Abstract
Tristetraprolin (TTP) is an inducible zinc finger AU-rich RNA binding protein essential for enforcing degradation of mRNAs encoding inflammatory chemokines and cytokines. Most studies on TTP center on the connection between mRNA half-life and inflammatory output, because loss of TTP amplifies inflammation by increasing stability of AU-rich mRNAs. Here we focused on how TTP controls cytokine and chemokine production in the non-resolving inflammation of cancer using tissue-specific approaches. By contrast to model in vitro macrophage systems, we found constitutive TTP expression in late stage tumor-associated macrophages (TAMs). However, TTP’s effects on AU-rich mRNA stability were negligible and limited by constitutive p38α MAP kinase activity which was the main driver of pro-inflammatory cytokine production in TAMs at the posttranscriptional level. Instead elimination of TTP caused excessive protein production of inflammatory mediators suggesting TTP-dependent translational suppression of AU-rich mRNAs. Manipulation of the p38α-TTP axis in macrophages has significant effects on the growth of tumors, and therefore represents a means to manipulate inflammation in the tumor microenvironment.
Keywords: Tumor-Associated Macrophages, Inflammation, Tumor Microenvironment, p38 MAPK, Tristetraprolin
Introduction
Non-resolving inflammation (NRI) underpins most chronic diseases, including obesity, auto-inflammatory diseases, atherosclerosis, chronic responses to implanted medical devices, chronic or latent infections, and cancer (1). NRI is caused by aberrant responses to a persisting entity. The persistence of malignant cells in cancer, for example, drives the production of hematopoietic cells from the bone marrow to seed a growing tumor (2). Therefore, a challenge of biology and medicine is to understand the cellular and molecular pathways underpinning NRI and how to mitigate their effects in chronic diseases.
In resolving inflammation triggered by TLR signaling, the TNF mRNA is rapidly produced from poised Pol II at the Tnf promoter (3–5). TNF protein is made and secreted, followed by a downregulation phase where the tristetraprolin (TTP) zinc finger protein, also induced in the early phase of TLR signaling, plays a decisive role in eliminating TNF mRNA (6). In model in vitro systems such as BMDMs the TNF-TTP pathway is controlled by two key factors, p38α MAPK and IL-10 (7–9). p38α activated by TLR signaling phosphorylates TTP there by blocking its function and sustaining TNF output (9, 10). In the resolution phase, DUSP proteins dephosphorylate p38α, which leads to dephosphorylation of TTP and a rise in TTP activity to eliminate TNF mRNA. IL-10, stimulated by TLR signaling to act in an autocrine way, further enhances removal of the TNF mRNA because it drives TTP and DUSP1 expression in a STAT3-dependent way (7, 11, 12).
To begin to understand the signaling pathways involved in regulating chronic inflammation in tumor-associated macrophages (TAMs) we used transplantable tumor models with predictable temporal inflammatory responses which can be tracked by flow cytometry and is amenable to the use of genetics to probe signaling pathways and how they become dysregulated. We focused on how TTP regulates cytokine and chemokine production because inflammatory cytokines are the key drivers of chronic inflammation such as cancer. A key tactic was using a conditional knockout allele of Zfp36 (encoding TTP) to avoid complications of systemic inflammation present in conventional Zfp36−/− mice early in development (13–15). Our results show TTP maintains macrophages on a ‘knife-edge’ between counter-regulation of inflammation and death. Our results also suggest TTP primarily regulates steps beyond mRNA stability such as mRNA translation in macrophages associated with cancer inflammation.
Materials and Methods
Mice
C57BL/6, Tie2-Cre (B6.Cg-Tg (Tek-cre)1Ywa/J), and LysM-Cre (Lyz2tm1(cre)Ifo/J) mice were obtained from Jackson Laboratories (Bar Harbor, ME). TTPKO (Zfp36−/−) mice were a gift of P. Blackshear (National Institute of Environmental Health Sciences, Research Triangle Park, NC, USA). The TTPΔM and Mapk14flox/floxmice have been described elsewhere (15, 16). Th-MYCN transgenic (tg) (17) and Rbflox/flox;p53flox/flox;Osx-Cre+ (mixed background) mice (18) were obtained from M. Dyer. For experiments, sex-matched mice were grouped and used between 6 and 10 weeks. All mice were on a C57BL/6 background unless indicated otherwise. Mice were used within the Animal Resource Center according to protocols approved by the IACUC of St. Jude Children’s Research Hospital.
Zymosan A induced peritonitis
Chronic peritonitis was induced by intraperitoneal injection of 10 mg/mouse of zymosan A (Sigma) as described (19) and CD11b+ cells purified by Miltenyi magnetic beads according to the manufacturer’s protocol.
Tumor associated macrophage (TAM) collection from solid tumors
Solid tumors were minced and digested with 5 ml of fresh Tumor Digestion Media as described in the Supplementary Materials and Methods. The suspension was then passed through a 70 μm strainer and cells were overlaid on a 35%/60% percoll gradient. After centrifugation cells were collected from the 35%/60% interphase and TAMs were isolated by CD11b purification using Miltenyi magnetic beads according to the manufacturer’s protocol.
Tumor models – in vivo
EG7 lymphoma line (ATCC) originating from the thymus (EL4), stably expressing OVA protein was tested for pathogen contamination prior to injection. 3x106 EG7 cells were injected subcutaneously into the flank of recipient mice and harvested 12 days later unless indicated otherwise. Orthotopically grown gliomas (20) and thymomas were harvested from mice between 1.5 and 2 weeks post-transplant. Spontaneous neuroblastomas from Th-MYCN transgenic (tg) and osteosarcomas collected from Rbflox/flox;p53flox/flox;Osx-Cre+ mice (18) were detected by ultrasound or visual inspection of tumors on the long bones or jaws, respectively.
Antibodies and immunoblotting
For flow cytometry: Anti-mouse MHC II (I-A/I-E; clone M5/114.15.2, cat. no. 12-5321-83), anti-mouse Ccl3 (clone DNT3CC, cat. no. 12-7532-80), anti-mouse IL-10 (clone Jess-16E3, cat. no. 11-7101-81), anti-mouse IL-1α (clone ALF 161, cat. no. 12-7011-81), anti-mouse IL-1β (clone NJTEN3, cat. no. 12-7117-80) all eBiosience, anti-mouse TNFα (cat. no. 554419, PharMingen), p-p38MAPK (T180/Y82; clone 3D7, cat. no. 9608S, Cell Signaling), anti-mouse/human CD11b (clone M1/70, cat. no. 101224, BioLegend), anti-mouse Ly6C (clone HK1.4, cat. no. 128016, BioLegend). For immunoblotting rabbit antibody to TTP (7). p-p38MAPK and p38MAPK were from Cell Signaling Technologies (T180/Y182; clone D3F9, cat. no. 4511S) and Santa Cruz Biotechnology (clone A-12, cat. no. sc-7972), respectively. Whole cell extracts were prepared and assayed by western blotting as described (7) using 4–15% Tris-HCl gels (Criterion, Bio-Rad). Equal loading was controlled by anti-mouse GRB2 antibody (cat. no. 610112, BD Bioscience).
Enzyme-linked Immunosorbent Assay (ELISA)
Murine TNF production was measured from supernatants of TAMs resting on tissue-culture plates for 6 hrs using pre-optimized Capture and Detection antibodies (both eBioscience).
Results
Tumor-associated macrophages produce pro-inflammatory cytokines during all stages of development
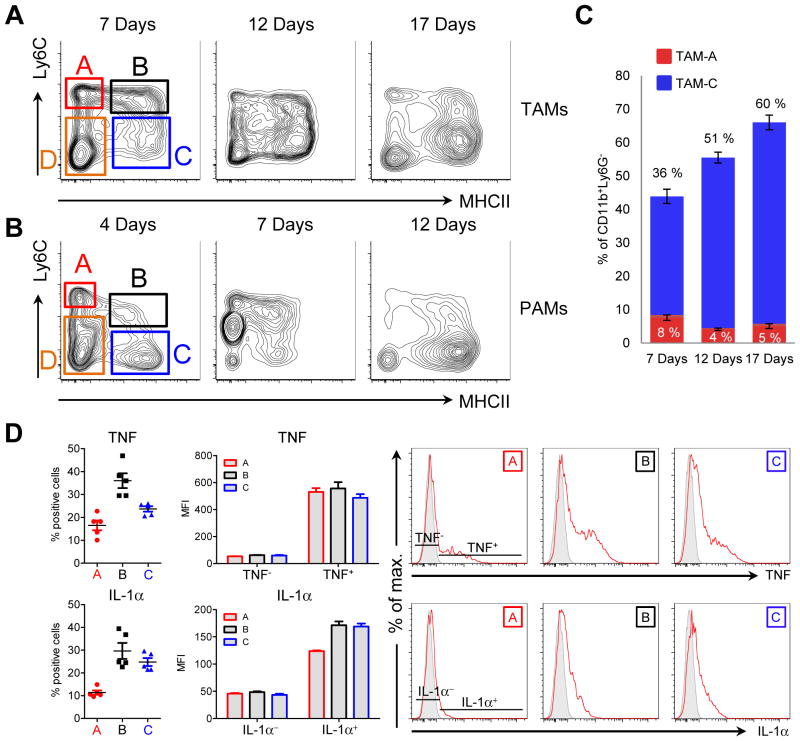
Blood-derived monocytes invade almost all types of solid tumors where they develop into macrophages. Macrophages are the most numerous myeloid cells in chronic inflammation, and the source of many key pro-inflammatory cytokines and chemokines that drive inflammation forward (21). To dissect macrophage function in cancer inflammation we developed methods to reliably distinguish macrophages at different points in their development from neutrophils and eosinophils. Building on a system developed by van Ginderachter and colleagues, we identified three monocyte-derived populations of macrophages in solid tumors as well as in chronic zymosan peritonitis, a complementary model for chronic inflammation where monocytes infiltrate the inflammatory site (Fig. 1A, B) (22). The ‘A’ population comprises inflammatory monocytes (Ly6Chi, MHCII−). Tracing tools argue that population ‘A’ gives rise to the Ly6C+, MHCII+ population ‘B’ which then develops into the mature ‘C’ fraction, consisting of cells that down-regulate Ly6C and increase expression of MHCII (22, 23). The ‘C’ macrophages increase in relative proportion over time (Fig. 1A–C). The remaining MHCII and Ly6C negative myeloid infiltrates consist of eosinophil-like cells maintained independently of monocytes (population D) (22). The developmental progression of tumor macrophages and macrophages from chronic peritonitis from monocytes to the C fraction is similar to the ‘waterfall’ model of in situ macrophage development observed in models where monocytes seed inflammatory sites, arguing for a general mode of macrophage development under inflammatory conditions (24). For all subsequent experiments we isolated TAMs from tumors at day 12, as at this stage all TAM populations were most abundant and amenable to detailed ex vivo analysis.
Figure 1. NRI associated macrophages share a common mode of development and express pro-inflammatory cytokines.
A and B, flow cytometry plots of CD11b+Ly6G− myeloid cells isolated from EG7 thymomas (A) or zymosan induced peritonitis (B) at different timepoints as indicated. Plots are representative for at least 2 experiments. Gates A, B, C and D indicate tumor-associated macrophage (TAM) or peritonitis-associated macrophage (PAM) populations used for sorting experiments or to measure cytokine production by intracellular flow cytometry. C, quantitative analysis of EG7 TAM populations A and C (gated as in A) over time. Data are values from 1 out of 2 independent experiments (n ≥ 4). Error bars, SEM. D, quantitative analysis of cytokine production by flow cytometry of EG7 TAM populations A, B and C gated as shown in 1A. Values from individual mice are either depicted as percent positive cells (left panels) or Median Fluorescence Intensity (MFI, middle panel) of TNF or IL-1α positive (TNF+, IL-1α+) compared to negative (TNF−, IL-1α−) macrophages with in population A, B and C. Example plots are shown (right panel) indicating the gating strategy for cells considered positive and negative. Unstained control is shown in grey. Data represent the mean +/− SEM of 2 independent experiments (n ≥ 2).
A hallmark of chronic inflammation is constitutive cytokine and chemokine production promoted by the presence of the inciting entity. A crucial question in understanding the relationships between cytokine and chemokine-producing cells in chronic inflammation is whether negative feedback loops counteracting disease-causing inflammation are inactive, and how they become dysregulated. We therefore asked if TAMs expressed inflammatory cytokines and whether their expression pattern was linked to the ‘A, B, C’ development pathway. Although we found highest expression of TNF and IL-1α in the ‘B’ fraction, all macrophage subsets expressed readily detectable amounts of both cytokines (Fig. 1D). For example, 36% of the ‘B’ fraction were TNF+(Fig. 1D).
AU-rich element binding proteins are highly expressed in TAMs
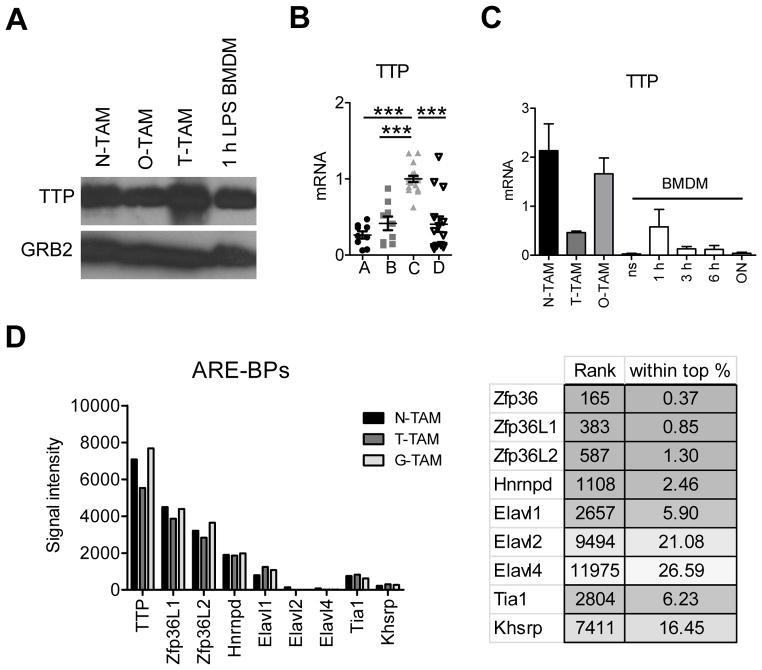
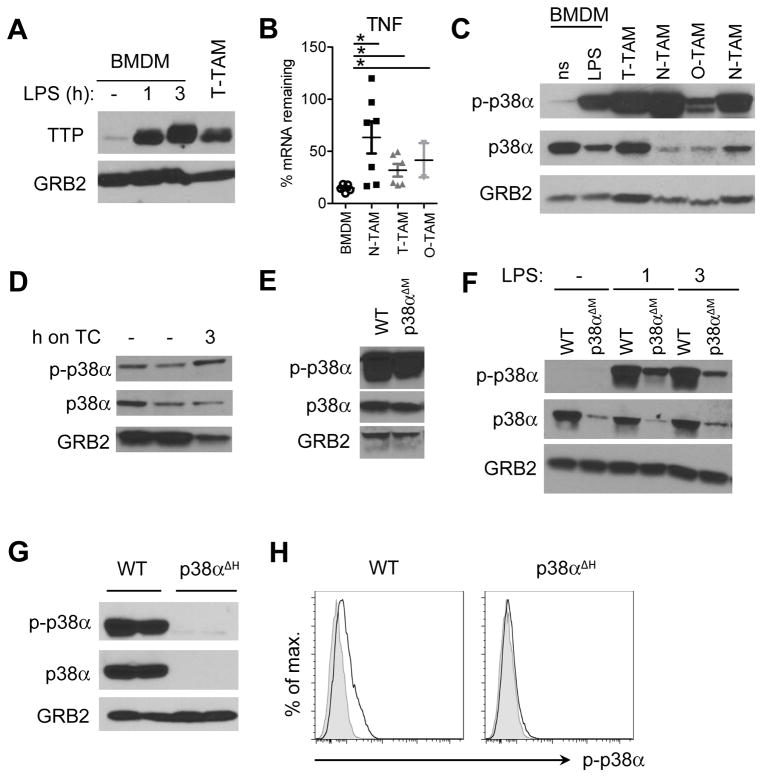
TTP is essential for mediating cytokine and chemokine mRNA decay in inflammation. Indeed, global absence of TTP in all cells causes systemic inflammatory pathology because multiple inflammatory cytokines including TNF and IL-23 are produced in excess (13, 14). TTP’s target mRNAs form a hierarchy dependent in part on the number and type of AU elements in the mRNA 3′ UTR (25). As both TNF and IL-1α mRNAs are TTP targets (6, 15, 26) but highly expressed in tumor inflammation-associated macrophages, we suspected sustained cytokine production by TAMs was caused by failure of TTP expression. Paradoxically, we found TTP mRNA and protein was highly expressed in TAMs, and expression increased as cells developed to the ‘C’ population (Fig. 2A, B). Although TTP was expressed, cytokine production was high (Fig. 1D). High expression of TTP was not an artifact of our experimental system because high amounts of the Zfp36 mRNA (encoding TTP) and TTP protein were detectable in macrophages from different cancer types, and were expressed at higher amounts than LPS-stimulated BMDMs (Fig. 2C). Interestingly, mRNAs encoding two TTP-related zinc finger mRNA destabilizing proteins, Zfp36L1 and Zfp36L2, were also highly expressed in tumor macrophages, being among the top 2% genes (Fig. 2D; Kratochvill et al. submitted, GEO accession number GSE59047). Given TTP, Zfp36L1 and Zfp36L2 were highly expressed in TAMs but cytokine expression was correspondingly high, we concluded that mRNA degradation processes are ineffective at eliminating inflammatory cytokine mRNAs in tumor macrophages.
Figure 2. TTP is highly expressed in selected TAM populations.
A, immunoblot analysis of TTP expression in neuroblastoma (N), osteosarcoma (O) and EG7 thymoma (T) TAMs compared to 1 hour LPS stimulated BMDMs. Blot was reprobed with anti-GRB2 to test for equal loading and represents 1 out of 3 experiments. B, TTP qRT-PCR of EG7 TAMs sorted for the indicated populations as in Figure 1A. Data from individual mice are expression values from 4 experiments (n ≥ 6). All values were first normalized to the corresponding GAPDH and then to the mean of TAM-C within each experiment. Data were analyzed by One-way Anova followed by Bonferroni Post Test. The mean per group is shown as black line. Error bars, SEM. C, qRT-PCR analysis of TTP in TAMs or BMDMs left untreated (ns) or stimulated with LPS + IFNγ over time. Data were normalized to the corresponding GAPDH for each value and represent the mean +/− SEM (n ≥ 3). D, microarray analysis of TAMs isolated from Neuroblastomas (N-TAM), EG7 Thymomas (T-TAM) and Gliomas (G-TAM) showing average signal intensities of selected AU-rich element binding proteins (ARE-BPs, n = 3 per TAM type). The right panel shows the rank of different ARE-BPs in N-TAMs together with the rank as percentage among all 45038 probe sets, respectively. The full microarray dataset has been submitted (GEO accession number GSE59047).
TTP limits the production of inflammatory cytokines in TAMs
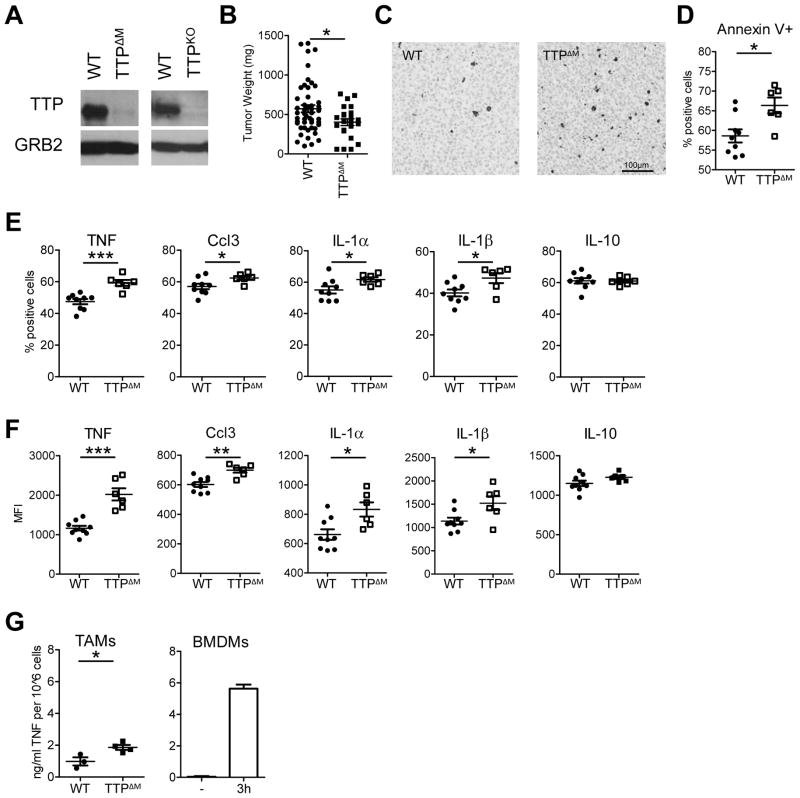
We next asked if TTP was required for regulating cytokine and chemokine production. To do this we generated TAMs from mice where TTP is specifically and efficiently eliminated from macrophages and neutrophils using the LysM-Cre deleter (called TTPΔM mice) (Fig. 3A). This tactic enabled us to avoid the effects of systemic inflammation in the Zfp36−/− mice where the absence of TTP increases numerous pro-inflammatory cytokines and results in early death (13, 14). We examined tumors in the TTPΔM mice and found they were smaller and contained more dying cells as compared to tumors from control mice (Fig. 3B–D). TAMs from TTPΔM mice had increased numbers of macrophages positive for TNF, Ccl3, IL-1α and IL-1β (Fig. 3E–F). In addition, TAMs isolated from TTPΔM animals secreted approximately 2.5-fold more TNF when rested on tissue culture dishes for 6 hours than WT TAMs (Fig. 3G). We further compared the TNF production in TAMs to resting or LPS stimulated BMDMs. TAMs produced more TNF than resting BMDMs and approximately ¼ of TNF secreted by an equivalent number of homogeneous BMDMs stimulated for 3 hrs with LPS (Fig. 3G). Production of the anti-inflammatory cytokine and TTP target IL-10 (27) was unaffected in terms of number of IL-10 producing macrophages or intensity of staining (Fig. 3E and F). Collectively, these data suggest TTP is required to temper the already high cytokine and chemokine production in the inflammatory milieu of chronic inflammation. When TTP is genetically eliminated in macrophages, cytokine and chemokine output further increases relative to the already high amounts in controls, which is linked to increased cell death and decreased tumor size.
Figure 3. Myeloid TTP counteracts inflammation and supports tumor growth.
A, immunoblot analysis using anti-TTP antibody of whole cell lysates of EG7 TAMs isolated from WT, TTPΔM or TTP−/− tumor bearing animals. Blots were re-probed using anti-GRB2 antibody to confirm equal loading and represent 1 out of at least 2 independent experiments (Note: the brighter band detected in TTPKO TAMs is as it appears on the blot and has not been manipulated). B, tumor mass of EG7 thymomas grown in WT (Zfp36flox/flox) and TTPΔM mice for 13 days. Data from individual mice from 3 independent experiments (n ≥ 23) are shown together with the mean and SEM. C, active Caspase 3 staining of representative EG7 tumors isolated from WT and TTPΔM animals. n = 3 animals per genotype. D, flow cytomery analysis of Annexin V+ cells in EG7 thymomas grown in WT and TTPΔM mice. Data represent values from individual mice as percent of total cells from 1 out of 3 experiments (n ≥ 6). E and F, cytokine production in EG7 TAMs (gated on CD11b+) measured by intracellular flow cytometry in WT (Zfp36flox/flox) or TTPΔM animals. Data are % positive cells of CD11b+ (E) or Median Fluorescence Intensity (MFI) of cytokine+CD11b+ cells (F) from individual mice from 1 out of 2–5 experiments (n ≥ 6). The mean is depicted as black line. Error bars, SEM. G, TNF cytokine production detected by ELISA in the supernatants of WT and TTPΔM TAMs after 6 hrs (right panel) or resting or 3 hrs LPS stimulated BMDMs (left panel). The mean is depicted as black line. Error bars, SEM. Values represent TAMs from individual mice (n ≥ 2) from 1 out of 2 experiments.
TTP-dependent mRNA decay is limited in TAMs
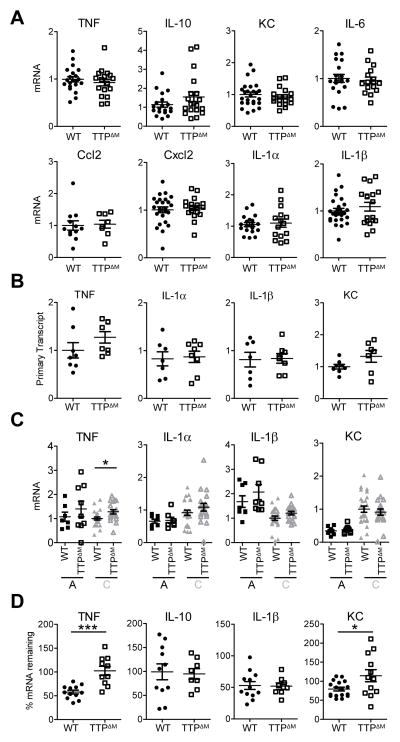
TTP increases the instability of AU-rich mRNAs (6). Given high expression of TTP in macrophages associated with chronic inflammation, we asked if TTP regulated AU-rich mRNA stability. We measured steady state mRNA amounts of AU-rich inflammatory mRNAs in the total CD11b+ myeloid fraction from tumors and found no differences between controls and TTPΔM cells (Fig. 4A). TTP has also been implicated in regulating the transcription status of certain genes, possibly compensating for alterations at the mRNA stability level (28). We therefore assessed transcription by detecting the unspliced mRNA primary transcripts of inflammatory mRNAs in WT and TTPΔM TAMs. Similar to total mRNA amounts we detected no significant changes between control and TTPΔM cells (Fig. 4B). Further, focusing on the TAM subsets where TTP expression is highest, we also failed to detect significant effects on total amounts of normally unstable TTP targets with the exception of the TNF mRNA, which is known to be affected by TTP-dependent mRNA decay even under conditions where TTP function is limited (Fig. 4C) (15). When we estimated the decay rates of TTP targets in control or TTPΔM TAMs we found significant effects on stability only for TNF and KC, the AU-rich mRNAs that show the highest effects of TTP activity in model systems (Fig. 4D) (15). However, the data in Fig. 4A, B and C indicate effects of TTP on mRNA stability were marginal, as the total quantity of TNF and KC mRNAs were equivalent between control and TTPΔM TAMs. Therefore, TTP controls chemokine and cytokine production at a level above transcription or mRNA stability. Thus tumor macrophages had high TTP protein, and TTP was required to inhibit inflammatory cytokine and chemokine production but not their cognate mRNA amounts or stability.
Figure 4. TTP-dependent mRNA decay is limited in TAMs.
A, qRT-PCR analysis of EG7 TAMs isolated from WT and TTPΔM mice. Data from at least 3 experiments were combined and values from individual mice (n ≥ 8) were normalized to the corresponding GAPDH followed by the mean WT within each experiment. The mean per group is shown as black line. Error bars, SEM. B, qRT-PCR analysis of primary transcripts of inflammatory mRNAs in WT and TTPΔM TAMs. Values from individual mice (n ≥ 7) were normalized to GAPDH and mean WT within each experiment and are combined from 2 experiments. Identical samples from each tumor were processed in the absence of reverse transcriptase to serve as controls for genomic DNA contamination. The mean per group is shown as black line. Error bars, SEM. C, qRT-PCR analysis of TTP target mRNAs in EG7 TAM population A and C isolated from WT and TTPΔM mice. Data represent the expression values of individual mice (n ≥ 7) combined from 2–4 experiments. Values were normalized to the corresponding GAPDH and subsequently to the mean of WT TAM-C within each experiment. Mean is shown as black line. Error bars, SEM. D, mRNA stability measured by qRT-PCR by comparing cells before and 90 minutes after transcriptional blockade by Actinomycin D in EG7 TAMs from WT and TTPΔM mice. Values were normalized to GAPDH and represent % cytokine mRNA remaining after Actinomycin D treatment as mean and SEM. Data are expression values from individual mice combined from 2–3 experiments (n ≥ 8). Mean is indicated by a black line. Error bars, SEM.
p38α is constitutively activated in non-resolving inflammation
If TTP is highly expressed in chronic inflammation, why is AU-rich mRNA degradation inefficient? To begin to answer this question we noted TTP activity towards its AU-rich mRNA substrates is negatively regulated by phosphorylation on specific serine and threonine residues (9). Phosphorylation of TTP could therefore account for why AU-rich mRNA quantity and stability were unaffected in macrophages. To test this, we assessed pTTP in TAMs and observed a characteristic ‘smear’ of TTP protein similar to that found in LPS-stimulated BMDMs (8) (Fig. 5A). In the latter, pTTP increases to stabilize inflammatory cytokine and chemokine mRNAs, enhancing their production after which active (unphosphorylated) TTP rises to eliminate AU-rich mRNAs to return macrophages to a ‘ground’ state (7, 10, 15). In accordance with high amounts of pTTP associated with inactive TTP-dependent mRNA decay, the stability of the TNF mRNA was increased in TAMs isolated from different tumor models compared to resting BMDMs where TTP activity is known to be high (Fig. 5B) (15).
Figure 5. Reduced TTP-dependent mRNA decay coincides with chronic p38α activation in TAMs and PAMs.
A, TTP expression analyzed by immunoblotting of whole cell lysates of EG7 TAMs and resting or LPS treated BMDMs. Blots were re-probed using anti-GRB2 antibody to confirm equal loading and represent 1 out of 3 experiments. B, TNF mRNA stability determined by qRT-PCR by comparing cells before and 90 minutes after transcriptional blockade by Actinomycin D in BMDMs and TAMs isolated from EG7 thymomas (T-TAM), neuroblastomas (N-TAM) or osteosarcomas (O-TAM). Values were normalized to GAPDH and represent % TNF mRNA remaining after Actinomycin D treatment as mean and SEM of at least 2 experiments (n ≥ 2). C, immunoblot analysis of total cell lysates of non-stimulated (ns) or 1hr LPS stimulated (LPS) BMDMs as well as TAMs isolated from different tumor models as in B, probed with anti-phospho-p38, anti-p38 or anti-GRB2 as loading control. The blot represents 1 out of 3 experiments. D, p38 phosphorylation analyzed by immunoblotting of whole EG7 TAM lysates from WT TAMs directly after isolation (−) or after resting them on tissue culture dishes (TC) for 3 hrs. E and F, immunoblot analysis of whole EG7 TAM lysates (E) or BMDMs left untreated or stimulated with LPS for 1 and 3 hrs (F) isolated from WT or p38αΔM animals. Blots represent 1 out of 2 experiments. G, Whole EG7 TAM lysates from WT or p38αΔH animals as in (E) analyzed for p38 and p-p38 protein expression by immunoblotting (n = 2). H, flow cytometry analysis of p-p38 in CD11b+ EG7 TAMs isolated from WT or p38αΔH animals (n ≥ 4). Representative plots are shown with unstained control as grey line.
TTP inhibition was reported to be mediated by p38α MAP kinase, the predominant MAP kinase in macrophages, and to be blocked by phosphatases, which act on p38α, and thus indirectly promote TTP function (26, 29). We therefore investigated how the p38α-TTP cycle works in chronic inflammation. First, we observed overall high p-p38α in tumor-derived macrophages despite varying degrees of total p38α (Fig. 5C). p38α activation was constitutive and not a transient effect from the isolation procedure as we could still detect strong p38α phosphorylation in TAMs rested on tissue culture dishes for 3 hours (Fig. 5D). Second, when we genetically eliminated p38α in macrophages with the LysM-Cre deleter (p38αΔM) as previously reported (30), we observed negligible depletion of p38α amounts, arguing conclusions drawn using this deleter system are difficult to interpret (Fig. 5E,F). Therefore, for subsequent experiments we adjusted our tactics and used the pan-hematopoietic deleter Tie2-Cre (p38αΔH), which produced a complete elimination of p38α (Fig. 5G, H).
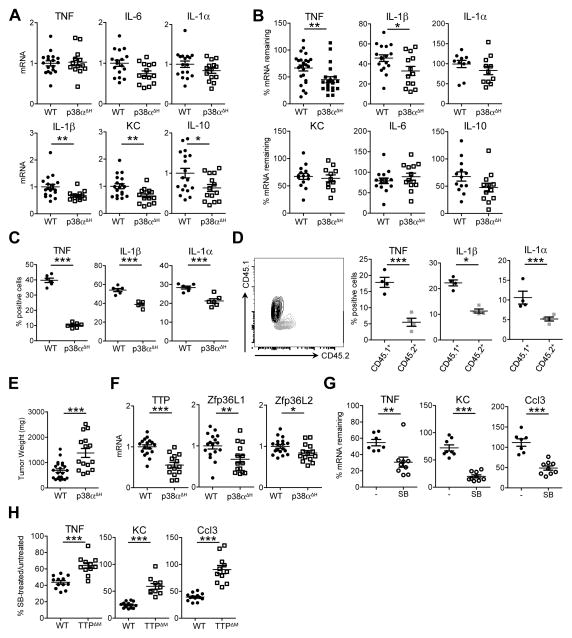
p38α drives chronic inflammation posttranscriptionally
When we measured inflammatory cytokine and chemokine mRNA amounts in p38α-deficient TAMs, we observed no differences between controls and knockouts for transcripts such as TNF, IL-1α and IL-6 and only minor differences for other selected transcripts (Fig. 6A). In the same system, mRNA stability in p38α-deficient TAMs was reduced for TNF and IL-1β mRNA only (Fig. 6B). However, despite unaltered mRNA amounts p38α-deficient cells had a substantial reduction in the amounts of certain inflammatory cytokines such as TNF and IL-1α, suggesting p38α is required for translation of inflammatory mediators (Fig. 6C, Supplementary Fig. S1). In order to exclude the possibility that the detected alterations are due to p38α deletion in non-myeloid cells caused by the use of Tie2-Cre, we performed mixed radiation chimera experiments with 1:1 mixtures of control and p38αΔH bone marrow. In chimeric mice only p38α-deficient macrophages had reduced cytokine production, confirming p38α primarily acts in a macrophage-autonomous way in our chronic inflammation models (Fig. 6D). Returning to the p38αΔH mice, we found the reduction in inflammatory cytokines made by p38α-deficient TAMs correlated with an increase in overall tumor size; the opposite of the phenotype observed in the TTPΔM mice (Fig. 6E compared to Fig. 3B).
Figure 6. p38α regulates TTP expression and activity in TAMs.
A, EG7 TAM RNA from WT or p38αΔH tumor bearing mice was analyzed by qRT-PCR. Expression values from individual mice combined from 3 experiments (n ≥ 13) were normalized to GAPDH and the mean WT within each experiment and are depicted as mean indicated by black lines. Error bars, SEM.
B and G, mRNA stability measured by qRT-PCR as in Figure 4Din EG7 TAMs from WT (Mapk14flox/flox) and p38αΔH animals (B) and in unstimulated (−) or SB203580 (SB) treated WT EG7 TAMs (G). SB treatment was started concomitant to Actinomycin D imposed transcriptional blockade. Data represent values from individual mice normalized to GAPDH from either 3–5 (n ≥ 10, B) or 1 out of 2 (n ≥ 7, G) experiments, respectively. Black lines indicate the mean per group. Error bars, SEM. C, cytokine expression analyzed by flow cytometry of EG7 TAMs (gated on CD11b+) isolated from WT or p38αΔH mice. Values from individual mice are percent of CD11b+ cells (n = 6) representing 1 out of 2 experiments with the mean depicted as black line. Error bars, SEM. D, cytokine production evaluated by intracellular flow cytometry in lethally irradiated mice reconstituted with a mixture of WT (CD45.1) and p38αΔH (CD45.2) bone marrow (n = 4). Depicted values represent % positive cells of CD11b+ TAMs gated for CD45.1 or CD45.2 as shown (left panel). Error bars, SEM. E, EG7 tumor mass 12days post transplantation of EG7 cells into WT or p38αΔH animals. Values from individual mice combined from 3 experiments are shown with the black line representing the mean. Error bars, SEM. F, qRT-PCR analysis of TTP and TTP family members in EG7 TAMs from WT and p38αΔH mice. Data represent expression values from individual mice (n ≥ 14) normalized to GAPDH and the corresponding mean WT from 3 independent experiments. The mean is shown as black line. Error bars, SEM. H, mRNA decay measured in WT and TTPΔM EG7 TAMs with and without SB treatment as in G. Values of individual mice are depicted as % of mRNA stability after SB treatment relative to the corresponding stability in untreated WT or TTPΔM TAMs. Values were normalized to GAPDH and the corresponding untreated TAM sample (n ≥ 9) and were combined from 2 experiments. The black line represents the mean. Error bars, SEM.
As mentioned above, the mRNA stability of several TTP-target transcripts was unaffected by p38α deletion (Fig. 6A, B). However, in accordance with in vitro studies (8), expression of TTP as well as the TTP related proteins ZFP36L1 and ZFP36L2 was p38α-dependent (Fig. 6F). Given a straightforward genetic means to address the role of p38α in regulating TTP activity was negated by the p38α dependence of TTP expression, we used a pharmacological approach to block p38α. When we blocked p38α activity using the kinase inhibitor SB203580, we found AU-rich mRNAs were rapidly degraded (Fig. 6G). In TTP-deficient cells SB203580 treatment only partially increased mRNA decay. For instance, in WT TAMs SB203580 treatment reduced TNF mRNA stability to approximately 43% of untreated TAMs compared to 64% in TTPΔM TAMs suggesting that p38α regulates mRNA stability in a TTP-dependent and independent manner (Fig. 6H). Two conclusions can be drawn from these experiments. First, TTP-dependent mRNA decay in TAMs is blocked by p38α, consistent with previous data generated from in vitro systems. However, TTP is still able to limit protein production of target mRNAs thereby counteracting inflammation. Thus, in tumor inflammation, TTP’s mRNA degrading function is negated by p38α, but TTP is still required to temper inflammatory mediator output. Second, p38α is a driver of chronic inflammation at the posttranscriptional level. Our data suggest that p38α is constitutively activated in TAMs and resistant to the known negative feedback mechanisms described for acute inflammatory responses (12, 31, 32).
Discussion
Genetic experiments have established the basic parameters of TNF production in scenarios where the inflammatory signal is limited. First, p38α cooperates within the TLR signaling pathway to initiate Tnf expression and TNF production (33). In the subsequent resolving phase, a step-wise process cooperates to extinguish TNF production once TLR signaling tapers: DUSP proteins dephosphorylate p38α; the decreased p38α activity releases TTP inhibition (induced by TLR signaling) to degrade the TNF mRNA and IL-10 (induced by TLR signaling) amplifies TTP expression because Zfp36 is a STAT3-dependent target of IL-10 signaling (7, 12, 31, 32). Using conditional knockout approaches to minimize the effects of systemic inflammation, we found that in TAMs none of the step-by-step processes in TNF regulation occurs. Instead, the tumor microenvironment enforces constitutive p38α activation, leading to blockade of TTP activity and abatement of TNF mRNA removal.
TTP is generally considered to primarily control AU-rich mRNA stability (34). Although TTP also contributes to blocking translation of some mRNAs (35), our experiments suggest these two functions are differentially regulated within the tumor microenvironment. We found mRNA stability and total amounts of TTP target mRNAs were marginally affected in the absence of TTP. These results contrast with in vitro studies where TTP influences the stability and amounts of hundreds of AU-rich mRNAs (15, 27). However, the absence of TTP caused inflammatory protein amounts to increase, leading to increased death in the tumor microenvironment and decreased tumor volume. Therefore, in TAMs we suggest that TTP mainly regulates steps beyond mRNA decay such as translation. A further implication of our studies is the two functions of TTP can be separated by the inhibitory actions of chronic p38α activity: TTP expression is constitutively high in TAMs and its effects on mRNA stability are blocked by p38α phosphorylation. However, in contrast to in vitro studies of acute inflammation where p38α blocks both TTP-dependent mRNA decay and translation (35), in TAMs TTP can retain its inhibitory activity on cytokine production. Our results therefore increase the understanding of gene and protein regulation in complex environments.
Our data indicate that p38α is constitutively active during NRI and drives the production of inflammatory cytokines at the posttranscriptional level. What signals lead to p38α activation in TAMs? Multiple NRI models have shown endogenous ‘alarmins’ drive TLR and other inflammatory signaling. Alarmins include S100a8/a9, IL-1α and HMGB1. Release of any of these can provoke TLR/IL-1R signaling leading to Tnf expression (36). However, a consequence of ongoing TLR/IL-1R is activation of p38α phosphorylation and subsequent TTP deactivation, leading to further TNF production and potential for additive TNFR signaling to perpetuate the overall response.
Our data implicate p38α activity as the nexus of inflammatory signaling in TAMs, regardless of what occurs downstream in the pathway. As long as p38α is active, all the subsequent steps leading to signaling abatement cannot occur in a step-wise manner. When we targeted p38α activity with the kinase inhibitor SB203580, inflammatory mRNAs were greatly reduced. Therefore, p38α is a central checkpoint in chronic inflammation. Given NRI inflammation occurs in virtually all chronic diseases, our results point to targeting two aspects of the disease: first the inciting entity must be reduced, modified or eliminated, coincident with targeting upstream inflammatory signaling at the level of p38α.
Supplementary Material
Acknowledgments
Financial Support: F.K. is supported by the Austrian Science fund [J3309-B19]. This work was supported by The Hartwell Foundation (PJM), Alex’s Lemonade Stand Foundation (PJM), National Institutes of Health grants CA189990 (PJM), CA138064 (JEQ), NCI grant P30 CA21765 and the American Lebanese Syrian Associated Charities.
We thank Perry J. Blackshear for generously providing the TTPKO and Michael A. Dyer for the Th-MYCN transgenic and Rbflox/flox;p53flox/flox;Osx-Cre+mouse stains. We thank Martine F. Roussel for providing the Glioma tumor model as well as Lidija Barbaric, Parker Ingle, Greig Lennon and Richard Cross for technical assistance and Derek Gilroy and Roland Lang for discussion and advice.
Footnotes
The authors disclose no potential conflicts of interest.
Author Contributions
F.K. designed the research, did and analyzed experiments and wrote the manuscript; N.G. and J.E.Q. did experiments; L.-A.V. assisted with flow cytometry and immunoblotting; H.C. provided research reagents; P.K. provided advice, animals and reagents; P.J.M conceived the study, designed experiments and wrote the manuscript with FK.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Nathan C, Ding A. Nonresolving inflammation. Cell. 2010;140(6):871–82. doi: 10.1016/j.cell.2010.02.029. [DOI] [PubMed] [Google Scholar]
- 2.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nature reviews Immunology. 2012;12(4):253–68. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138(1):129–45. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, et al. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Molecular cell. 2013;50(2):212–22. doi: 10.1016/j.molcel.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150(2):279–90. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281(5379):1001–5. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 7.Schaljo B, Kratochvill F, Gratz N, Sadzak I, Sauer I, Hammer M, et al. Tristetraprolin is required for full anti-inflammatory response of murine macrophages to IL-10. Journal of immunology. 2009;183(2):1197–206. doi: 10.4049/jimmunol.0803883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahtani KR, Brook M, Dean JL, Sully G, Saklatvala J, Clark AR. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Molecular and cellular biology. 2001;21(19):6461–9. doi: 10.1128/MCB.21.9.6461-6469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, et al. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. The EMBO journal. 2004;23(6):1313–24. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brook M, Tchen CR, Santalucia T, McIlrath J, Arthur JS, Saklatvala J, et al. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Molecular and cellular biology. 2006;26(6):2408–18. doi: 10.1128/MCB.26.6.2408-2418.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaba A, Grivennikov SI, Do MV, Stumpo DJ, Blackshear PJ, Karin M. Cutting edge: IL-10-mediated tristetraprolin induction is part of a feedback loop that controls macrophage STAT3 activation and cytokine production. Journal of immunology. 2012;189(5):2089–93. doi: 10.4049/jimmunol.1201126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammer M, Mages J, Dietrich H, Servatius A, Howells N, Cato AC, et al. Dual specificity phosphatase 1 (DUSP1) regulates a subset of LPS-induced genes and protects mice from lethal endotoxin shock. The Journal of experimental medicine. 2006;203(1):15–20. doi: 10.1084/jem.20051753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4(5):445–54. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 14.Molle C, Zhang T, Ysebrant de Lendonck L, Gueydan C, Andrianne M, Sherer F, et al. Tristetraprolin regulation of interleukin 23 mRNA stability prevents a spontaneous inflammatory disease. The Journal of experimental medicine. 2013;210(9):1675–84. doi: 10.1084/jem.20120707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kratochvill F, Machacek C, Vogl C, Ebner F, Sedlyarov V, Gruber AR, et al. Tristetraprolin-driven regulatory circuit controls quality and timing of mRNA decay in inflammation. Molecular systems biology. 2011;7:560. doi: 10.1038/msb.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishida K, Yamaguchi O, Hirotani S, Hikoso S, Higuchi Y, Watanabe T, et al. p38alpha mitogen-activated protein kinase plays a critical role in cardiomyocyte survival but not in cardiac hypertrophic growth in response to pressure overload. Molecular and cellular biology. 2004;24(24):10611–20. doi: 10.1128/MCB.24.24.10611-10620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. The EMBO journal. 1997;16(11):2985–95. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes & development. 2008;22(12):1662–76. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stables MJ, Shah S, Camon EB, Lovering RC, Newson J, Bystrom J, et al. Transcriptomic analyses of murine resolution-phase macrophages. Blood. 2011;118(26):e192–208. doi: 10.1182/blood-2011-04-345330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawauchi D, Robinson G, Uziel T, Gibson P, Rehg J, Gao C, et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer cell. 2012;21(2):168–80. doi: 10.1016/j.ccr.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Movahedi K, Laoui D, Gysemans C, Baeten M, Stange G, Van den Bossche J, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C (high) monocytes. Cancer research. 2010;70(14):5728–39. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 23.O’Sullivan T, Saddawi-Konefka R, Vermi W, Koebel CM, Arthur C, White JM, et al. Cancer immunoediting by the innate immune system in the absence of adaptive immunity. The Journal of experimental medicine. 2012;209(10):1869–82. doi: 10.1084/jem.20112738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bain CC, Scott CL, Uronen-Hansson H, Gudjonsson S, Jansson O, Grip O, et al. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal immunology. 2013;6(3):498–510. doi: 10.1038/mi.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai WS, Carrick DM, Blackshear PJ. Influence of nonameric AU-rich tristetraprolin-binding sites on mRNA deadenylation and turnover. The Journal of biological chemistry. 2005;280(40):34365–77. doi: 10.1074/jbc.M506757200. [DOI] [PubMed] [Google Scholar]
- 26.Tudor C, Marchese FP, Hitti E, Aubareda A, Rawlinson L, Gaestel M, et al. The p38 MAPK pathway inhibits tristetraprolin-directed decay of interleukin-10 and pro-inflammatory mediator mRNAs in murine macrophages. FEBS letters. 2009;583(12):1933–8. doi: 10.1016/j.febslet.2009.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, et al. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. The Journal of biological chemistry. 2008;283(17):11689–99. doi: 10.1074/jbc.M709657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schichl YM, Resch U, Hofer-Warbinek R, de Martin R. Tristetraprolin impairs NF-kappaB/p65 nuclear translocation. The Journal of biological chemistry. 2009;284(43):29571–81. doi: 10.1074/jbc.M109.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammer M, Mages J, Dietrich H, Schmitz F, Striebel F, Murray PJ, et al. Control of dual-specificity phosphatase-1 expression in activated macrophages by IL-10. European journal of immunology. 2005;35(10):2991–3001. doi: 10.1002/eji.200526192. [DOI] [PubMed] [Google Scholar]
- 30.Chakraborty M, Lou C, Huan C, Kuo MS, Park TS, Cao G, et al. Myeloid cell-specific serine palmitoyltransferase subunit 2 haploinsufficiency reduces murine atherosclerosis. The Journal of clinical investigation. 2013;123(4):1784–97. doi: 10.1172/JCI60415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chi H, Barry SP, Roth RJ, Wu JJ, Jones EA, Bennett AM, et al. Dynamic regulation of pro- and anti-inflammatory cytokines by MAPK phosphatase 1 (MKP-1) in innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2274–9. doi: 10.1073/pnas.0510965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Q, Wang X, Nelin LD, Yao Y, Matta R, Manson ME, et al. MAP kinase phosphatase 1 controls innate immune responses and suppresses endotoxic shock. The Journal of experimental medicine. 2006;203(1):131–40. doi: 10.1084/jem.20051794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nature reviews Cancer. 2009;9(8):537–49. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 34.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Molecular and cellular biology. 1999;19(6):4311–23. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, et al. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS genetics. 2012;8(9):e1002977. doi: 10.1371/journal.pgen.1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan JK, Roth J, Oppenheim JJ, Tracey KJ, Vogl T, Feldmann M, et al. Alarmins: awaiting a clinical response. The Journal of clinical investigation. 2012;122(8):2711–9. doi: 10.1172/JCI62423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.