Abstract
Recombination is a central process to stably maintain and transmit a genome through somatic cell divisions and to new generations. Hence, recombination needs to be coordinated with other events occurring on the DNA template, such as DNA replication, transcription, and the specialized chromosomal functions at centromeres and telomeres. Moreover, regulation with respect to the cell-cycle stage is required as much as spatiotemporal coordination within the nuclear volume. These regulatory mechanisms impinge on the DNA substrate through modifications of the chromatin and directly on recombination proteins through a myriad of posttranslational modifications (PTMs) and additional mechanisms. Although recombination is primarily appreciated to maintain genomic stability, the process also contributes to gross chromosomal arrangements and copy-number changes. Hence, the recombination process itself requires quality control to ensure high fidelity and avoid genomic instability. Evidently, recombination and its regulatory processes have significant impact on human disease, specifically cancer and, possibly, neurodegenerative diseases.
The process of recombination requires quality control to ensure high fidelity and avoid genomic instability. It must be coordinated with other events occurring on the DNA template, during the cell cycle, and within the nucleus.
Homologous recombination (HR) is a high-fidelity DNA-damage-repair and tolerance pathway that is critical for complex DNA damage such as DNA double-stranded breaks (DSB) or interstrand cross-links (Krogh and Symington 2004; Wyman and Kanaar 2006; Heyer 2007; Filippo et al. 2008; Deans and West 2011). Moreover, recombination is inextricably linked to DNA replication, acting on stalled or broken replication forks to repair replication-associated single-stranded DNA (ssDNA) gaps and one-sided DSBs as well as to tolerate base damage that stalls the progress of replicative DNA polymerases (Cox et al. 2000; Wu and Hickson 2006; Li and Heyer 2008; Malkova and Haber 2012). Finally, HR is essential for meiotic chromosome segregation by generating crossovers, which impart a physical linkage between homologous chromosomes that is critical for accurate chromosome segregation in the first meiotic division` (Zickler and Kleckner 1999; Hunter 2007; Sasaki et al. 2010; Baudat et al. 2013).
REGULATION WITH COMPETING EVENTS ON CHROMOSOMES
HR does not happen in a vacuum and the cell requires coordination of different DNA-repair pathways targeting the same DNA lesion as well as coordination between DNA repair and other processes using the same chromosomal template (Symington and Gautier 2011; Chapman et al. 2012; Aguilera and Garcia-Muse 2013).
Template Choice and Pathway Control
Role of Cohesion on Template Choice
HR is a template-dependent process and in somatic cells there is a significant bias toward the sister chromatid (Kadyk and Hartwell 1992), although there is evidence for HR between homologs in the G1 phase of the cell cycle (Fabre 1978). In meiosis, the primary objective is to establish crossover between homologs, although many meiotic DSBs do not lead to crossover formation and may ultimately be repaired by sister chromatid template HR (see section Genome Maintenance and Human Disease) (Hunter 2007). Sister chromatid cohesion ensures the spatial proximity of the template and additional cohesion complexes are loaded after DSB formation, likely to further favor and stabilize sister interactions (Ström et al. 2004, 2007; Ünal et al. 2004, 2007; Ström and Sjogren 2005; Cortes-Ledesma and Aguilera 2006). Replication-associated cohesion involves acetylation of the Smc3 subunit of cohesion by Eco1, which is induced in S phase and suppressed outside S phase (Ünal et al. 2008). However, also DSB-triggered cohesion loading is Eco1 dependent and occurs outside S phase (Ünal et al. 2007). Suppression of cohesion loading outside S phase may be partly overcome by the ubiquitous DSB mark γ-H2AX (Ünal et al. 2004) and also involves phosphorylation of Ser83 of the Kleisin subunit Scc1 (Mdc1) by the DNA damage response kinase Chk1 (Heidinger-Pauli et al. 2008). Sister chromatid preference and its enforcement by sister cohesion also counteracts ectopic recombination, which could lead to genomic rearrangements.
DSB-Repair Pathway Choice or How Resection Commits to HR
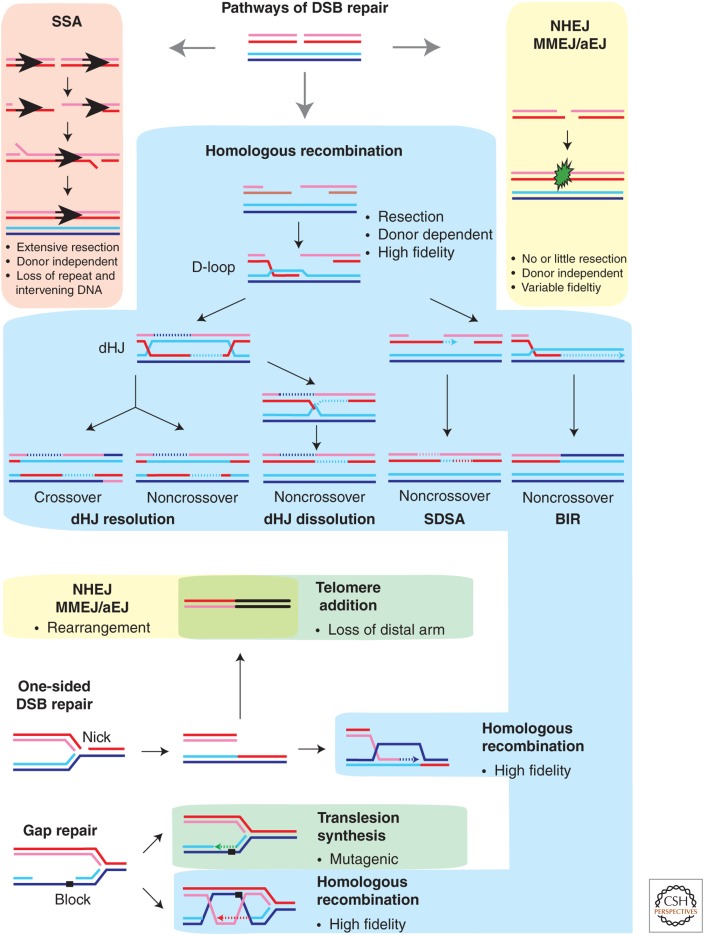
The cell-cycle-specific regulation of HR to enforce utilization of the sister chromatid as template involves multiple mechanisms. Besides the DNA damage-induced loading of cohesins discussed above, control of DSB end resection appears to be a critical component. The complexity of the DSB resection machineries has only recently been established by genetic and biochemical approaches (see Symington 2014). A key conserved target for cell-cycle-dependent kinases in the end-resection machinery was identified as yeast Sae2 and its mammalian homolog CtIP (Huertas et al. 2008; Huertas and Jackson 2009). Sae2/CtIP cooperate with the Mre11/Rad50/Xrs2 (mammalian MRE11/RAD50/NBS1) complex to provide the initial resection of DSBs. It is unclear whether Sae2/CtIP phosphorylation affects the MRE11-associated exo- and endonuclease activities, which differentially affect pathway choice in DSB repair (Lengsfeld et al. 2007; Cannavo and Cejka 2014; Shibata et al. 2014). In addition, yeast Dna2, a nuclease/helicase that cooperates with Sgs1 and RPA in long-range resection, is recruited to DSBs in a CDK-phosphorylation-dependent manner (Chen et al. 2011). Among the DSB-repair pathways, HR- and single-strand annealing (SSA) require extensive resection, whereas Nonhomologous endjoining (NHEJ) and alternative endjoining/microhomology-mediated endjoining (aEJ/MMEJ) require little or no resection (Fig. 1).
Figure 1.
Homologous recombination (HR) and pathway choices. The pathways of double-strand break (DSB) repair include nonhomologous endjoing (NHEJ), alternative endjoining (aEJ, also called microhomology-mediated endjoining), which are differentiated whether the joint involves no or few nucleotides (1–5 nt) or greater (∼5–25 nt) homology. Single-strand annealing (SSA) is possible when the DSB is flanked by direct repeat sequences and requires extensive resection. HR includes several subpathways including break-induced replication (BIR), which leads to loss of heterozygosity, synthesis-dependent strand-annealing (SDSA), which leads to a noncrossover outcome, and the double Holliday junction pathway (dHJ), which through nucleolytic resolution of dHJs generates crossover and noncrossovers outcomes. Dissolution of dHJs, in contrast, leads exclusively to noncrossover products. One-sided DSBs result from passing of a replication fork through a nicked template. DSB repair by NHEJ/aEJ will lead to a genomic rearrangement, whereas HR repairs such damage with high fidelity. Addition of telomeres to any DSB leads to loss of the distal chromosomal DNA. Replication-associated DNA gaps can result from lesions that block DNA polymerases. Bypass of such lesions by translesion DNA polymerases (translesion synthesis) is mutagenic, whereas HR provides lesion tolerance with high fidelity. For replication forks with their leading strand blocked, fork regression (formation of a chicken foot) and the associated template switch provides another pathway of lesion bypass (not shown). MMEJ, Microhomology-mediated endjoining.
Elaborate control of DSB end resection appears to be the critical point of DSB pathway choice. As detailed in Symington (2014), this regulation involves extensive chromatin remodeling, histone modifications, the Ku70–80 heterodimer, the checkpoint adaptor Rad9 (yeast) or its homologs 53BP1 (mammals), and RIF1. It appears that control of DSB end resection represents a key difference between DSB repair in yeast and mammalian cells, although the end resection machineries are largely conserved. Although in yeast, DSB end resection is primarily controlled by cell-cycle signals, mammalian cells subject this decision to additional levels of regulation including cell/tissue specific as well as spatial cues (see below for heterochromatin). On resected DSB ends, it is unclear how pathway control between HR and SSA is achieved. In a plasmid-based system with a DSB in the ADE2 gene with 15 bp complementary overhangs and no available homologous template, Nej1, a DNA ligase 4 cofactor involved in NEHJ and aEJ/MMEJ (XLF1/cernunnos in humans) (Deriano and Roth 2013), was found to recruit Srs2 to dissociate Rad51 filaments and bias repair toward Rad52-dependent SSA (Carter et al. 2009). Dun1-mediated phosphorylation on Nej1-S297/S298 was required for this effect (Carter et al. 2009). This suggests the involvement of an appreciable DNA-damage response in repair of the DSB in this system, unlike DSBs that can be repaired off a template by HR in which repair is not accompanied by full activation of the DNA damage response (Vaze et al. 2002; Harrison and Haber 2006). The kinetic delay resulting from unproductive homology search may have triggered the DNA damage response that favors as a consequence SSA after having attempted HR. Interestingly, Nej1 is the target of transcriptional down-regulation to inhibit NHEJ in diploid MATa/α budding yeast cells (Frank-Vaillant and Marcand 2001; Kegel et al. 2001; Valencia et al. 2001). As Nej1 acts late in the reaction at the ligation step in conjunction with DNA ligase 4 and its cofactor Lif1 (XRCC4 in humans) (Callebaut et al. 2006; Chen and Tomkinson 2011), it may suggest that earlier end-joining steps do not preclude subsequent HR. In sum, end resection appears to be the critical regulation in DSB-repair pathway control (Symington and Gautier 2011). An additional and poorly understood feature appears to be DSB-repair kinetics and the lifetime of the DNA damage. Real-time assays rather than genetic endpoint assays will be critical to elucidate the mechanisms determining the kinetic hierarchy in DSB repair (Uphoff and Kapanidis 2014).
Replication forks encountering a nick will generate one-sided DSBs (Fig. 1), a particularly pernicious lesion, as repair by NHEJ or aEJ/MMEJ will inevitably lead to genomic rearrangements. Interestingly, the Fanconi pathway (Kim and D’Andrea 2012) appears to be involved in inhibiting NHEJ in S phase (Adamo et al. 2010; Pace et al. 2010). This activity was unveiled by finding that Caenorhabditis elegans or human FANCD2-deficient cells are substantially suppressed for their interstrand cross-link sensitivity by eliminating NHEJ. Although this is likely not the only function of the Fanconi pathway, these findings underline the importance of pathway choice in DSB repair. The precise mechanism, by which the Fanconi pathway suppresses NEHJ during DNA replication, still needs to be established.
Template lesions on the leading or lagging strand can lead to the formation of replication-associated gaps (Fig. 1). The blocking lesion can be tolerated and the gap repaired postreplicationally by either specialized DNA polymerases performing translesion DNA synthesis or HR (Li and Heyer 2008). Translesion DNA synthesis accounts for a good proportion of the spontaneous single-base mutations in yeast, indicated by the lower spontaneous and DNA damage–induced mutation rates of rev3 mutants defective for the catalytic subunit of DNA polymerase ζ compared with wild type (Lemontt 1971; Lawrence and Christensen 1976; Nelson et al. 1996; Sakamoto et al. 2007). HR repairs gaps with high fidelity using the sister chromatid as a template. However, DSB repair by HR is not error free and is associated with mutagenesis (Strathern et al. 1995; Yang et al. 2008; Malkova and Haber 2012). Pathway choice between translesion DNA synthesis, template switch by fork regression or HR appears to be controlled by several pathways. First, monoubiquitylation of proliferating cell nuclear antigen (PCNA) on K164 by the Rad6–Rad18 complex provides an additional binding site for translesion DNA polymerases; for example, for DNA polymerase η, favoring the exchange between a replicative DNA polymerase (δ or ɛ) and a translesion DNA polymerase (Hoege et al. 2002; Stelter and Ulrich 2003; Prakash et al. 2005; Moldovan et al. 2007; Sale et al. 2012). Second, further ubiquitylation of this residue by Mms2-Ubc13-Rad5 complex favors a template-switching pathway by fork regression (Hoege et al. 2002; Stelter and Ulrich 2003). Third, the antirecombinase Srs2 is recruited to PCNA, when it is sumoylated on K127 (and K164), to actively repress HR by disassembling the Rad51-ssDNA nucleoprotein filament (Schiestl et al. 1990; Krejci et al. 2003; Veaute et al. 2003; Papouli et al. 2005; Pfander et al. 2005). Hence, mutants unable to ubiquitylate PCNA display high sensitivity to fork-stalling agents, which can be substantially suppressed by derepressing HR through elimination of Srs2 (Schiestl et al. 1990; Papouli et al. 2005; Pfander et al. 2005). These genetic observations may suggest that, in yeast, translesion synthesis or fork regression is a preferred pathway choice over HR at stalled forks or replication-associated gaps.
In sum, template choice (i.e., sister chromatid preference) in somatic cells and DSB-repair pathway choice implicate the same key step, DSB end-resection, to repress HR in the G1 phase of the cell cycle and control DSB pathway choice throughout the cell cycle. Regulation of pathway choice is critical to control mutagenesis and avoid genomic rearrangements.
Transcription and Connections to RNA Processing and Nuclear Export
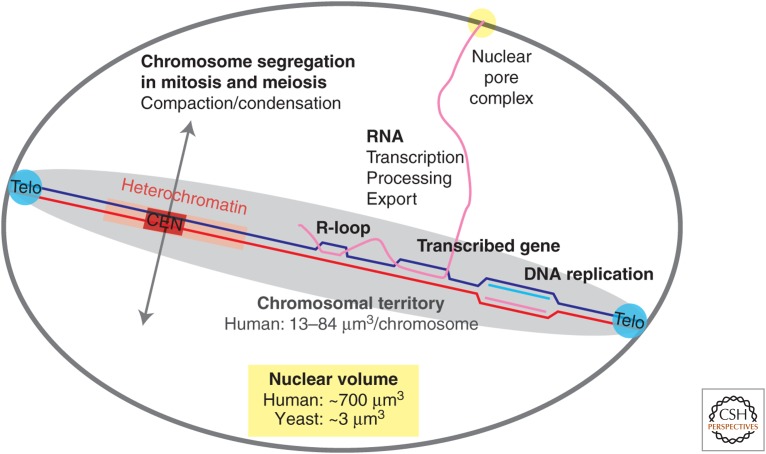
Transcription shares the same substrate with HR (Fig. 2), and the mechanisms by which transcription induces HR are authoritatively discussed by Aguilera and Gaillard (2014). A major source for recombinogenic structures (single-strand gaps or DSBs) appears to derive from encounters of replication forks with transcription complexes. The R-loop, a structure in which a single-stranded RNA displaces one of the two strands in the duplex DNA to generate a structure similar to a D-loop (Figs. 1 and 2), is considered to be the key intermediate. R-loops are more stable than their DNA counterpart because of the increased stability of the RNA:DNA hybrid compared with the same sequence duplex DNA (Roberts and Caruthers 1992). Although previously considered to be a rare side product, R-loops are common during transcription and have also been found to be involved in epigenetic regulation affecting DNA methylation (Ginno et al. 2012). The continuum of RNA transcription, processing, and nuclear export poses additional topological problems that affect genomic maintenance (Bermejo et al. 2011; Kim and Jinks-Robertson 2012).
Figure 2.
Homologous recombination (HR) and other nuclear processes. Other processes compete with HR on the chromosomal template (shown are the Watson and Crick strands of a single chromosome) including RNA transcription, DNA replication, and other DNA-repair pathways (not shown). Regions, such as telomeres (Telo), centromeres (CEN), and heterochromatin, pose challenges for the accessibility by the HR machinery. Chromosome compaction and condensation during mitotic and meiotic chromosome segregation likely interfere with HR. Chromosomes reside in a specific territory of 1.8%–12% of the nuclear volume (human HT-1080 cells [Müller et al. 2010]). The nuclear volume estimate for yeast is from Jorgensen et al. (2007) and for human HeLa cells from Monier et al. (2000).
DNA Replication and Recombination
The intimate linkage between DNA replication and recombination was first appreciated in bacteriophage T4 by Mosig (1998), and the fundamental importance of recombination and HR factors in DNA replication is now generally accepted as elegantly detailed in Syeda et al. (2014). Stalling of replication forks, conflicts with transcription (see above) (Aguilera and Gaillard 2014), and certain chromosomal regions that are “difficult” to replicate (e.g., fragile sites [Barlow et al. 2013] and telomeres [see below]) constitute recombinogenic events that generate gross chromosomal rearrangements (Kolodner et al. 2002). A particular problem specific to eukaryotes and organisms with linear genomes is replication termination. Although circular genomes have defined termination sites and dedicated mechanisms (Neylon et al. 2005), equivalent systems are lacking in eukaryotes. On one hand, this provides greater flexibility in eukaryotes, as replication forks will eventually converge. On the other hand, in chromosomal areas with sparse origins of replication, this leads to problems of unreplicated regions at the time of chromosome segregation resulting in anaphase bridges (Chan et al. 2009). Such sites correspond to common fragile sites that are strongly associated with genetic instability (Letessier et al. 2011).
Telomere Function and Recombination
Telomeres are multifunctional protein–DNA assemblies composed of telomeric repeat DNA and proteins collectively called the shelterin complex. The evolution of linear genomes requires mechanisms to distinguish the natural DSBs at the end of chromosomes from accidental DSBs, which represent DNA damage. Doksani and de Lange (2014) provide a comprehensive discussion of how telomeres shield DNA ends from NHEJ to avoid genomic rearrangements, control HR to maintain telomere length homeostasis, prevent DNA damage signaling to preclude improper signaling, and protect chromosome ends from end resection. Of particular interest is the involvement of HR in the telomerase-independent maintenance of telomeres, termed “alternative lengthening of telomeres” (ALT), which maintains telomeres in a subset of human cancers (Bryan et al. 1997). The mechanism has the hallmarks of the break-induced replication (BIR) pathway of HR (Fig. 1). Cells of the human osteosarcoma cell line U2OS maintain their telomeres by ALT, and in these cells the MUS81 gene, encoding the catalytic subunit of MUS81-EME1 structure-selective endonuclease (see Wyatt and West 2014), becomes essential for growth (Zeng et al. 2009). This may indicate a role of MUS81-EME1 in ALT directly, but could also be related to replication problems associated with the larger telomeres often found associated with ALT (Bryan et al. 1995). Replication through telomeric repeats and subtelomeric DNA has been shown to be slow and prone to stalling requiring the Rrm3 helicase (Ivessa et al. 2002). In fact, in mammals, telomeres are considered fragile sites (Sfeir et al. 2009). The importance of telomere function and its associated proteins is further underlined by the finding that elimination of the Pif1 helicase, which restricts telomerase activity (Schulz and Zakian 1994; Vannier et al. 2012), leads to rampant telomere addition at interstitial DSBs induced by endonuclease cleavage (see Fig. 1) (Myung et al. 2001).
Centromere Function and Recombination in Heterochromatin
Centromeres are large protein–DNA complexes that are essential for mitotic and meiotic chromosome segregation (Talbert and Henikoff 2010). In most organisms, such as humans, but not in the budding yeast Saccharomyces cerevisiae, centromeres are associated with large expanses of satellite repeat DNA (100–250 Mbp in humans) that is packaged into constitutive heterochromatin (Talbert and Henikoff 2010). Centromeres were found to block crossovers, as noticed in the earliest genetic mapping studies (Sax 1930; Beadle 1932). Because HR between repeated DNA can lead to genomic rearrangements (George and Alani 2012), it was assumed that centromeres and heterochromatic repeat regions were devoid of HR, but recent discoveries provide intriguing new insights. First, centromeres do participate in HR but centromeric HR is specifically restricted to gene conversion (Shi et al. 2010). Second, it was shown that DSB repair in heterochromatic regions was specifically channeled to HR-mediated repair (Chiolo et al. 2011). This seminal counterintuitive finding discovered that the broken chromosome (and presumably its unbroken sister chromatid template) is extruded from the heterochromatic subcompartment to insulate the repair process from other repeat templates to avoid genomic rearrangements. The factors that govern HR in heterochromatin, the control to isolate the broken chromosome and its sister template, and the process to reassemble heterochromatin after DSB repair are presently poorly understood and represent exciting opportunities for new discoveries (Soria et al. 2012).
REGULATION AND COORDINATION WITH NUCLEAR STRUCTURE AND FUNCTION
The discovery that RAD51 and other HR proteins assemble into microscopically visible foci in response the DNA damage paired with genetic analysis enabled to develop a spatiotemporal “choreographic” view of HR as brilliantly discussed by Lisby and Rothstein (2015). The single-cell resolution complements the analysis of genetic endpoints, such as survival or recombination rates and in vivo biochemical approaches, demonstrating that not only assembly but also disassembly of HR intermediates is critical (Symington and Heyer 2006). Tagging HR proteins and the DNA damage sites they act on revealed unanticipated dynamics not only of the damaged chromosome but also undamaged chromosomes in response to DSB formation (Dion et al. 2012; Krawczyk et al. 2012; Mine-Hattab and Rothstein 2012). The mechanisms enabling this dynamics are not yet understood, and it is unclear how these processes relate to chromosomal territories (Fig. 2). The significance of overall nuclear architecture is also illustrated by the observation that tethering of telomeres and centromeres reduces the efficiency of recombination between distant genomic loci, reducing the risk of ectopic recombination (Agmon et al. 2013). The nuclear pore complex has been identified as a hub involved in organizing genome architecture, affecting DNA topology, and sequestering long-lived DSBs (Fig. 2) (Bermejo et al. 2012). New optical and cell biological techniques discussed by Lisby and Rothstein (2015) will further push the resolution limit and provide insights into HR in specific nuclear subcompartments, such as heterochromatin (see above).
The regulation of HR in the context of the nuclear space and function faces specific challenges during chromosome segregation in the mitotic and meiotic divisions. The typical human chromosome is compacted ∼10,000-fold to allow spindle-based chromosome transport (Hirano 2000; Luger et al. 2012). It appears unlikely that HR can occur on such highly condensed substrates. However, the discovery the HR may be the predominant DSB-repair pathway for heterochromatic DSBs (Goodarzi et al. 2010; Chiolo et al. 2011) suggests that specific mechanisms may exist for HR to act on highly condensed substrates. Although chromosome condensation may provide an intuitive barrier to HR, in turn, HR has been shown to affect chromosome condensation. Human cells with defects in processing HR-dependent joint molecules (e.g., BLM-deficient cells with concurrent depletion of structure-selective endonucleases) show defects in chromosome condensation (Wechsler et al. 2011; Wyatt and West 2014). It is unclear presently which mechanisms are at work, but one would suspect the three classes of SMC protein complexes to be involved (cohesins, condensins, and the Smc5/Smc6 complex). In particular, the Smc5–Smc6 complex plays an essential role in HR, which is currently poorly understood (Ström and Sjogren 2007; Torres-Rosell et al. 2007; Copsey et al. 2013; Xaver et al. 2013). As discussed by Lisby and Rothstein (2015), the cell biological analysis of HR is still in its early stages and full of promise to elucidate the mechanisms controlling HR dynamics and its spatial control.
PATHWAY CONTROL OF HOMOLOGOUS RECOMBINATION
Homologous Recombination Subpathway Control
HR competes with other pathways to repair/tolerate DNA damage, and as discussed above, DSB resection appears to be a critical control point. Subpathway control within the HR process differentiates between BIR, synthesis-dependent strand-annealing (SDSA), and mechanisms involving double Holliday junction (dHJ), which determines crossover or noncrossover outcome (Fig. 1) (see Mehta and Haber 2014). Although dHJs are formed during somatic and meiotic HR, their processing differs, leading to crossover in meiosis and both outcomes in somatic cells (Schwacha and Kleckner 1995; Bzymek et al. 2010; Bizard and Hickson 2014; Wyatt and West 2014; Yim et al. 2014). It is unclear, presently, what determines whether the initial D-loop intermediate is processed to a dHJ or to enter the SDSA subpathway. BIR is not or rarely observed in the repair of a frank (two-sided) DSB, precluding loss-of-heterozygosity associated with the process (Fig. 1). Experiments in yeast have shown that the absence of a second DSB end causes a long kinetic delay in the onset of recombination-associated DNA synthesis (Jain et al. 2009). The mechanisms involved remain to be determined. A role of Sgs1 has been identified in suppressing BIR (Jain et al. 2009), which may involve signaling to the DNA polymerase(s) or a change in the invasion intermediate.
Quality Control by Pathway Reversibility
The functions of HR in DNA repair, DNA damage tolerance, DNA replication, and meiotic recombination support genome stability during ontogenic development and in the propagation of the genome through generations. However, it has also been recognized that HR also has the potential to induce genome instability, acting inappropriately or in an unregulated fashion (Elliott and Jasin 2002; Kolodner et al. 2002). For example, ectopic recombination between repeated DNA is a potent driver of chromosomal rearrangements, as suspected from genomic analysis and established in genetic model systems (Szankasi et al. 1986; Cooper et al. 1997; Agarwal et al. 2006; Haber 2006; Weinstock et al. 2006; Putnam et al. 2009; Song et al. 2014). Furthermore, the identification of synthetic lethality in double mutants that depend on HR, called recombination-dependent lethality, shows that uncontrolled HR leads to potentially toxic intermediates and cell death (Heude et al. 1995; Schild 1995; Gangloff et al. 2000; Fabre et al. 2002; Bastin-Shanower et al. 2003).
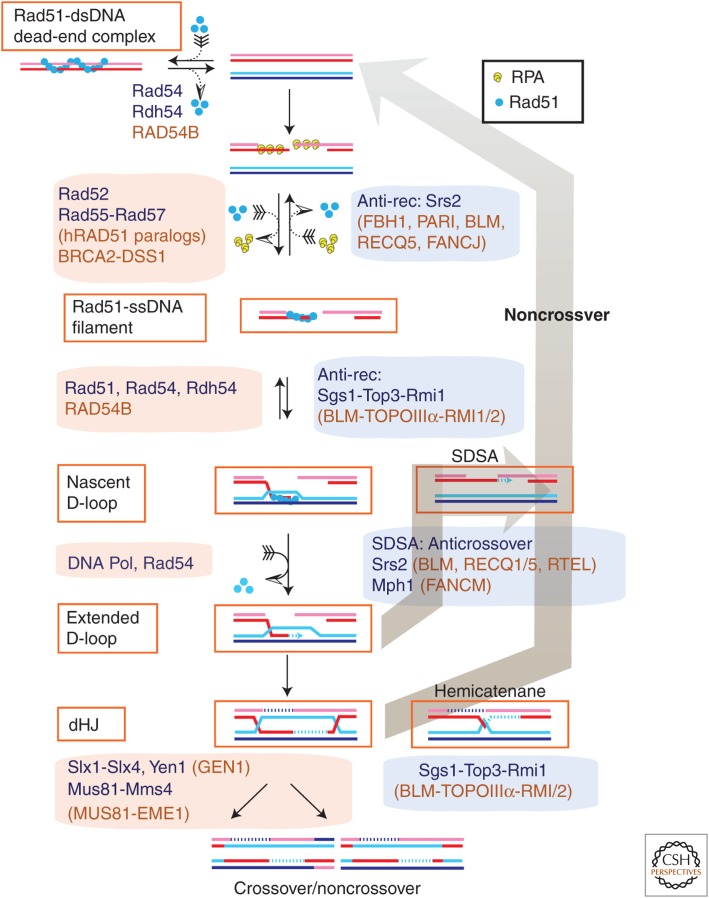
Two critical HR intermediates are subject to reversal and exist in a balance of the forward and backward reactions: the Rad51–ssDNA filament and the nascent D-loop (Fig. 3). Genetic analysis implicates one of these or both as the potentially lethal HR intermediate. Targeting these early HR intermediates avoids recombination-associated DNA synthesis, which could lead to genomic rearrangements or mutations.
Figure 3.
Pathway reversibility in homologous recombination. Homologous recombination (HR) involves key intermediates that are reversible and metastable including (1) the Rad51–ssDNA filament, (2) the initial displacement-loop (D-loop), (3) the extended D-loop, and (4) the double Holliday junction (dHJ). The dead-end complex of Rad51/Dmc1 with dsDNA, although not an HR intermediate, can be added to this list of reversible HR protein–DNA complexes (Holzen et al. 2006). Enzymes acting in the forward reactions are shown with red shading and enzymes active in the backward reactions are shown with blue shading. The yeast enzymes are shown in black, and different or unique human enzymes are shown in orange. ssDNA, Single-stranded DNA; SDSA, synthesis-dependent strand annealing; anti-rec, anti-recombination. (From Heyer et al. 2010; adapted and modified, with permission, from the author.)
Reversibility at the Rad51 Nucleoprotein Filament Stage: Restricting HR
The assembly of the Rad51–ssDNA filaments is a surprisingly complex reaction that involves replacement of the ssDNA-binding protein (SSB, RPA) by Rad51 assisted by mediator proteins (see Zelensky et al. 2014). The budding yeast Srs2 helicase has set the paradigm for reversing HR by dissociating Rad51 from ssDNA, providing a satisfying mechanism for the antirecombination function of Srs2 (Aboussekhra et al. 1989, 1992; Schiestl et al. 1990; Krejci et al. 2003; Veaute et al. 2003). The Rad51 paralog complex Rad55–Rad57 was found to counteract Srs2-mediated disruption of the Rad51–ssDNA filament (Liu et al. 2011a), suggesting a dynamic balance between the forward and backward reactions that may additionally be modulated by PTMs. Several mammalian proteins have been implicated in dissociating RAD51 from ssDNA (Heyer et al. 2010). The human FBH1 helicase contains a UvrD helicase-related motor domain like Srs2 and can functionally replace budding yeast Ssr2 in its antirecombination function (Chiolo et al. 2007). Curiously, this required the F-box domain of FBH1, which functions as an ubiquitin ligase, but is absent in Srs2 (Chiolo et al. 2007). Also genetic results in the fission yeast Schizosaccharomyces pombe support a role of its FBH1 homolog in antirecombination dissociating Rad51 from ssDNA and triggering Rad51 degradation (Osman et al. 2005; Tsutsui et al. 2014). Depletion of FBH1 in human cells or disruption of the gene in mice led to an accumulation of RAD51 on chromatin and hyperrecombination, whereas overexpression impaired RAD51 recruitment and suppressed HR (Fugger et al. 2009; Simandlova et al. 2013). Also, direct biochemical evidence supports the similarity between Srs2 and FBH1, showing that human FBH1 displays translocase-dependent disruption of RAD51–ssDNA filaments (Simandlova et al. 2013). Another UvrD-helicase domain-containing protein, PARI, has been found to destabilize RAD51–ssDNA filaments and, like Srs2, interacts with PCNA (Papouli et al. 2005; Pfander et al. 2005; Moldovan et al. 2012). Although PARI itself lacks translocase activity and appears to act by competitive binding with RAD51 (Moldovan et al. 2012), it may act in conjunction with another motor protein. Human BLM has been shown to dissociate RAD51 from ssDNA, specifically after ATP turnover was triggered by the chelation of Ca2+, which renders the filament nonfunctional for homology search and DNA-strand invasion (Bugreev et al. 2007). The FANCJ helicase inhibits RAD51-mediated D-loop formation by dissociating RAD51 from ssDNA (Sommers et al. 2009). Likewise, the RECQ5 helicase was found to dissociate RAD51 from ssDNA in an ATP-dependent fashion and involving protein interaction with RAD51 (Hu et al. 2007; Schwendener et al. 2010). The biological relevance and specificity of the biochemical activity dissociating RAD51 from ssDNA still needs to be established for some of these proteins. The number of proteins suggests additional complexity in humans compared with budding yeast, in which only a single protein, Srs2, has been described to dissociate Rad51 from ssDNA.
Some of this complexity could be the result of a second function of the RAD51–ssDNA filaments in vertebrate cells during DNA replication. Experiments in Xenopus cell extracts showed that formation of RAD51 filaments at stalled replication protects from degradation by the Mre11 nuclease (Hashimoto et al. 2010). Similar observations were made in BRCA2-deficient mammalian cells, in which fork protection was found to be independent of RAD54, suggesting that it is the RAD51–ssDNA filament structure and not its ability for DNA strand invasion that protects from Mre11 degradation (Schlacher et al. 2011). The exact mechanism of fork protection from Mre11 nuclease and how it might enable fork restart remains to be established.
Finally, the DNA-binding properties of the two major eukaryotic DNA-strand exchange proteins, Rad51 and its meiosis-specific paralog Dmc1, pose a challenge. Both proteins form the active nucleoprotein filaments on ssDNA but also bind dsDNA with an affinity that is only slightly less than their ssDNA binding (see Morrical 2014; Brown and Bishop 2015). Thus, Rad51 and Dmc1 form dead-end complexes on duplex DNA, unless they are constantly dissociated by the motor proteins Rad54 and Rdh54/Tid1 (Solinger et al. 2002; Holzen et al. 2006; Shah et al. 2010). Although such dead-end complexes are not direct intermediates in the HR pathway, they illustrate the need for reversibility of DNA complexes formed by HR proteins.
Reversibility of Nascent D-Loops: A Mechanism of Antirecombination
HR is exquisitely sensitive to heterology between the two recombining DNA molecules. Spies and Fishel (2015) provide a comprehensive discussion of heteroduplex rejection, the process by which mismatch repair enforces interaction between HR partners of sufficient homology. Already a single mismatch was found to reduce HR in a mismatch-repair-dependent fashion (Datta et al. 1997). Use of the identical sister chromatid template in somatic cells allows setting a low threshold for heteroduplex rejection in mitotically growing cells. During meiotic HR, the genetic differences between homologous chromosomes may require a different limit, but it is unknown whether such a difference exists. It is presently unknown how the mismatch-repair machinery that recognizes the mismatches during HR interfaces with proteins/mechanisms that can disrupt the D-loop before DNA synthesis (see Fig. 3) (see also George and Alani 2012).
Reversibility of Extended D-Loops: A Step in SDSA and a Mechanism of Anticrossover
As discussed for heteroduplex rejection (Spies and Fishel 2015), the nascent D-loop containing mismatches is the target for this quality-control mechanism. Although disruption of the nascent D-loop before DNA synthesis from the invading 3′-OH end is a mechanism of antirecombination, the disruption of the extended D-loop is a required step for SDSA and a mechanism of anticrossover (see Fig. 3). The profoundly different outcomes of the processing of seemingly related structures suggest the existence of subtle substrate discrimination mechanisms, involving both the heteroduplex DNA (quality of homology) and the protein environment presence of PCNA, Rad51, etc. There are several candidate proteins for both mechanisms targeting either nascent or extended D-loops.
Interestingly, both yeast Srs2 and mammalian RECQ5 have been implicated in antirecombination (see above) and anticrossover pathways (Aylon et al. 2003; Ira et al. 2003; Paliwal et al. 2014). Although the mechanisms involved still need to be established, it is possible that removal of Rad51 from ssDNA is involved in both. Dissociation of Rad51 from the invading end avoids HR (antirecombination), whereas dissociation of Rad51 from the second, noninvading end could enable strand annealing during SDSA (anticrossover). This interpretation is consistent with genetic and biochemical evidence showing that Rad51 impedes ssDNA annealing mediated by Rad52 (Dupaigne et al. 2008; Wu et al. 2008; Carter et al. 2009; Mitchel et al. 2013; Miura et al. 2013). RTEL1, a DNA helicase found in metazoans but not fungi, displays genetic properties reminiscent of yeast srs2 mutants including DNA damage sensitivity, hyperrecombination, and colethality with mutations in SGS1/BLM (Barber et al. 2008). During meiosis, RTEL1 in C. elegans is required for crossover homeostasis and interference (Youds et al. 2010), which may be related to its biochemical ability to dissociate D-loops (Barber et al. 2008).
FANCM protein in plants and its homologs in fission (Fml1) and budding yeast (Mph1) have been implicated in anticrossover control during meiotic HR and in vegetative cells, respectively (Prakash et al. 2009; Crismani et al. 2012; Lorenz et al. 2012). Budding yeast Mph1 has been shown to dissociate nascent D-loops in reconstituted reactions with Rad51 and Rad54 as well as protein-free D-loops in a helicase-dependent manner (Prakash et al. 2009). Further analysis will be required to determine the exact mechanism used by these enzymes to avoid crossovers.
Human BLM and its yeast homolog Sgs1 have been implicated in anticrossover through their activity dissolving dHJs in conjunction with TOPOIIIα-RMI1/2 and Top3-Rmi1, respectively (Wu and Hickson 2003; Cejka et al. 2010; see Bizard and Hickson 2014). BLM (van Brabant et al. 2000; Bachrati et al. 2006) and Sgs1 (Fasching et al. 2015) have also been shown to dissociate protein-free D-loops, providing the potential for a second way these enzymes may act as antirecombinases and/or enforce a noncrossover outcome by dissociating D-loops.
Although not strictly an HR intermediate, invasions by the 5′-end could be viewed as dead-end invasion products, because they cannot prime DNA synthesis. However, such 5′-end invasion could also stabilize DNA ends for later processing. The human RecQ-family helicase RECQ1 was found to specifically dismantle D-loops resulting from 5′-end invasions (Bugreev et al. 2008). It is unclear how this biochemical activity relates to the enhanced sister chromatid-exchange phenotype found in RECQ1-depleted cells (Sharma and Brosh 2007).
Concluding Remarks on HR Pathway Reversibility
The above discussion illustrates that HR involves a number of metastable, reversible intermediates, the Rad51/Dmc1–ssDNA filament, the nascent D-loop, the extended D-loop, and the double Holliday junction, as well as dead-end complexes, such as 5′-end invasion or Rad51/Dmc1–dsDNA complexes (Fig. 3). Reversibility of these intermediates ensures that HR once initiated by Rad51-filament formation does not have to blindly proceed until the formation of recombinants but can be regulated at several pathway steps. Such quality control ensures flexibility and, ultimately, pathway robustness (Symington and Heyer 2006; Kanaar et al. 2008; Heyer et al. 2010; Zinovyev et al. 2013).
Genetic Implications of Reversible Pathways—An Alternative Mechanism to Generate Synthetic Lethality
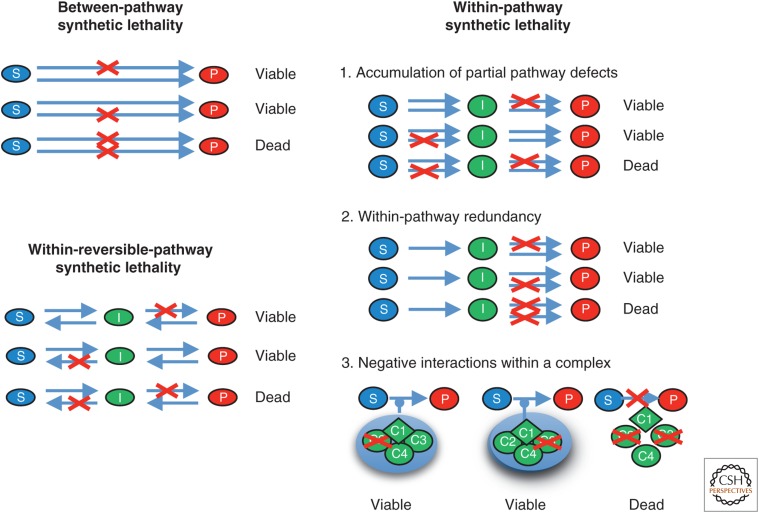
Synthetic lethality or synthetic sickness has become an important genetic phenomenon to elucidate gene function (Dobzhansky 1946; Schuldiner et al. 2006; Boone et al. 2007). Targeted or systematic double mutant analysis as well as chemical ablation in single mutant backgrounds not only developed into a favorite genetic discovery tool but also into a novel approach to cancer therapy (Hartwell et al. 1997). The acute sensitivity of BRCA1- or BRCA2-deficient cells to inhibition of poly(ADP-ribose) polymerase (PARP) showcases this application (Bryant et al. 2005; Farmer et al. 2005). Mechanistically, synthetic lethality/sickness can be explained in several different ways, including the existence of two parallel and competing pathways or the synergistic degradation of an “essential” pathway and complex (Fig. 4) (Boone et al. 2007; Bandyopadhyay et al. 2008; Baryshnikova et al. 2010; Costanzo et al. 2011). Pathway reversibility adds an additional potential mechanistic interpretation of synthetic lethality in that two mutations may trap a potential toxic intermediate or trap a vital resource for the cell (Fig. 4) (Zinovyev et al. 2013). One illustration of this type of within-reversible-pathway synthetic lethality is exemplified by the synthetic lethality of budding yeast srs2 rad54 double mutants and its suppression by any mutation blocking Rad51–ssDNA filament formation (Palladino and Klein 1992; Schild 1995). As discussed above, Srs2 dissociates the Rad51–ssDNA filament, whereas Rad54 is required for DNA strand invasion by the Rad51–ssDNA filament and recombination-associated DNA synthesis (Petukhova et al. 1998; Li and Heyer 2009). Hence, in an srs2 rad54 double mutant, either the Rad51–ssDNA filament or the nascent DNA strand invasion intermediate might be trapped. Any mutation preventing Rad51–ssDNA filament formation (rad51, rad52, rad55, or rad57) rescues this recombination-dependent lethality (Schild 1995). As discussed in detail in Zinovyev et al. (2013), this concept can be applied to many other synthetic lethal/sick interactions, in particular involving reversible PTMs. Moreover, this model provides a rationale for synthetic lethality between gene mutations in genes whose products physically interact. Such an observation would not be expected for proteins acting in parallel, competing pathways. Hence, specific defects in HR (and likely in other reversible pathways) in cancer cells coupled with targeted chemical ablation may create tumor-cell-specific synthetic lethality with potential therapeutic relevance.
Figure 4.
Mechanistic models for synthetic lethality between two mutations. “Between-pathway synthetic lethality” represents the canonical interpretation of a synthetic lethal relationship between two mutations in genes, whose products act in parallel, competing pathways. “Within-pathway synthetic lethality” involves internal redundancy within a single, essential pathway or protein complex, in which pathway flux drops below a minimal threshold in the double mutant. “Within-reversible-pathway synthetic lethality” can generate synthetic lethality between two mutations, whose products function in single nonessential pathway by accumulating a toxic intermediate or depletion of an essential resource. Red crosses define single mutations. S, substrate; I, intermediate; P, product; C1–4, components of a protein complex (for additional discussion, see Zinovyev et al. 2013).
Regulation by Substrate Modification
Chromatin is the substrate for HR and chromatin can be modified by PTMs of histones, including phosphorylation, acetylation, methylation, ubiquitylation, and sumoylation (Strahl and Allis 2000). In addition, ATP-dependent chromatin remodelers change chromatin structure by sliding or evicting nucleosomes or exchanging histones in nucleosomes (Clapier and Cairns 2009). Both types of substrate modifications have been implicated in regulating HR, as discussed for meiotic recombination in Székvölgyi et al. (2015). The seminal discovery that the histone H3 lysine 4 methyltransferase, PRDM9, controls meiotic recombination hotspot activity provides an impressive example of how chromatin modification intersects with HR activity (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010). Histone modifications play a major role in the DNA-damage response providing binding sites for signaling and repair proteins in somatic cells, for example, γ-H2AX for the Rad50-Mre11-Xrs2/NBS complex (Rogakou et al. 1998; Kobayashi et al. 2002). A specific role for the chromatin remodeler, Fun30, was identified in long-range end resection of DSB by Exo1 or Sgs1-Dna2 (Chen et al. 2012; Costelloe et al. 2012; Eapen et al. 2012). Recent reviews provide excellent overviews of the effects of chromatin modifications and remodeling in response to DNA damage (Polo and Jackson 2011; Soria et al. 2012; Price and D’Andrea 2013; Seeber et al. 2013; Tsabar and Haber 2013).
Direct Regulation of HR Pathway Components by PTMs
PTMs of proteins is a well-established mechanism to regulate proteins, including their activity, stability, localization, and protein interactions. Not surprisingly, many HR factors display PTMs in response to cell-cycle cues, DNA damage, and possibly other signals (Heyer et al. 2010; Krejci et al. 2012; Jackson and Durocher 2013). The technical progress in identifying PTMs has been impressive, but it remains difficult to establish the biological relevance and mechanistic consequences of specific PTMs. In fact, several well-documented cases show that not a given specific PTM is critical but, rather, an accumulation of modifications on a given protein or multiple proteins. For example, individual SUMO modifications can act redundantly or additively, or may mediate dedicated functions as well (Jentsch and Psakhye 2013). Likewise phosphorylation, where multi-site phosphorylation can set a threshold to engage an E3 ubiquitin ligase for targeted degradation dependent on charge density but not on any given individual phosphorylation site (Nash et al. 2001). A number of protein PTMs have drastic effects on the regulation and mechanism of HR and can be analyzed by endpoint assays, including the ubiquitylation of PCNA and the CDK-mediated phosphorylation of CtIP/Sae2, as discussed above. Other PTMs may impart only kinetic changes for which a biological effect will be more difficult to document, likely requiring real-time kinetic assays. Biochemical assays to efficiently measure and simultaneously compare differently modified isoforms of a protein are fraught with the difficulty of purifying sufficient amounts of homogeneously modified proteins. Recent technical developments to mimic tyrosine phosphorylation by amber suppressor-mediated incorporation of a phosphotyrosine analog show promise (Honda et al. 2011; O’Donoghue et al. 2013).
GENOME MAINTENANCE AND HUMAN DISEASE
Most, if not all, forms of cancer are associated with genomic rearrangements and/or genomic instability (Lengauer et al. 1998). Prakash et al. (2015) focuses on key HR genes, BRCA1, BRCA2, PALB2, and the RAD51, RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3, in an illuminating discussion of the roles of these genes in human cancer predisposition. Drawing on mouse models as well as human genetics and pathology, Prakash et al. (2015) also emphasize the developmental defects caused by HR deficiency. These defects are possibly the consequence of premature senescence and/or apoptosis in certain cell populations that affect ontogenic development, which may form the basis for the involvement of HR in other human diseases and syndromes (see below).
A particularly vexing problem in genome maintenance is the identification of mechanisms involved in gene duplications/amplifications and the mechanisms underlying complex genomic rearrangements causing massive genomic instability observed in some cancer cells, such as chromothrypsis (Hastings et al. 2009; Liu et al. 2011b, 2012; Forment et al. 2012). Drawing on their insightful analysis of gene duplications and amplifications in bacteria, Reams and Roth (2015) provide compelling arguments for a dynamic process leading to stable duplications/amplifications that involve multiple stages of selection defying one-step models. This may involve processes related to BIR, termed microhomology/microsatellite-induced replication, which was shown to lead to segmental duplications in yeast independent of HR or NHEJ (Payen et al. 2008). The importance of determining the mechanisms of duplications and amplification has gained additional significance by the realization that copy-number variations (CNVs) are the most common changes in human genomes (Stankiewicz and Lupski 2010). Moreover, de novo point mutant and CNVs hotspots have been identified in patients with autism spectrum disorder as well as bipolar disease and schizophrenia (Malhotra et al. 2011; Michaelson et al. 2012). It is presently unclear whether these changes are causative for the syndrome or which mechanisms are involved. An exciting intersection between HR and autism occurs in the tumor suppressor phosphatase and tensin homolog (PTEN). PTEN-negative cells are HR deficient (Shen et al. 2007; Mendes-Pereira et al. 2009; Bassi 2013). The mechanisms involved are not understood and may be cell-type or tissue specific. Importantly, PTEN is associated with a subset of individuals with autism spectrum disorders (Butler et al. 2005; Varga et al. 2009; McBride et al. 2010). This may be related to the function of PTEN in neuronal stem cells (Amiri et al. 2012). It is unknown what role HR plays in neuronal stem-cell development, and HR-deficient viable mice have not been tested for neurodevelopmental defects. Interestingly, BRCA1, a key HR component, is involved in hair follicle stem-cell development (Sotiropoulou et al. 2013). These emerging results indicate a potential role of HR in human diseases and syndromes other than cancer.
CONCLUDING REMARKS
The integration of diverse experimental approaches ranging from single-molecule biophysics, biochemistry, cell biology, and genetics and the parallel analysis of multiple systems and organisms (phage, bacteria, fungi, flies, worms, mammals, and humans) has generated a wealth of insights into the mechanisms and regulation of HR and has resulted in first clinical applications (e.g., Fong et al. 2009). These efforts have generated consensus models for HR, such as shown in Figures 1 and 3 (see Mehta and Haber 2014). Such positivistic projections of data and interpretations from diverse models and organisms into a single model have accelerated progress in the field but also bear the danger of ignoring informative species-specific differences. The recent literature highlights the tremendous progress in our understanding of recombination and illuminates a vibrant field that is in the process of making growing contributions in the diagnosis and treatment of human disease, specifically cancer (Barker and Powell 2010; Moynahan and Jasin 2010; Krawczyk et al. 2011; Chernikova et al. 2012; Carvalho and Kanaar 2014).
ACKNOWLEDGMENTS
I am grateful to all members of my laboratory, past and present, for their contributions and dedication, especially Aurèle Piazza for comments on the manuscript. Big thanks go to my coeditors Steve Kowalczykowski and Neil Hunter for years of fruitful discussions, helpful encouragement, and constructive comments on our work. The support by the National Institutes of Health (GM58015, CA92776, and CA154920) and the Department of Defense (BC133980) is gratefully acknowledged. I apologize to those whose work could not be cited because of space constraints.
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Aboussekhra A, Chanet R, Zgaga Z, Cassier Chauvat C, Heude M, Fabre F. 1989. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair characteristics of radH mutants and sequence of the gene. Nucleic Acids Res 17: 7211–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboussekhra A, Chanet R, Adjiri A, Fabre F. 1992. Semi-dominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to prokaryotic RecA protein. Mol Cell Biol 12: 3224–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, Martinez-Perez E, Boulton SJ, La Volpe A. 2010. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell 39: 25–35. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Tafel AA, Kanaar R. 2006. DNA double-strand break repair and chromosome translocations. DNA Repair 5: 1075–1081. [DOI] [PubMed] [Google Scholar]
- Agmon N, Liefshitz B, Zimmer C, Fabre E, Kupiec M. 2013. Effect of nuclear architecture on the efficiency of double-strand break repair. Nat Cell Biol 15: 694–7281. [DOI] [PubMed] [Google Scholar]
- *.Aguilera A, Gaillard H. 2014. Transcription and recombination: When RNA meets DNA. Cold Spring Harb Perspect Biol 6: a016543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Garcia-Muse T. 2013. Causes of genome instability. Annu Rev Genet 47: 1–32. [DOI] [PubMed] [Google Scholar]
- Amiri A, Cho W, Zhou J, Birnbaum SG, Sinton CM, McKay RM, Parada LF. 2012. Pten deletion in adult hippocampal neural stem/progenitor cells causes cellular abnormalities and alters neurogenesis. J Neurosci 32: 5880–5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Bitan-Banin G, Kupiec M. 2003. Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol Biol Cell 23: 1403–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati CZ, Borts RH, Hickson ID. 2006. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res 34: 2269–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Kelley R, Krogan NJ, Ideker T. 2008. Functional maps of protein complexes from quantitative genetic interaction data. PLoS Comp Biol 4: e1000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MIR, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. 2008. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 135: 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker CA, Powell SN. 2010. Enhancing radiotherapy through a greater understanding of homologous recombination. Sem Rad Oncol 20: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Faryabi RB, Callen E, Wong N, Malhowski A, Chen HT, Gutierrez-Cruz G, Sun HW, McKinnon P, Wright G, et al. 2013. Identification of early replicating fragile sites that contribute to genome instability. Cell 152: 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryshnikova A, Costanzo M, Kim Y, Ding H, Koh J, Toufighi K, Youn JY, Ou J, San Luis BJ, Bandyopadhyay S, et al. 2010. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods 7: 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi C. 2013. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science 341: 1064–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin-Shanower SA, Fricke WM, Mullen JR, Brill SJ. 2003. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol Cell Biol 23: 3487–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B. 2013. Meiotic recombination in mammals: Localization and regulation. Nat Rev Genet 14: 794–806. [DOI] [PubMed] [Google Scholar]
- Beadle GW. 1932. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc Natl Acad Sci 18: 160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Jossen R, Colosio A, Frattini C, Carotenuto W, Cocito A, Doksani Y, Klein H, Gomez-Gonzalez B, et al. 2011. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell 146: 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Kumar A, Foiani M. 2012. Preserving the genome by regulating chromatin association with the nuclear envelope. Trends Cell Biol 22: 465–473. [DOI] [PubMed] [Google Scholar]
- *.Bizard AH, Hickson ID. 2014. The dissolution of double Holliday junctions. Cold Spring Harb Perspect Biol 6: a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone C, Bussey H, Andrews BJ. 2007. Exploring genetic interactions and networks with yeast. Nat Rev Genet 8: 437–449. [DOI] [PubMed] [Google Scholar]
- *.Brown MS, Bishop DK. 2015. DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb Perspect Biol 7: a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S, Reddel RR. 1995. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J 14: 4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med 3: 1271–1274. [DOI] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV. 2007. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev 21: 3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Brosh RM, Mazin AV. 2008. RECQ1 possesses DNA branch migration activity. J Biol Chem 283: 20231–20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, Miles JH, Wang CH, Stratton R, Pilarski R, et al. 2005. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet 42: 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. 2010. Double Holliday junctions are intermediates of DNA break repair. Nature 464: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, de Villartay JP. 2006. Cernunnos interacts with the XRCC4 center dot DNA-ligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1. J Biol Chem 281: 13857–13860. [DOI] [PubMed] [Google Scholar]
- Cannavo E, Cejka P. 2014. Sae2 promotes dsDNA endonuclease activity within Mre11–Rad50–Xrs2 to resect DNA breaks. Nature 514: 122–125. [DOI] [PubMed] [Google Scholar]
- Carter SD, Vigasova D, Chen J, Chovanec M, Astrom SU. 2009. Nej1 recruits the Srs2 helicase to DNA double-strand breaks and supports repair by a single-strand annealing-like mechanism. Proc Natl Acad Sci 106: 12037–12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho JFS, Kanaar R. 2014. Targeting homologous recombination-mediated DNA repair in cancer. Expert Opin Ther Targets 18: 427–458. [DOI] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. 2010. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat Struct Mol Biol 17: 1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying SM, Hickson ID. 2009. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11: 753–760. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47: 497–510. [DOI] [PubMed] [Google Scholar]
- Chen X, Tomkinson AE. 2011. Yeast Nej1 is a key participant in the initial end binding and final ligation steps of nonhomologous end joining. J Biol Chem 286: 4931–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XF, Niu HY, Chung WH, Zhu Z, Papusha A, Shim EY, Lee SE, Sung P, Ira G. 2011. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol 18: 1015–U1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XF, Cui DD, Papusha A, Zhang XT, Chu CD, Tang JW, Chen KF, Pan XW, Ira G. 2012. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489: 576–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernikova SB, Game JC, Brown JM. 2012. Inhibiting homologous recombination for cancer therapy. Cancer Biol Ther 13: 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Saponaro M, Baryshnikova A, Kim JH, Seo YS, Liberi G. 2007. The human F-Box DNA helicase FBH1 faces Saccharomyces cerevisiae Srs2 and postreplication repair pathway roles. Mol Cell Biol 27: 7439–7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. 2011. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304. [DOI] [PubMed] [Google Scholar]
- Cooper JP, Nimmo ER, Allshire RC, Cech TR. 1997. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 385: 744–747. [DOI] [PubMed] [Google Scholar]
- Copsey A, Tang SM, Jordan PW, Blitzblau HG, Newcombe S, Chan ACH, Newnham L, Li ZB, Gray S, Herbert AD, et al. 2013. Smc5/6 coordinates formation and resolution of joint molecules with chromosome morphology to ensure meiotic divisions. PLoS Genet 9: e1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Ledesma F, Aguilera A. 2006. Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep 7: 919–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Myers CL, Andrews B, Boone C. 2011. Charting the genetic interaction map of a cell. Curr Opin Biotechnol 22: 66–74. [DOI] [PubMed] [Google Scholar]
- Costelloe T, Louge R, Tomimatsu N, Mukherjee B, Martini E, Khadaroo B, Dubois K, Wiegant WW, Thierry A, Burma S, et al. 2012. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 489: 581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. 2000. The importance of repairing stalled replication forks. Nature 404: 37–41. [DOI] [PubMed] [Google Scholar]
- Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R. 2012. FANCM limits meiotic crossovers. Science 336: 1588–1590. [DOI] [PubMed] [Google Scholar]
- Datta A, Hendrix M, Lipsitch M, Jinks-Robertson S. 1997. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci 94: 9757–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans AJ, West SC. 2011. DNA interstrand crosslink repair and cancer. Nat Rev Cancer 11: 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Roth DB. 2013. Modernizing the nonhomologous end-joining repertoire: Alternative and classical NHEJ share the stage. Annu Rev Genet 47: 433–455. [DOI] [PubMed] [Google Scholar]
- Dion V, Kalck V, Horigome C, Towbin BD, Gasser SM. 2012. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat Cell Biol 14: 502–509. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. 1946. Genetics of natural populations. XIII: Recombination and variability in populations of Drosophila pseudoobscura. Genetics 31: 269–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Doksani Y, de Lange T. 2014. The role of double-strand break repair pathways at functional and dysfunctional telomeres. Cold Spring Harb Perspect Biol 6: a016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupaigne P, Le Breton C, Fabre F, Giangloff S, Le Cam E, Veaute X. 2008. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: Implications for crossover incidence during mitotic recombination. Mol Cell 29: 243–254. [DOI] [PubMed] [Google Scholar]
- Eapen VV, Sugawara N, Tsabar M, Wu WH, Haber JE. 2012. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol Cell Biol 32: 4727–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott B, Jasin M. 2002. Double-strand breaks and translocations in cancer. Cell Mol Life Sci 59: 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F. 1978. Induced intragenic recombination in yeast can occur during the G1 mitotic phase. Nature 272: 795–797. [DOI] [PubMed] [Google Scholar]
- Fabre F, Chan A, Heyer WD, Gangloff S. 2002. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci 99: 16887–16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt ANJ, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921. [DOI] [PubMed] [Google Scholar]
- Fasching CL, Cejka P, Kowalczykowski SC, Heyer WD. 2015. Top3-Rmi1 dissolve Rad51-mediated D loops by a topoisomerase-based mechanism. Mol Cell Biol 57: 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippo JS, Sung P, Klein H. 2008. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257. [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, et al. 2009. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361: 123–134. [DOI] [PubMed] [Google Scholar]
- Forment JV, Kaidi A, Jackson SP. 2012. Chromothripsis and cancer: Causes and consequences of chromosome shattering. Nat Rev Cancer 12: 663–670. [DOI] [PubMed] [Google Scholar]
- Frank-Vaillant M, Marcand S. 2001. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev 15: 3005–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugger K, Mistrik M, Danielsen JR, Dinant C, Falck J, Bartek J, Lukas J, Mailand N. 2009. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J Cell Biol 186: 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, Soustelle C, Fabre F. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet 25: 192–194. [DOI] [PubMed] [Google Scholar]
- George CM, Alani E. 2012. Multiple cellular mechanisms prevent chromosomal rearrangements involving repetitive DNA. Crit Rev Biochem Mol Biol 47: 297–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. 2012. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45: 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Jeggo P, Lobrich M. 2010. The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax. DNA Repair 9: 1273–1282. [DOI] [PubMed] [Google Scholar]
- Haber JE. 2006. Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair 5: 998–1009. [DOI] [PubMed] [Google Scholar]
- Harrison JC, Haber JE. 2006. Surviving the breakup: The DNA damage checkpoint. Annu Rev Genet 40: 209–235. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. 1997. Integrating genetic approaches into the discovery of anticancer drugs. Science 278: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V. 2010. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17: 1305–U1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. 2009. Mechanisms of change in gene copy number. Nat Rev Genet 10: 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Guacci V, Koshland D. 2008. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol Cell 31: 47–56. [DOI] [PubMed] [Google Scholar]
- Heude M, Chanet R, Fabre F. 1995. Regulation of the Saccharomyces cerevisiae Srs2 helicase during the mitotic cell cycle, meiosis and after irradiation. Mol Gen Genet 248: 59–68. [DOI] [PubMed] [Google Scholar]
- Heyer WD. 2007. Biochemistry of eukaryotic homologous recombination. In Molecular genetics of recombination (ed. Aguilera A, Rothstein R), pp. 95–133. Springer, Berlin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J. 2010. Regulation of homologous recombination in eukaryotes. Annu Rev Genet 44: 113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. 2000. Chromosome cohesion, condensation, and separation. Annu Rev Biochem 69: 115–144. [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141. [DOI] [PubMed] [Google Scholar]
- Holzen TM, Shah PP, Olivares HA, Bishop DK. 2006. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev 20: 2593–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M, Okuno Y, Yoo J, Ha T, Spies M. 2011. Tyrosine phosphorylation enhances RAD52-mediated annealing by modulating its DNA binding. EMBO J 30: 3368–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. 2007. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev 21: 3073–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP. 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem 284: 9558–9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. 2007. Meiotic recombination. In Homologous recombination (ed. Aguilera A, Rothstein R), pp. 381–441. Springer, Berlin. [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev 16: 1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Durocher D. 2013. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell 49: 795–807. [DOI] [PubMed] [Google Scholar]
- Jain S, Sugawara N, Lydeard J, Vaze M, Tanguy Le Gac N, Haber JE. 2009. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev 23: 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S, Psakhye I. 2013. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu Rev Genet 47: 167–186. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B. 2007. The size of the nucleus increases as yeast cells grow. Moll Biol Cell 18: 3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH. 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar R, Wyman C, Rothstein R. 2008. Quality control of DNA break metabolism: In the “end,” it’s a good thing. EMBO J 27: 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel A, Sjostrand JO, Astrom SU. 2001. Nej1p, a cell type-specific regulator of nonhomologous end joining in yeast. Curr Biol 11: 1611–1617. [DOI] [PubMed] [Google Scholar]
- Kim H, D’Andrea AD. 2012. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev 26: 1393–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S. 2012. Transcription as a source of genome instability. Nat Rev Genet 13: 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K. 2002. NBS1 localizes to γ-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol 12: 1846–1851. [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Putnam CD, Myung K. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552–557. [DOI] [PubMed] [Google Scholar]
- Krawczyk PM, Eppink B, Essers J, Stap J, Rodermond H, Odijk H, Zelensky A, van Bree C, Stalpers LJ, Buist MR, et al. 2011. Mild hyperthermia inhibits homologous recombination, induces BRCA2 degradation, and sensitizes cancer cells to poly (ADP-ribose) polymerase-1 inhibition. Proc Natl Acad Sci 108: 9851–9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk PM, Borovski T, Stap J, Cijsouw T, ten Cate R, Medema JP, Kanaar R, Franken NA, Aten JA. 2012. Chromatin mobility is increased at sites of DNA double-strand breaks. J Cell Sci 125: 2127–2133. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309. [DOI] [PubMed] [Google Scholar]
- Krejci L, Altmannova V, Spirek M, Zhao X. 2012. Homologous recombination and its regulation. Nucleic Acids Res 40: 5795–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. 2004. Recombination proteins in yeast. Annu Rev Genet 38: 233–271. [DOI] [PubMed] [Google Scholar]
- Lawrence CW, Christensen R. 1976. UV mutagenesis in radiation-sensitive strains of yeast. Genetics 82: 207–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemontt JF. 1971. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics 68: 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. 1998. Genetic instabilities in human cancers. Nature 396: 643–649. [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. 2007. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28: 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M. 2011. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470: 120–123. [DOI] [PubMed] [Google Scholar]
- Li X, Heyer WD. 2008. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18: 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD. 2009. RAD54 controls access to the invading 3-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res 37: 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lisby M, Rothstein R. 2015. Cell biology of mitotic recombination. Cold Spring Harb Perspect Biol 7: a016535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer W-D. 2011a. Rad51 paralogs Rad55–Rad57 balance the anti-recombinase function of Srs2 in Rad51 pre-synaptic filament formation. Nature 479: 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PF, Erez A, Nagamani SCS, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. 2011b. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146: 888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PF, Carvalho CMB, Hastings PJ, Lupski JR. 2012. Mechanisms for recurrent and complex human genomic rearrangements. Curr Op Genet Dev 22: 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Osman F, Sun W, Nandi S, Steinacher R, Whitby MC. 2012. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science 336: 1585–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, Tremethick DJ. 2012. New insights into nucleosome and chromatin structure: An ordered or a disordered affair? Nat Rev Mol Cell Biol 13: 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, McCarthy S, Michaelson JJ, Vacic V, Burdick KE, Yoon S, Cichon S, Corvin A, Gary S, Gershon ES, et al. 2011. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron 72: 951–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Haber JE. 2012. Mutations arising during repair of chromosome breaks. Annu Rev Genet 46: 455–473. [DOI] [PubMed] [Google Scholar]
- McBride KL, Varga EA, Pastore MT, Prior TW, Manickam K, Atkin JF, Herman GE. 2010. Confirmation study of PTEN mutations among individuals with autism or developmental delays/mental retardation and macrocephaly. Autism Res 3: 137–141. [DOI] [PubMed] [Google Scholar]
- *.Mehta A, Haber JE. 2014. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol 6: a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, Waldman T, Lord CJ, Ashworth A. 2009. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med 1: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D, Jin X, Jian M, Liu G, Greer D, Bhandari A, et al. 2012. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151: 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine-Hattab J, Rothstein R. 2012. Increased chromosome mobility facilitates homology search during recombination. Nat Cell Biol 14: 510–517. [DOI] [PubMed] [Google Scholar]
- Mitchel K, Lehner K, Jinks-Robertson S. 2013. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS Genet 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Shibata T, Kusano K. 2013. Putative antirecombinase Srs2 DNA helicase promotes noncrossover homologous recombination avoiding loss of heterozygosity. Proc Natl Acad Sci 110: 16067–16072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. 2007. PCNA, the maestro of the replication fork. Cell 129: 665–679. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, Dejsuphong D, Petalcorin MIR, Hofmann K, Takeda S, Boulton SJ, D’Andrea AD. 2012. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell 45: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier K, Armas JC, Etteldorf S, Ghazal P, Sullivan KF. 2000. Annexation of the interchromosomal space during viral infection. Nat Cell Biol 2: 661–665. [DOI] [PubMed] [Google Scholar]
- *.Morrical SW. 2014. DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb Perspect Biol 6: a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosig G. 1998. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu Rev Genet 32: 379–413. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 11: 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller I, Boyle S, Singer RH, Bickmore WA, Chubb JR. 2010. Stable morphology, but dynamic internal reorganization, of interphase human chromosomes in living cells. PLoS ONE 5: e11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD. 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411: 1073–1076. [DOI] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall M, Sicheri F, Pawson T, Tyers M. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414: 514–521. [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. 1996. Thymine–thymine dimer bypass by yeast DNA polymerase ζ. Science 272: 1646–1649. [DOI] [PubMed] [Google Scholar]
- Neylon C, Kralicek AV, Hill TM, Dixon NE. 2005. Replication termination in Escherichia coli: Structure and antihelicase activity of the Tus–Ter complex. Microbiol Mol Biol Rev 69: 501–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue P, Ling J, Wang YS, Soll D. 2013. Upgrading protein synthesis for synthetic biology. Nat Chem Biol 9: 594–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Dixon J, Barr AR, Whitby MC. 2005. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol Cell Biol 25: 8084–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. 2010. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science 329: 219–223. [DOI] [PubMed] [Google Scholar]
- Paliwal S, Kanagaraj R, Sturzenegger A, Burdova K, Janscak P. 2014. Human RECQ5 helicase promotes repair of DNA double-strand breaks by synthesis-dependent strand annealing. Nucleic Acids Res 42: 2380–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F, Klein HL. 1992. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics 132: 23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli E, Chen SH, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. 2005. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell 19: 123–133. [DOI] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G. 2008. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet 4: e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova G, Stratton S, Sung P. 1998. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature 393: 91–94. [DOI] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433. [DOI] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. 2011. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev 25: 409–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L. 2005. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem 74: 317–353. [DOI] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. 2009. Yeast Mph1 helicase dissociates Rad51-made D-loops: Implications for crossover control in mitotic recombination. Genes Dev 23: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Prakash R, Zhang Y, Feng W, Jasin M. 2015. Homologous recombination and human health: The roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol 7: a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, D’Andrea AD. 2013. Chromatin remodeling at DNA double-strand breaks. Cell 152: 1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hayes TK, Kolodner RD. 2009. Specific pathways prevent duplication-mediated genome rearrangements. Nature 460: U984–U965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Reams AB, Roth JR. 2015. Mechanisms of gene duplication and amplification. Cold Spring Harb Perspect Biol 7: a016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RW, Caruthers DM. 1992. Stability and properties of double and triple helices: Dramatic effects of RNA or DNA backbone composition. Science 258: 1463–1466. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868. [DOI] [PubMed] [Google Scholar]
- Sakamoto AN, Stone JE, Kissling GE, McCulloch SD, Pavlov YI, Kunkel TA. 2007. Mutator alleles of yeast DNA polymerase ζ. DNA Repair 6: 1829–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R. 2012. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol 13: 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S. 2010. Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol 11: 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sax K. 1930. Chromosome structure and mechanism of crossing over. J Arnold Arb 11: 193–220. [Google Scholar]
- Schiestl RH, Prakash S, Prakash L. 1990. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124: 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild D. 1995. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics 140: 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Weissman JS, Krogan NJ. 2006. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods 40: 344–352. [DOI] [PubMed] [Google Scholar]
- Schulz VP, Zakian VA. 1994. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76: 145–155. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. 1995. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83: 783–791. [DOI] [PubMed] [Google Scholar]
- Schwendener S, Raynard S, Paliwal S, Cheng A, Kanagaraj R, Shevelev I, Stark JM, Sung P, Janscak P. 2010. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J Biol Chem 285: 15739–15745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeber A, Hauer M, Gasser SM. 2013. Nucleosome remodelers in double-strand break repair. Curr Opin Genet Dev 23: 174–184. [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Zheng XZ, Epshtein A, Carey JN, Bishop DK, Klein HL. 2010. Swi2/Snf2-Related translocases prevent accumulation of toxic Rad51 complexes during mitotic growth. Mol Cell 39: 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Brosh RM Jr. 2007. Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS ONE 2: e1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, Yin Y. 2007. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 128: 157–170. [DOI] [PubMed] [Google Scholar]
- Shi J, Wolf SE, Burke JM, Presting GG, Ross-Ibarra J, Dawe RK. 2010. Widespread gene conversion in centromere cores. PLoS Biol 9: e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, et al. 2014. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell 53: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simandlova J, Zagelbaum J, Payne MJ, Chu WK, Shevelev I, Hanada K, Chatterjee S, Reid DA, Liu Y, Janscak P, et al. 2013. FBH1 helicase disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells. J Biol Chem 288: 34168–34180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Kiianitsa K, Heyer W-D. 2002. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell 10: 1175–1188. [DOI] [PubMed] [Google Scholar]
- Sommers JA, Rawtani N, Gupta R, Bugreev DV, Mazin AV, Cantor SB, Brosh RM. 2009. FANCJ uses its motor ATPase to destabilize protein-DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. J Biol Chem 284: 7502–7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Dominska M, Greenwell PW, Petes TD. 2014. Genome-wide high-resolution mapping of chromosome fragile sites in Saccharomyces cerevisiae. Proc Natl Acad Sci 111: 2210–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Polo SE, Almouzni G. 2012. Prime, repair, restore: The active role of chromatin in the DNA damage response. Mol Cell 46: 722–734. [DOI] [PubMed] [Google Scholar]
- Sotiropoulou PA, Karambelas AE, Debaugnies M, Candi A, Bouwman P, Moers V, Revenco T, Rocha AS, Sekiguchi K, Jonkers J, et al. 2013. BRCA1 deficiency in skin epidermis leads to selective loss of hair follicle stem cells and their progeny. Genes Dev 27: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Spies M, Fishel R. 2015. Mismatch repair during homologous and homeologous recombination. Cold Spring Harb Perspect Biol 7: a022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. 2010. Structural variation in the human genome and its role in disease. Annu Rev Med 61: 437–455. [DOI] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD. 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403: 41–45. [DOI] [PubMed] [Google Scholar]
- Strathern JN, Shafer BK, Mcgill CB. 1995. DNA synthesis errors associated with double-strand-break repair. Genetics 140: 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ström L, Sjogren C. 2005. DNA damage-induced cohesion. Cell Cycle 4: 536–539. [DOI] [PubMed] [Google Scholar]
- Ström L, Sjogren C. 2007. Chromosome segregation and double-strand break repair—A complex connection. Curr Opin Cell Biol 19: 344–349. [DOI] [PubMed] [Google Scholar]
- Ström L, Lindroos HB, Shirahige K, Sjögren C. 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell 16: 1003–1015. [DOI] [PubMed] [Google Scholar]
- Ström L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C. 2007. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317: 242–245. [DOI] [PubMed] [Google Scholar]
- *.Syeda AH, Hawkins M, McGlynn P. 2014. Recombination and replication. Cold Spring Harb Perspect Biol 6: a016550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Symington LS. 2014. End resection at double-strand breaks: Mechanism and regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271. [DOI] [PubMed] [Google Scholar]
- Symington LS, Heyer WD. 2006. Some disassembly required: Role of DNA translocases in the disruption of recombination intermediates and dead-end complexes. Genes Dev 20: 2479–2486. [DOI] [PubMed] [Google Scholar]
- Szankasi P, Gysler C, Zehntner U, Leupold U, Kohli J, Munz P. 1986. Mitotic recombination between dispersed but related tRNA genes of Schizosaccharomyces pombe generates a reciprocal translocation. Mol Gen Genet 202: 394–402. [Google Scholar]
- *.Székvölgyi L, Ohta K, Nicolas A. 2015. Initiation of meiotic homologous recombination: Flexibility, impact of histone modifications, and chromatin remodeling. Cold Spring Harb Perspect Biol 7: a016527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. 2010. A possible influence of the spindle fibre on crossing-over in Drosophila. PLoS Biol 8: e1000326.20231873 [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M. 2007. The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol 9: 923–931. [DOI] [PubMed] [Google Scholar]
- Tsabar M, Haber JE. 2013. Chromatin modifications and chromatin remodeling during DNA repair in budding yeast. Curr Opin Genet Dev 23: 166–173. [DOI] [PubMed] [Google Scholar]
- Tsutsui Y, Kurokawa Y, Ito K, Siddique MSP, Kawano Y, Yamao F, Iwasaki H. 2014. Multiple regulation of Rad51-mediated homologous recombination by fission yeast Fbh1. PLoS Genet 10: e1004542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ünal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell 16: 991–1002. [DOI] [PubMed] [Google Scholar]
- Ünal E, Heidinger-Pauli JM, Koshland D. 2007. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317: 245–248. [DOI] [PubMed] [Google Scholar]
- Ünal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. 2008. A molecular determinant for the establishment of sister chromatid cohesion. Science 321: 566–569. [DOI] [PubMed] [Google Scholar]
- Uphoff S, Kapanidis AN. 2014. Studying the organization of DNA repair by single-cell and single-molecule imaging. DNA Repair 20: 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia M, Bentele M, Vaze MB, Herrmann G, Kraus E, Lee SE, Schar P, Haber JE. 2001. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 414: 666–669. [DOI] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German JL, Ellis NA, Holloman WK. 2000. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry 39: 14617–14625. [DOI] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MIR, Ding H, Boulton SJ. 2012. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149: 795–806. [DOI] [PubMed] [Google Scholar]
- Varga EA, Pastore M, Prior T, Herman GE, McBride KL. 2009. The prevalence of PTEN mutations in a clinical pediatric cohort with autism spectrum disorders, developmental delay, and macrocephaly. Genet Med 11: 111–117. [DOI] [PubMed] [Google Scholar]
- Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE. 2002. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell 10: 373–385. [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312. [DOI] [PubMed] [Google Scholar]
- Wechsler T, Newman S, West SC. 2011. Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature 471: 642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Richardson CA, Elliott B, Jasin M. 2006. Modeling oncogenic translocations: Distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair 5: 1065–1074. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Hickson ID. 2003. The Bloom’s syndrome helicase suppresses crossing-over during homologous recombination. Nature 426: 870–874. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. 2006. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu Rev Genet 40: 279–306. [DOI] [PubMed] [Google Scholar]
- Wu Y, Kantake N, Sugiyama T, Kowalczykowski SC. 2008. Rad51 protein controls Rad52-mediated DNA annealing. J Biol Chem 283: 14883–14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wyatt HDM, West SC. 2014. Holliday junction resolvases. Cold Spring Harb Perspect Biol 6: a023192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman C, Kanaar R. 2006. DNA double-strand break repair: All’s well that ends well. Annu Rev Genet 40: 363–383. [DOI] [PubMed] [Google Scholar]
- Xaver M, Huang LZ, Chen D, Klein F. 2013. Smc5/6-Mms21 prevents and eliminates inappropriate recombination intermediates in meiosis. PLoS Genet 9: e1004067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. 2008. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet 4: e1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim E, O’Connell KE, St Charles J, Petes TD. 2014. High-resolution mapping of two types of spontaneous mitotic gene conversion events in Saccharomyces cerevisiae. Genetics 198: 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, Oneil NJ, Rose AM, West SC, Meyer BJ, Boulton SJ. 2010. RTEL-1 enforces meiotic crossover interference and homeostasis. Science 327: 1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zelensky A, Kanaar R, Wyman C. 2014. Mediators of homologous DNA pairing. Cold Spring Harb Perspect Biol 6: a016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng SC, Xiang T, Pandita TK, Gonzalez-Suarez I, Gonzalo S, Harris CC, Yang Q. 2009. Telomere recombination requires the MUS81 endonuclease. Nat Cell Biol 11: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 1999. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet 33: 603–754. [DOI] [PubMed] [Google Scholar]
- Zinovyev A, Kuperstein I, Barillot E, Heyer WD. 2013. Synthetic lethality between gene defects affecting a single non-essential molecular pathway with reversible steps. PLoS Comp Biol 9: e1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]