impact [1]. It is characterized by alterations in bone-cartilage homeostasis leading to progressive degeneration of synovial joints [2]. OA etiology is largely unknown; however it has been associated to susceptibility factors such as age, prior joint injury and other alterations linked to genetic and epigenetic factors [3, 4]. Currently, no cure exists for OA, beyond pain relief. In this context, cell therapies are being studied as an alternative for OA treatment [5]. Therapeutic approaches using MSCs (Mesenchymal Stem in Cells) are considered one of the best options for future treatment of damaged OA cartilage and bone. This approach is based on the potential of MSCs to differentiate into chondrocytes and osteoblasts and their immunomodulatory properties.
Biochemical mechanisms involved in OA initiation and progression are modulated by a fine-tuned crosstalk between different signaling pathways, most of them converging in the WNT signaling pathway [6]. WNT signaling is involved into a range of biological processes essential for maintenance of joint homeostasis, including cell proliferation, cell-fate determination and differentiation [7–9]. In addition to cell signaling, another important regulatory mechanism of cell function is the post-transcriptional modulation of gene expression by microRNAs (miRNAs), a family of small noncoding RNAs. MiRNAs have been described as important players interfering in the expression of multiple target genes and thus modulating their biological functions, e.g. chondrogenesis and osteogenesis [10–12].
Dysfunction of canonical Wnt/β-catenin signaling has been implicated in OA pathogenesis [13]. A number of components including Wnts, frizzled, secreted frizzled-related protein (sFRP), Dickkopf and LRPs (LDL-receptor-related protein) play crucial roles during cartilage and bone development and joint maintenance. Moreover, increased levels of β-catenin have been observed in degenerative cartilage, suggesting that continuous Wnt signaling might contribute to cartilage loss. This evidence was supported by the fact that a polymorphism in the SFRP3 is associated with a reduced ability to limit β-catenin signaling and with an increased susceptibility to OA development [14].
Together, these evidences suggest that deregulation of signaling pathways by miRNAs and their relationship may provide clues to understand the OA pathogenesis. Indeed, Wnt β-catenin signaling pathway has been described to be susceptible of modulation by targeted repression of translation of specific pathway components [15, 16]. For example, increased expression levels of miR-335-5p regulate bone development promoting osteogenic differentiation by downregulating DKK1 and thus activating Wnt signaling [17]. On the other hand, some miRNAs are also specifically expressed under regulation of Wnt/β-catenin in a regulatory feedback loop [18, 19]. Moreover, tissue damage or proinflammatory signals may also cause miR-335 downregulation which in turn activates the proliferative, migratory and differentiation capacities of MSCs [20].
Given that miRNAs are important post-transcriptional regulators of gene expression, variations in miRNA expression levels are likely involved in molecular changes occurring during joint formation and/or remodeling [21]. To describe the role of miRNAs in OA pathophysiology, we aimed to evaluate the existence of a differential expression signature of miRNAs in bone marrow MSCs comparing OA and healthy donors during in vitro induced osteogenesis. This knowledge is essential, both for better understand the regulatory mechanisms taking place in OA pathophysiology and for development of new therapeutic approaches based in selective targeting of key molecules.
Methods
Patients and specimens
Bone marrow aspirates were obtained from patients undergoing total hip arthroplasty of OA patients (n = 10) (71.2 ± 9.2 years) and control subjects (n = 10) with femoral neck fracture (82.0 ± 9.5 years). OA diagnosis was established according to the ACR criteria [22]. Control subjects, evaluated by an independent observer, did not show OA radiographic changes and their clinical records did not evidenced signs of osteoporosis assessed by densitometric T-score > −2.5 SD. All samples were processed after written informed consent was obtained. The study was approved by our institutional ethics committee (CEIC Hospital Clínico San Carlos) according to the principles expressed in the Declaration of Helsinki.
BM-MSCs cell cultures
MSCs were obtained from 5 to 10 ml of bone marrow aspirates. Briefly, each aspirate was diluted 1:1 with Dulbecco’s modified Eagle’s medium (DMEM) (Promocell GmbH, Heilderberg, Germany) and layered over an equal volume of Ficoll. After centrifugation, at 900 × g for 30 min, the mononuclear cell layer was recovered, washed with DMEM, and re-suspended in DMEM supplemented with 10 % fetal bovine serum 100 U/ml penicillin, 100 mg/ml streptomycin, and 2 mM L-glutamine. The cultures were washed to remove the non-adherent cells and further expanded until approximately 80 % confluence. Cells at confluence were detached with 0.25 % trypsin-EDTA for 5 min at 37 °C and replated for continued passaging at dilution 1:2. Cells at the third passage were used for experiments.
BM-MSCs characterization by flow cytometry and lineage multipotential
Phenotypic characterization of isolated MSCs was performed by flow citometry in a Gallios flow cytometer (Beckman Coulter). Cells recovered at the third passage were washed with PBS prior incubation during 30 min at 4 °C with specific phycoerythrin conjugated antibodies and their isotype matched antibodies (all from Miltenyi Biotech). Expression of CD105, CD73 and CD90, and lack of expression of CD45, CD34, and CD14 were the minimal criteria required prior to evaluation of their differentiation potential towards chondrogenic, adipogenic and osteogenic lineages, assessed by histochemical stainings.
Induction of osteogenic, adipogenic and chondrogenic differentiation
Cultures for histochemical analysis were performed in 24-well tissue culture plates at a seeding density of (30,000 cells/cm2) using commercial culture media with supplements according to manufacturer instructions (Promocell Cat# C-28013 for osteogenesis, Cat# C-28012 for chondrogenesis and Cat# 28011 for adipogenesis).
The degree of mineralization in osteogenic cultures was assessed after staining for 20 min with 2 % Alizarin red S (Sigma). Adipogenic lineage commitment was evaluated in adipogenic induced cultures, by staining for 5 min with an Oil red O working solution (3 parts of 0.5 % Oil Red O in isopropanol diluted with 2 parts of distilled water). Chondrogenic potential was evaluated by measuring proteoglycan synthesis in chondrogenic induced cultures. Staining was performed for 20 min with 1 % Alcian Blue. Visualization of cells was performed under phase contrast microscopy (Leica 4000b DMI, Leica. Germany).
Extraction of miRNAs and microarray analysis
Total small RNA was recovered at days 1, 10 and 21 of osteogenically induced MSCs. RNA was solated using the mirVana PARIS kit (Ambion, Grand Island, NY, USA) according to the manufacturer’s instructions. MiRNA quality was assessed using Agilent’s 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Total miRNA profiles were generated using an ABI Taqman OpenArray MicroRNA pools A and B to measure the expression of 754 well-characterized miRNA sequences from the Sanger miRBase v14.
Briefly, total small RNA isolated from cells was divided equally for each sample and used with TaqMan Megaplex RT primer pools A or B to generate cDNA which was subsequently amplified using the corresponding Megaplex PreAmp Primers (pools A or B, respectively) following the manufacturer’s instructions. Real-time PCR was performed on the Taqman Open Array MicroRNA plates using the Applied Biosystem Open Array Real-Time PCR system. Data were further exported for analysis (included in Additional file 1).
Total RNA isolation, cDNA synthesis and RT-PCRs
For miRNA RT-PCR, total RNA was isolated with miRCURY isolation kit (Exiqon) and cDNA Synthesis was carried out using 10 ng of RNA and the Universal cDNA synthesis kit II (Exiqon). Following reverse transcription, cDNAs were 1:100 diluted and loaded onto the real time PCR plates using a 1:1 mixture of Exilent SYBR Green master mix 2× (Exiqon). Real Time PCR reactions were performed in a 7900HT Fast Real time PCR System (Applied Biosystems) under the following conditions: 95 °C 10 min, 45 cycles of 95 °C 10s and 60 °C 1 min. After run completion, Ct values were calculated using the SDS 2.4 software (Applied Biosystems) and data were analyzed using the StatMiner® software (Integromics). The data sets supporting the results of this article are available in the GEO repository, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70856.
Total RNA was isolated from cultured cells with a miRNeasy Mini Kit (Qiagen, Valencia, CA, USA). Human miR-335 and MEST, were quantified by real-time RT-PCR using the corresponding TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA, USA). RNU6B and GAPDH were used as endogenous normalization controls for miRNAs and MEST, respectively.
For PCR amplification of DKK1 and SFRP1, RNA from cultures was pre-amplified using the RT2 Nano PreAMP cDNA Synthesis Kit (Cat# C06, SABiosciences, Qiagen).
cDNA conversion was performed with the Maxima H Minus Kit (Cat#K1682. Fermentas) and the Maxima Sybr/R QPCR kit (Cat# K0221. Fermentas) was used for PCR amplification in a Mastercycler realplex4 (Eppendorf) using the following conditions: 95 °C, 10 min; 40 cycles of (95 °C, 15 s; and 60 °C, 60 s). Forward and Reverse primer sequences used were respectively: ACTB (β-actin), 5′-CGC CCC AGG CAC CAG GCG-3′; 5′-GCT GGG GTG TTG AAG GT-3′. DKK1: 5′-GTG CAA ATC TGT CTC GCC TG-3′; 5′-GCA CAG TCT GAT GAC CGG AG-3′ and SFRP1: 5′-TTT GAG GAG AGC ACC CTA GGC-3′; 5′-TGT GTA TCT GCT GGC AAC AGG-3′. ACTB was used as internal reference gene. Amplification of the relevant amplicon sizes (284, 266 and 75 bp respectively) was confirmed by electrophoresis on a 2 % agarose gel stained with Midori Green (Nippon Genetics Europe GmbH) and visualized under ultraviolet light.
Data analysis and statistical methods
Microarray data preprocessing was done with the R programming language. MiRNAs with Ct values higher than 35 were considered as undetected. Data were normalized using a mean-centering restricted (MCR) strategy as described by Wylie et al. [23], which uses miRNAs expressed in all samples for data normalization. The mean of these fully expressed miRNAs in a given sample is subtracted from all miRNAs in that sample. After normalization, statistical analysis was performed via custom scripts based on the R/Bioconductor package LIMMA (Linear Models for Microarray) [24]. Comparisons between experimental groups were performed using a moderated t-test from LIMMA and P-values were adjusted for multiple testing using the Benjamini-Hochberg method [25]. Adjusted P-values < 0.05 were considered statistically significant. Unless specified, median and interquartile rank (IQR) values were calculated. P values less than 0.05 were considered significant. Data were analyzed using Graph-Pad Prism 6.01 (GraphPad Software, San Diego, CA).
Results
Characterization of bone marrow MSCs
The phenotypic uniformity of BM-MSCs used in the study was assessed according to several minimal criteria: these include the plastic-adherence in culture, positivity for expression of CD90, CD73, CD105 and absence of hematopoietic surface markers CD34, CD45 and CD14 expression and their osteogenic and chondrogenic lineage potential. The expression pattern of the cells used was similar in all samples (Fig. 1a and b) In addition, the multilineage differentiation potential of MSCs used in the study was evaluated. All samples were able to differentiate into osteogenic, chondrogenic and adipogenic lineages under the appropriate stimulatory conditions (Fig. 1a and b).
Fig. 1.
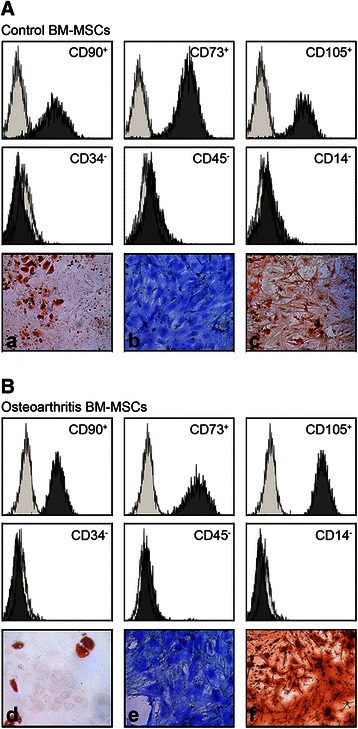
Characterization of BM-MSCs by Flow Citometry and in vitro potential of differentiation. Representative flow Cytometry analysis of bone marrow MSCs obtained from one Control (non-OA) donor (a). and one osteoarthritic patient (b). MSCs at third passage were positive for MSC specific markers (CD90, CD73, CD105 and negative for CD34, CD45 and CD14). Figure represents an overlay image of antibody isotype controls (light gray histogram) and specific marker antibodies (dark gray histogram). Histochemical staining of In vitro potential of differentiation through the adipogenic, chondrogenic and osteogenic lineages measured at day 21 (adipogenesis and osteogenesis) or 28 (chondrogenesis) for Control BM-MSCs (a, b and c respectively) and OA BM-MSCs (d, e and f). Magnification × 100
MiRNA expression profiles during osteogenic induction using microarray
To address the hypothesis that OA- or Control-MSCs have a different epigenetic signature of miRNAs during osteogenic differentiation. Expression levels of 754 miRNAs were assessed using miRNA microarray expression chips. Raw data were preprocessed and filtered using R routines. A total of 246 miRNAs with at least two observations for each combination of status (OA or Control) and differentiation days (t = 0, 10, 21) were further processed. The variability introduced by single individuals was detected using statistical random effects models. Given that no significant effect was detected, data were finally analyzed using fixed effect models.
Combinations of variables which best explained the data were detected after a linear discriminant analysis (LDA). Four miRNAs; hsa-miR-197, hsa-miR-320, hsa-miR-616 and hsa-miR-99b_ showed differences during differentiation indicating the existence of an interaction between status and differentiation variables (Additional file 2). On the other hand, 15 miRNAs did not show any interaction (that is, were independent of cell status) but showed differences during osteogenic progression (Additional file 3). Similarly, 30 miRNAs were independent during differentiation times analyzed but showed differences between OA and control samples (Additional file 4). However, although all the differences noted were statistically significant after multiple test correction this significance was lost. The data set supporting these results are included in Additional file 1.
MiRNA validation by quantitative PCR
A set of miRNAs with consistent differential expression in our microarray was further validated by q-PCR. This included a total of 21 miRNAs selected according to their lower q values (Table 1). Distribution of miRNAs analyzed and their overlapping among different comparisons are provided in Additional file 5.
Table 1.
miRNAs with consistent differential expression in the microarray and their target sequences, validated by q-PCR. This included a total of 21 miRNAs selected according to their lower q values
| microRNA name | Target sequence |
|---|---|
| hsa-miR-103a-3p | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-134 | UGUGACUGGUUGACCAGAGGGG |
| hsa-miR-181a-5p | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-191-5p | CAACGGAAUCCCAAAAGCAGCUG |
| hsa-miR-193b-3p | AACUGGCCCUCAAAGUCCCGCU |
| hsa-miR-197-3p | UUCACCACCUUCUCCACCCAGC |
| hsa-miR-210 | CUGUGCGUGUGACAGCGGCUGA |
| hsa-miR-222-3p | AGCUACAUCUGGCUACUGGGU |
| hsa-miR-222-5p | CUCAGUAGCCAGUGUAGAUCCU |
| hsa-miR-24-2-5p | UGCCUACUGAGCUGAAACACAG |
| hsa-miR-27b-3p | UUCACAGUGGCUAAGUUCUGC |
| hsa-miR-296-5p | AGGGCCCCCCCUCAAUCCUGU |
| hsa-miR-30c-5p | UGUAAACAUCCUACACUCUCAGC |
| hsa-miR-335-5p | UCAAGAGCAAUAACGAAAAAUGU |
| hsa-miR-370 | GCCUGCUGGGGUGGAACCUGGU |
| hsa-miR-379-3p | UAUGUAACAUGGUCCACUAACU |
| hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| hsa-miR-433 | AUCAUGAUGGGCUCCUCGGUGU |
| hsa-miR-497-5p | CAGCAGCACACUGUGGUUUGU |
| hsa-miR-539-5p | GGAGAAAUUAUCCUUGGUGUGU |
| hsa-miR-543 | AAACAUUCGCGGUGCACUUCUU |
| hsa-miR-628-3p | UCUAGUAAGAGUGGCAGUCGA |
| hsa-miR-628-5p | AUGCUGACAUAUUUACUAGAGG |
Two miRNAs, hsa-miR-210 and hsa-miR-335-5p out of the 21 used for validation showed a significative downregulated expression during osteogenesis. More specifically, hsa-miR-210 expression pattern was quite similar in OA and control MSCs, which is reflected by an average downregulation of 5.5 times between days [0–10] and 9.5 times between days [0–21]. A downregulatory trend also occurs during osteogenesis for miR-335-5p, which was down-regulated an average of 2 and 12 fold respectively, although not significant in the case of OA-MSCs (Fig. 2).
Fig. 2.
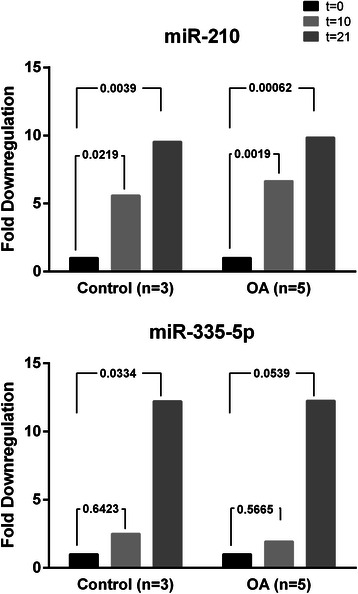
Relative Quantification of comparisons between t = 10 and t = 0; t = 21 and t = 0 of significant miRNAs in OA-MSCs and Control-MSCs. Expression of miR210 and miR-335-5p follow a similar downregulation trend during osteogenic differentiation of MSCs, both in OA (n = 5) or in Control subjects (n = 3). Project was normalised using: hsa-miR-103a-3p; hsa-miR-191-5p; hsa-miR-30c-5p; SNORD49A. Test Type used: Parametric test (Limma). FDR method: Benjamini-Hochberg Adjusted p-value threshold: 0.05
BM-MSCs from OA patients express lower levels of miR-335 than control subjects
Current findings have described miR-335 as a critical regulator in bone homeostasis and MSCs commitment. Besides, it has also been described that expression levels of mature miR-335 in human MSCs correlate with those of its host coding-gene MEST (mesoderm-specific transcript homolog). Therefore, we aimed to determine whether expression of miR-335-5p correlated with OA status measuring the expression of miR-335-5p and MEST in undifferentiated BM-MSC populations from OA and control subjects.
Our results showed lower expression levels of miR-335 in BM-MSCs obtained from OA donors (Fig. 3a). Additionally, although a positive correlation between miR-335-5p and MEST expression was detected (Spearman correlation coefficient, r = 0.59; p = 0.0961 in OA and r = 0.6; p = 0.0968 in controls) MEST expression was similar between OA and control MSCs (Fig. 3b). These results clearly indicated that downregulation of miR-335-5p was an inherent feature of OA-MSCs.
Fig. 3.
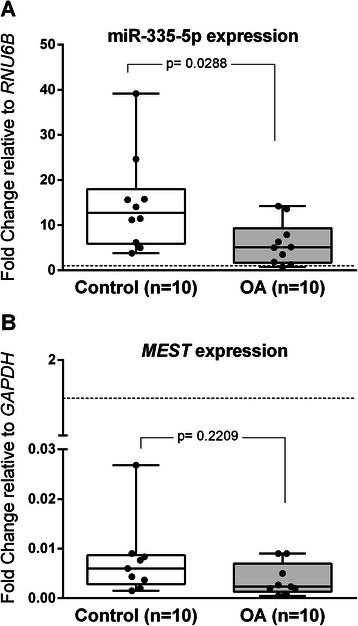
miR-335-5p and MEST expression in OA-MSCs and Control-MSCs. a. Box and wiskers plot of miR-335-5p expression in OA-MSCs and Control-MSCs relative to the RNU6B internal control gene. b. MEST expression in human in OA-MSCs and Control-MSCs relative to the GAPDH reference gene. A positive correlation between miR-335-5p and MEST expression was detected (Spearman correlation coefficient, r = 0.59; p = 0.0961 in OA and r = 0.6; p = 0.0968 in controls
Wnt antagonists DKK1 and SFRP1 are not differentially inhibited by miR-335-5p
Increased bone formation is one of the main OA features. In addition osteoinduction through Wnt signaling can be promoted by inhibition of Wnt antagonists such as DKK1 and SFRP1, two known predicted targets of miR-335-5p. We next aimed to confirm if differential expression of miR-335-5p in OA-MSCs correlated with a reduced expression of: DKK1 and SFRP1. Our data revealed that independently of miR-335-5p expression, mRNA levels of antagonists were similar both in undifferentiated OA- or Control-MSCs (Fig. 4).
Fig. 4.
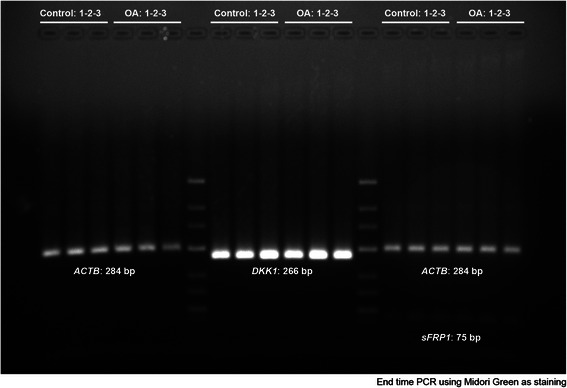
Two percent agarose gel of DKK1 and SFRP amplification products from OA-MSCs and Control-MSCs. After synthesizing the first strand cDNA, it was amplified using PCR. The PCR product was analyzed by agarose gel electrophoresis. PCR reactions (25ul) were loaded as follow: Lanes [1–6] ACTB [8–13] DKK1. [15–20] ACTB + SFRP1. Lanes 7 and 14 correspond to the molecular markers (GeneRuler™ Low Range DNA Ladder, Fermentas). This result shows that single specific PCR product was obtained with expected molecular amplicon sizes. No differences were found in band intensity between OA-MSCs and Control-MSCs
Discussion
Bone homeostasis, which largely depends on the appropriate differentiation of chondro- and osteo-progenitor MSCs, is susceptible of disturbances leading to skeletal dysplasias or articular diseases such as osteoarthritis [26], however it has been described that during OA progression bone marrow OA-MSCs exhibit different properties that from healthy age-matched MSCs [27]. These alterations are characterized by a lower proliferative capacity, lower adipogenesis and chondrogenesis and increased osteogenesis in OA-MSCs. These changes in OA-MSCs can be attenuated or reversed by particular supplementation of culture media [28]. This could explain why in our in vitro cultures cell proliferation and differentiation was quite similar.
Although the role of MSCs still remains unknown, and the exact pathogenetic mechanisms of OA are unravelled, the central role of WNT signaling pathway is considered essential [6]. The Wnt signalling participates during skeletogenesis and regulation of bone remodelling processes in adult tissues through the expression of target genes during endochondral ossification [29]. However, the role of Wnt signalling in osteogenesis is by far controversial. While some authors have reported that its activation promotes osteogenesis [30, 31], others argued the detrimental effects in osteogenesis upon Wnt activation [32, 33]. Moreover, Liu et al. [34] recently described that Wnt promoted the osteogenesis in normal medium while inhibited it the osteogenic differentiation medium. This dual effect was attributed to a regulatory loop between miR-17 and its target (TCF3). Taken together, these contradictory results suggest the critical effect of experimental conditions or microenvironmental and epigenetic factors rather than genetic differences. In this regard, a recent report described that reduced methylation in the osteoarthritic bone, compared to osteoporotic bone, results in an upregulated expression of genes and increased WNT pathway activity [35].
As occurs with other signalling pathways, regulation takes place at different levels. Post-transcriptional regulation by miRNAs have emerged as key modulators of signal transmission and cellular response to extracellular signals [36]. In this study we performed a comparative profiling of miRNA expression during in vitro osteogenic differentiation of BM-MSCs isolated from OA and Control subjects. We aimed to describe a signature of miRNAs potentially involved in the deregulated osteogenesis occurring in OA.
The role of miR-335-5p on Wnt signalling has been described primarily in cancer. Its implication in bone development has been only partially evidenced. At least 30 genes, related or belonging to the Wnt pathway, have been previously described with varying degrees of evidence as validated targets of this miRNA [17, 37–40] (Table 2). In our study, after filtering of 754 initially screened miRNAs and further validation of 21 of them, only hsa-miR-210 and hsa-miR-335-5p showed significative differences in expression during osteogenesis. Subsequently, using the latest miRTarBase database, we analyzed if these miRNAs showed experimentally validated miRNA-target interactions in the Wnt pathway. Given that only miR-335-5p showed the existence of direct potential targets we focused our attention on this miRNA. (Additional file 6).
Table 2.
Genes implicated in WNT pathway, matrix and cytoskeleton described as targets of hsa-miR-335-5p
| WNT related genes | Other | ||||
|---|---|---|---|---|---|
| Gene | EntrezID | Reference | Gene | EntrezID | Reference |
| AXIN1 | 8312 | 39 | RUNX2 | 860 | 20 |
| CCND2 | 894 | 39 | COL1A1 | 1277 | 40 |
| CTNNBIP1 | 56998 | 39 | COL10A1 | 1300 | 39 |
| DAAM1 | 23002 | 39 | COL11A1 | 1301 | 39 |
| DKK1 | 22943 | 17 | COL15A1 | 1306 | 39 |
| DKK2 | 27123 | 39 | COL21A1 | 81578 | 39 |
| DKK4 | 27121 | 39 | COL3A1 | 1281 | 39 |
| FGF4 | 2249 | 39 | COL4A5 | 1287 | 39 |
| FZD1 | 8321 | 39 | COL6A1 | 1291 | 39 |
| FZD10 | 11211 | 39 | COL6A5 | 256076 | 39 |
| FZD8 | 8325 | 39 | COL6A6 | 131873 | 39 |
| LRP5 | 4041 | 39 | COL8A2 | 1296 | 39 |
| LRP6 | 4040 | 39 | COL9A2 | 1298 | 39 |
| MYC | 4609 | 37, 39 | COL9A3 | 1299 | 39 |
| NFAT5 | 10725 | 39 | |||
| PPP1R1A | 5502 | 39 | |||
| PPP2R5A | 6422 | 39 | |||
| PPP3R2 | 5528 | 39 | |||
| PRICKLE2 | 166336 | 39 | |||
| PRKCG | 5582 | 39 | |||
| SFRP1 | 6422 | 39 | |||
| SMAD3 | 4088 | 39 | |||
| TBL1X | 6907 | 39 | |||
| VANGL2 | 57216 | 39 | |||
| WIF1 | 11197 | 39 | |||
| WNT1 | 7471 | 39 | |||
| WNT10B | 7480 | 39 | |||
| WNT3 | 7473 | 39 | |||
| WNT7B | 7477 | 39 | |||
| WNT9A | 7483 | 39 | |||
In the present study, we have demonstrated the existence of a downregulation of miR-335-5p in OA-MSCs compared to Control-MSCs. Moreover, we demonstrated the existence of a similar and progressive downregulatory trend of this miRNA along the osteogenic differentiation Control-MSCs not significant in the case of OA-MSCs. Decreased expression of Wnt antagonists such as DKK1 and SFRP1 are likely a major cause of increased Wnt/β-catenin signalling. In this particular, expression of miR-335-5p has been previously described as a key regulator of bone development promoting the downregulation of DKK1 at initial stages of osteogenesis [17]. This modulatory effect significantly persist at later stages of osteogenesis in Control-MSCs, likely to prevent excessive mineralization by promoting elevated levels of DKK1 and/or SFRP1 through dowregulation of miR-335-5p. Although post-transcriptional regulation by miRNAs usually results in gene silencing by reduction of translation and mRNA levels, we found that miR-335-5p did not exert this inhibitory effect on DKK1 or SFRP1 mRNA levels. This result, however, is consistent with evidences suggesting that under particular conditions, such as stress, inhibition of translation initiation and mRNA stabilization can occur simultaneously [41].
Previous observations of Tomé et al. [20] proposed the key regulatory role of miR-335 in MSCs biology, concluding that upregulation of miR-335 impairs the MSC reparative phenotype. Here we provide new insights about the putative role of miR-335 in Wnt signalling throughout osteogenesis in OA pathology suggesting a more active Wnt signalling and thus increased osteogenesis and mineralization in OA. In addition to the modulatory role of WNT pathway by miR-335-5p, several other genes involved in extracellular matrix formation have been also described as targets of this miRNA, in particular collagen genes. Some of them, previously described by our group, were clearly downregulated in OA-MSCs. In particular COL10A1 [42].
Conclusions
Collectively, these evidences indicate the existence of a correlation between miR-335-5p expression and OA indicating a putative role of this miRNA. These features are of particular interest given that subchondral and periarticular bone remodelling as well as establishment of a proper subchondral cartilage plays a role in the progression of osteoarthritis (OA). These data contribute to our understanding of the molecular mechanisms involved in MSCs mediating homeostatic control in OA pathophysiology.
Acknowledgments
This work was supported in part by institutional grants from the Instituto de Salud Carlos III CP10/00346 and PI10/00178 and the Spanish society of orthopaedics surgery and traumatology (SECOT).
J.R. Lamas is supported by the Miguel Servet program from ISCIII-Fondo Investigación Sanitaria-Spain (CP10/00346).
We thanks the Hospital Clínico San Carlos orthopaedic surgeons for providing the samples used in the study.
Abbreviations
- BM-MSCs
Bone marrow mesenchymal stem cells (hereinafter referred as to MSCs)
- OA-MSCs
Bone marrow mesenchymal stem cells from osteoarthritis patients. hsa-miR-335-5p (hereinafter referred as to miR-335-5p)
Additional files
Data analysis of the expression profile of miRNA sequences, from the Sanger miRBase v14 using the Taqman OpenArray MicroRNA pools A and B. (XLS 2463 kb)
miRNAs showing interactions between status (OA and Control) and differentiation time points (p values <0.05 without multiple test correction applied. (TIFF 82 kb)
miRNAs without status and differentiation interaction that show differential expression between osteogenic differentiation time points (p value <0.05 without multiple test correction applied). (TIFF 123 kb)
miRNAs without status and differentiation interaction that show differential expression between OA and Control samples (p value <0.05 without multiple test correction applied). (TIFF 157 kb)
Venn diagrams showing the distribution of miRNAs analyzed and their overlapping among different status and osteogenic time points. (PDF 19 kb)
Extraction of all the experimentally validated microRNA-target interactions for hsa-miR-210 and hsa-miR-335-5p from the miRTarBase database. An extraction of miR-335 target genes related to the WNT pathway was also included. These belong to the Wnt signaling pathway, according to the enriched annotations provided in the Kyoto Encyclopedia of Genes and Genomes (KEGG) 04310. (XLS 2121 kb)
Footnotes
Benjamín Fernández-Gutiérrez and José Ramón Lamas are share senior authorship.
Competing interests
The corresponding author declares the absence of any conflict of interest regarding the submitted manuscript. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Authors’ contributions
PT performed the isolation of cell, culture, expansion, cytometry analysis and helped to draft the manuscript. LR carried out the miRNAs isolation from samples and adquired miRNA data. LA performed the statistical analysis of data and participated in the design of the study. MT carried out the molecular genetic studies. PLR performed the microarray statistical analysis and interpretation of results. EH performed the cell culture differentiations, histochemistry and obtained total RNA from cultures. MAG participated in the design of the study. FM and EM participated in the design of the study, performed the surgery and provided the surgical samples. BF participated in the design of the study, coordination of the study and helped to revise the manuscript. JRL performed data analysis, conceived and coordinated the study and draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Pilar Tornero-Esteban, Email: mptornero@gmail.com.
Luis Rodríguez-Rodríguez, Email: lrodriguezr.hcsc@salud.madrid.org.
Lydia Abásolo, Email: lydia.abasolo@salud.madrid.org.
María Tomé, Email: mtome@cnb.csic.es.
Pedro López-Romero, Email: plromero1@gmail.com.
Eva Herranz, Email: evaherranzdlp@hotmail.com.
Manuel A. González, Email: maglez75@hotmail.com
Fernando Marco, Email: fmarco@med.ucm.es.
Enrique Moro, Email: enrmoro@hotmail.com.
Benjamín Fernández-Gutiérrez, Email: benjamin.fernandez@salud.madrid.org.
José Ramón Lamas, Phone: +34 91 3303615, Email: jrlamas@gmail.com.
References
- 1.Bitton R. The economic burden of osteoarthritis. Am J Manag Care. 2009;15(8 Suppl):S230–5. [PubMed] [Google Scholar]
- 2.Goldring MB, Goldring SR. Articular cartilage and subchondral bone in the pathogenesis of osteoarthritis. Ann N Y Acad Sci. 2010;1192:230–7. doi: 10.1111/j.1749-6632.2009.05240.x. [DOI] [PubMed] [Google Scholar]
- 3.Blagojevic M, Jinks C, Jeffery A, Jordan KP. Risk factors for onset of osteoarthritis of the knee in older adults: a systematic review and meta-analysis. Osteoarthritis Cartilage. 2010;18(1):24–33. doi: 10.1016/j.joca.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Reynard LN, Loughlin J. Insights from human genetic studies into the pathways involved in osteoarthritis. Nat Rev Rheumatol. 2013;9(10):573–83. doi: 10.1038/nrrheum.2013.121. [DOI] [PubMed] [Google Scholar]
- 5.Diekman BO, Guilak F. Stem cell-based therapies for osteoarthritis: challenges and opportunities. Curr Opin Rheumatol. 2013;25(1):119–26. doi: 10.1097/BOR.0b013e32835aa28d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med. 2013;19(2):179–92. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
- 7.Miki T, Yasuda SY, Kahn M. Wnt/beta-catenin signaling in embryonic stem cell self-renewal and somatic cell reprogramming. Stem Cell Rev. 2011;7(4):836–46. doi: 10.1007/s12015-011-9275-1. [DOI] [PubMed] [Google Scholar]
- 8.Zimmerman ZF, Moon RT, Chien AJ. Targeting Wnt pathways in disease. Cold Spring Harb Perspect Biol. 2012;4(11). [DOI] [PMC free article] [PubMed]
- 9.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5(9):691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 10.Dong S, Yang B, Guo H, Kang F. MicroRNAs regulate osteogenesis and chondrogenesis. Biochem Biophys Res Commun. 2012;418(4):587–91. doi: 10.1016/j.bbrc.2012.01.075. [DOI] [PubMed] [Google Scholar]
- 11.Bakhshandeh B, Soleimani M, Paylakhi SH, Ghaemi N. A microRNA signature associated with chondrogenic lineage commitment. J Genet. 2012;91(2):171–82. doi: 10.1007/s12041-012-0168-0. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145) J Biol Chem. 2012;287(2):916–24. doi: 10.1074/jbc.M111.302430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corr M. Wnt-beta-catenin signaling in the pathogenesis of osteoarthritis. Nat Clin Pract Rheumatol. 2008;4(10):550–6. doi: 10.1038/ncprheum0904. [DOI] [PubMed] [Google Scholar]
- 14.Loughlin J, Dowling B, Chapman K, Marcelline L, Mustafa Z, Southam L, et al. Functional variants within the secreted frizzled-related protein 3 gene are associated with hip osteoarthritis in females. Proc Natl Acad Sci U S A. 2004;101(26):9757–62. doi: 10.1073/pnas.0403456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Huang T, Zhao X, Cheng L. MicroRNAs modulate the Wnt signaling pathway through targeting its inhibitors. Biochem Biophys Res Commun. 2011;408(2):259–64. doi: 10.1016/j.bbrc.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Su J, Zhang A, Shi Z, Ma F, Pu P, Wang T, et al. MicroRNA-200a suppresses the Wnt/beta-catenin signaling pathway by interacting with beta-catenin. Int J Oncol. 2012;40(4):1162–70. doi: 10.3892/ijo.2011.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Tu Q, Bonewald LF, He X, Stein G, Lian J, et al. Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J Bone Miner Res. 2011;26(8):1953–63. doi: 10.1002/jbmr.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang K, Zhang JX, Han L, You YP, Jiang T, Pu PY, et al. MicroRNA roles in beta-catenin pathway. Mol Cancer. 2010;9:252. doi: 10.1186/1476-4598-9-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou AD, Diao LT, Xu H, Xiao ZD, Li JH, Zhou H, et al. beta-Catenin/LEF1 transactivates the microRNA-371-373 cluster that modulates the Wnt/beta-catenin-signaling pathway. Oncogene. 2011. [DOI] [PubMed]
- 20.Tome M, Lopez-Romero P, Albo C, Sepulveda JC, Fernandez-Gutierrez B, Dopazo A, Bernad A, Gonzalez MA. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011;18(6):985–95. doi: 10.1038/cdd.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen ED, Gopalakrishnan R, Westendorf JJ. Regulation of gene expression in osteoblasts. Biofactors. 2010;36(1):25–32. doi: 10.1002/biof.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31(3):315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 23.Wylie D, Shelton J, Choudhary A, Adai AT. A novel mean-centering method for normalizing microRNA expression from high-throughput RT-qPCR data. BMC Res Notes. 2011;4:555. doi: 10.1186/1756-0500-4-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth GK. limma: Linear Models for Microarray Data. In: Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc. 1995;B57(1):289–300. [Google Scholar]
- 26.Kawaguchi H. Endochondral ossification signals in cartilage degradation during osteoarthritis progression in experimental mouse models. Mol Cells. 2008;25(1):1–6. [PubMed] [Google Scholar]
- 27.Murphy JM, Dixon K, Beck S, Fabian D, Feldman A, Barry F. Reduced chondrogenic and adipogenic activity of mesenchymal stem cells from patients with advanced osteoarthritis. Arthritis Rheum. 2002;46(3):704–13. doi: 10.1002/art.10118. [DOI] [PubMed] [Google Scholar]
- 28.Im GI, Jung NH, Tae SK. Chondrogenic differentiation of mesenchymal stem cells isolated from patients in late adulthood: the optimal conditions of growth factors. Tissue Eng. 2006;12(3):527–36. doi: 10.1089/ten.2006.12.527. [DOI] [PubMed] [Google Scholar]
- 29.Lu C, Wan Y, Cao J, Zhu X, Yu J, Zhou R, et al. Wnt-mediated reciprocal regulation between cartilage and bone development during endochondral ossification. Bone. 2013;53(2):566–74. doi: 10.1016/j.bone.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 30.Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS, et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280(39):33132–40. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- 31.Liu G, Vijayakumar S, Grumolato L, Arroyave R, Qiao H, Akiri G, et al. Canonical Wnts function as potent regulators of osteogenesis by human mesenchymal stem cells. J Cell Biol. 2009;185(1):67–75. doi: 10.1083/jcb.200810137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Boer J, Siddappa R, Gaspar C, van Apeldoorn A, Fodde R, van Blitterswijk C. Wnt signaling inhibits osteogenic differentiation of human mesenchymal stem cells. Bone. 2004;34(5):818–26. doi: 10.1016/j.bone.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Khavandgar Z, Lin SH, Murshed M. Lithium chloride attenuates BMP-2 signaling and inhibits osteogenic differentiation through a novel WNT/GSK3- independent mechanism. Bone. 2011;48(2):321–31. doi: 10.1016/j.bone.2010.09.033. [DOI] [PubMed] [Google Scholar]
- 34.Liu W, Liu Y, Guo T, Hu C, Luo H, Zhang L, et al. TCF3, a novel positive regulator of osteogenesis, plays a crucial role in miR-17 modulating the diverse effect of canonical Wnt signaling in different microenvironments. Cell Death Dis. 2013;4:e539. doi: 10.1038/cddis.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Ibarbia C, Delgado-Calle J, Casafont I, Velasco J, Arozamena J, Perez-Nunez MI, et al. Contribution of genetic and epigenetic mechanisms to Wnt pathway activity in prevalent skeletal disorders. Gene. 2013;532(2):165–72. doi: 10.1016/j.gene.2013.09.080. [DOI] [PubMed] [Google Scholar]
- 36.Hong E, Reddi AH. MicroRNAs in chondrogenesis, articular cartilage, and osteoarthritis: implications for tissue engineering. Tissue Eng Part B Rev. 2012;18(6):445–53. doi: 10.1089/ten.teb.2012.0116. [DOI] [PubMed] [Google Scholar]
- 37.Heyn H, Engelmann M, Schreek S, Ahrens P, Lehmann U, Kreipe H, et al. MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int J Cancer. 2011;129(12):2797–806. doi: 10.1002/ijc.25962. [DOI] [PubMed] [Google Scholar]
- 38.Iliopoulos D, Bimpaki EI, Nesterova M, Stratakis CA. MicroRNA signature of primary pigmented nodular adrenocortical disease: clinical correlations and regulation of Wnt signaling. Cancer Res. 2009;69(8):3278–82. doi: 10.1158/0008-5472.CAN-09-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egeblad M, Shen HC, Behonick DJ, Wilmes L, Eichten A, Korets LV, et al. Type I collagen is a genetic modifier of matrix metalloproteinase 2 in murine skeletal development. Dev Dyn. 2007;236(6):1683–93. doi: 10.1002/dvdy.21159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huch S, Nissan T. Interrelations between translation and general mRNA degradation in yeast. Wiley Interdiscip Rev RNA. 2014;5(6):747–63. doi: 10.1002/wrna.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamas JR, Rodriguez-Rodriguez L, Vigo AG, Alvarez-Lafuente R, Lopez-Romero P, Marco F, et al. Large-scale gene expression in bone marrow mesenchymal stem cells: a putative role for COL10A1 in osteoarthritis. Ann Rheum Dis. 2010;69(10):1880–5. doi: 10.1136/ard.2009.122564. [DOI] [PubMed] [Google Scholar]


