Highlight
Dysfunction of the Pep-PEPR system and its interplay with JA signalling results in increased plant susceptibility towards herbivore attack indicating that endogenous signalling also contributes to herbivore defence.
Key words: DAMP, herbivory, jasmonic acid, oral secretions, Pep, PEPR, Spodoptera littoralis.
Abstract
A number of plant endogenous elicitors have been identified that induce pattern-triggered immunity upon perception. In Arabidopsis thaliana eight small precursor proteins, called PROPEPs, are thought to be cleaved upon danger to release eight peptides known as the plant elicitor peptides Peps. As the expression of some PROPEPs is induced upon biotic stress and perception of any of the eight Peps triggers a defence response, they are regarded as amplifiers of immunity. Besides the induction of defences directed against microbial colonization Peps have also been connected with herbivore deterrence as they share certain similarities to systemins, known mediators of defence signalling against herbivores in solanaceous plants, and they positively interact with the phytohormone jasmonic acid. A recent study using maize indicated that the application of ZmPep3, a maize AtPep-orthologue, elicits anti-herbivore responses. However, as this study only assessed the responses triggered by the exogenous application of Peps, the biological significance of these findings remained open. By using Arabidopsis GUS-reporter lines, it is now shown that the promoters of both Pep-receptors, PEPR1 and PEPR2, as well as PROPEP3 are strongly activated upon herbivore attack. Moreover, pepr1 pepr2 double mutant plants, which are insensitive to Peps, display a reduced resistance to feeding Spodoptera littoralis larvae and a reduced accumulation of jasmonic acid upon exposure to herbivore oral secretions. Taken together, these lines of evidence extend the role of the AtPep-PEPR system as a danger detection mechanism from microbial pathogens to herbivores and further underline its strong interaction with jasmonic acid signalling.
Introduction
Plants use sophisticated perception and signalling systems to detect biotic dangers, such as microbial pathogens or feeding herbivores and, subsequently, to induce an efficient defence response against these threats. In the case of microbial pathogens, several membrane-bound pattern recognition receptors (PRRs) have been characterized that specifically detect conserved microbial structures (referred to as microbe-associated molecular patterns—MAMPs) and eventually trigger a set of defence responses. This mechanism is commonly referred to as pattern-triggered immunity (PTI) (Boller and Felix, 2009; Schwessinger and Ronald, 2012; Macho and Zipfel, 2014).
In the case of herbivorous insects, plants rely on similar detection systems to induce defence signalling and, eventually, herbivore deterrence. The initial recognition of herbivore attack is at least partially achieved by the detection of elicitor compounds in insect oral secretions (Howe and Jander, 2008; Xu et al., 2015) and is potentially mediated by a set of membrane-bound receptors similar to MAMP recognition (Schmelz et al., 2009).
In addition to mechanisms for the detection of exogenous danger, plants also rely on endogenous signalling molecules that are capable of eliciting defence responses (Boller and Felix, 2009; Albert, 2013). Whereas some of these so-called danger-associated molecular patterns (DAMPs), such as cell wall fragments and cutin monomers, are derived from the degradation of the plant cell wall caused by invading pathogens (Sieber et al., 2000; D’Ovidio et al., 2004), others, like the peptides of the systemin family in solanaceous plants, are actively produced by the plant upon the detection of danger (Ryan and Pearce, 2003). Interestingly, systemins have been postulated to be involved in both the deterrence of microbes and herbivores as they have been shown not only to trigger PTI-like responses but also specific defence responses against herbivory. The latter include the biosynthesis of proteinase inhibitors (PI) and the emission of volatile compounds to attract herbivore predators (Ryan and Pearce, 2003; Degenhardt et al., 2010; Sun et al., 2011). However, since the systemin receptor(s) have yet to be fully identified or are under dispute (Holton et al., 2008; Lanfermeijer et al., 2008; Malinowski et al., 2009), the assessment of the biological relevance of systemins to defence signalling has remained difficult.
More recently, a family of endogenous elicitor peptides has been discovered in Arabidopsis thaliana, referred to as AtPeps (Huffaker and Ryan, 2007; Bartels et al., 2013). Like systemins, AtPeps are small peptides (23–29 amino acids long) derived from the C-terminal ends of larger precursor proteins, the PROPEPs (Yamaguchi and Huffaker, 2011). In contrast to the still contested perception mechanism of systemins, AtPeps have been shown to be perceived by two membrane-based receptors referred to as PEP-Receptor 1 (PEPR1) and PEPR2 (Yamaguchi et al., 2006, 2010; Krol et al., 2010). Upon AtPep perception, both PEPRs trigger PTI-like defence responses reminiscent of the ones elicited by well-known MAMPs, such as flg22 or elf18 (Yamaguchi et al., 2010; Bartels et al., 2013; Flury et al., 2013). Given this similarity between MAMP and AtPep-induced responses and the fact that PROPEP/AtPep expression is induced upon biotic stress, AtPeps are believed to function as amplifiers of the initial defence response. In addition they might also be involved in spreading the signal of danger from the damaged or infected area to distal, not yet infected parts of the plant (Boller and Felix, 2009; Yamaguchi and Huffaker, 2011; Ross et al., 2014). A variety modes of amplification of defence signalling by AtPeps have recently been proposed, either by interacting with defence-related plant hormones (Liu et al., 2013; Tintor et al., 2013; Ross et al., 2014) or by amplifying the production of reactive oxygen species upon previous MAMP detection (Flury et al., 2013; Klauser et al., 2013).
Further support for a role of AtPeps as amplifiers of defence responses came from the fact that the exogenous application of AtPeps has been shown to enhance immunity against the hemibiotrophic pathogens Pseudomonas syringae (Yamaguchi et al., 2010) and the necrotrophic pathogen Botrytis cinerea (Liu et al., 2013). Moreover, the application of ZmPep1, an AtPep homologue in Zea mays has been shown to induce resistance against Cochliobolis heterostrophus and Colletotrichum graminicola (Huffaker et al., 2011). However, despite the apparent similarities to systemin, it was only very recently that the exogenous application of ZmPep3 has been shown to induce herbivore defence signalling, including the production of plant volatile emissions, insect deterrent metabolites, and defence-mediating phytohormones, rendering treated maize plants more resistant to the generalist herbivore Spodoptera exigua (Huffaker et al., 2013).
However, since only the exogenous application of Peps has so far been shown to induce an increased resistance against herbivore feeding, the contribution of endogenous Pep-signalling to herbivore deterrence has largely remained elusive. Using promPROPEP and promPEPR reporter lines driving a β-glucuronidase (GUS) reporter gene, the expression patterns of both PEPRs, as well as PROPEPs, were investigated here upon feeding by caterpillars of the noctuid moth Spodoptera littoralis. Using mutant plants insensitive to AtPeps, the contribution of endogenous AtPep-signalling to herbivore deterrence was also investigated and our observations were linked to specific hormone signalling cascades involved in mediating defence responses against herbivores.
Materials and methods
Plant material
Arabidopsis plants of the indicated phenotypes were grown individually in small pots at 21 °C with a 10h photoperiod for 4–5 weeks. T-DNA insertion mutants for the pepr1 pepr2 mutants are in a Col-0 background and were obtained from Birgit Kemmerling (University of Tübingen). The promPEPR::GUS and promPROPEP::GUS reporter lines used are described in Bartels et al. (2013).
Elicitor peptides and insect oral secretions
Peptides of flg22 (QRLSTGSRINSAKDDAAGLQIA) and AtPep1 (ATKVKAKQRGKEKVSSGRPGQHN) obtained from EZBiolabs were dissolved in a solution containing 1mg ml–1 bovine serum albumin and 0.1M NaCl.
Oral secretions of Spodoptera littoralis larvae (obtained from Syngenta Crop Protection, Switzerland) were obtained by gently pushing the forehead region of third and fourth instar larvae as described by Turlings et al. (1993). Until use, the oral secretions were stored at –20 °C.
GUS staining
Plant leaves were either wounded using sterile cork borers, exposed to feeding herbivores as indicated, or treated with 1 μl of Spodoptera littoralis oral secretions (by applying two droplets on the upper leaf surface). After 12h, leaves were harvested and the tissue was fixed in ice-cold 90% acetone for 20min, washed with water and then placed in GUS staining buffer (1mM 5-bromo-4-chloro-3-indolyl β-d-glucuronidase (Gold BioTechnology, St Louis, Missouri, USA), 100mM sodium phosphate (pH 7.5), 0.5mM potassium ferricyanide, 0.5mM potassium ferrocyanide, 10mM EDTA, and 0.1% (v/v) Triton X-100) at 37 °C for 12h. Plant tissue was cleared with 70% (v/v) ethanol and photographed using an Olympus SZX12 binocular microscope in combination with an Olympus DP72 camera and the CellSens imaging software (Olympus America, Pennsylvania, USA).
Gene expression analysis
Total RNA was extracted from Arabidopsis leaves using the NucleoSpin RNA plant extraction kit (Macherey-Nagel) and treated with rDNase according to the manufacturer’s specifications. AMV reverse transcriptase together with oligo(dT) primers were used to synthesize cDNA. Quantitative RT-PCR was performed in a 96-well format using a LightCycler® 480 Instrument (Roche). Based on the obtained CT values, normalized expression to the reference gene UBQ10 (AT4G05320) was calculated using the qGene protocol (Muller et al., 2002). The gene-specific primers used were as follows: UBQ10 (AT4G05320) with UBQ_fw (5′-GGCCTTGTATAATCCCTGATGAATAAG) and UBQ_rev (5′-AAAGAGATAACAGGAACGGAAACATAG), PEPR1 (AT1G73080) with PEPR1_qRT-fw (5′-ATTCCTATTGAGATA TGGAAGAG) and PEPR1_qRT_rv (5′-CCTCTTCTAAGCTGC TGTTCAC), PEPR2 (AT1G17750) with PEPR2_qRT_fw (5′-ACCA ATAATTCACCGCGACATC) and PEPR2_qRT_rv (5′-CGCATTT TCTGGTGCAATGTAC), PROPEP1 (AT5G64900) with PP1_qRT_fw (5′-ATCAGATAGACGAAGCGAAG) and PP1_qRT_rv (5′-CTAATTATGTTGGCCAGGAC), and PROPEP3 (AT5G64905) with PP3_qRT_fw (5′-CAACGATGGAGAATCTCAGA) and PP3_qRT_rv (5′-CTAATTGTGTTTGCCTCCTTT).
Microarray data analysis
Data from two recent microarrays depicting gene expression patterns after either Spodoptera littoralis feeding for 8 d (Schweizer et al., 2013) or the exogenous application of AtPep2 (Ross et al., 2014) were compared to identify similarly induced genes. This was done by cross-referencing the 50 most strongly up-regulated genes after herbivore feeding to the 1000 most strongly induced genes 2h after the application of 1 μM AtPep2.
Herbivore feeding assays
Adult plants in the vegetative stage were separately exposed to Spodoptera littoralis first instar larvae (10 per plant) for 10 d. Larvae were weighed at the beginning and at the end of the experiment to assess the mass gained, and live larvae were counted to assess survival at the end of the assay. Differences between treatments were then analysed using one-way ANOVA (α=0.05, JMP9). Weight data were square root transformed to meet the assumptions of the model. A total of 15 plants of each of the two Arabidopsis lines tested were used.
Plant hormone analysis
Several leaf discs (90mg fresh weight) were cut from leaves treated by applying 1 μl of insect oral secretions on to the upper leaf surface. Leaf tissue samples were then flash-frozen in liquid nitrogen and stored at –80 °C until hormone level quantification. Hormone extraction and analysis was performed as described by Glauser et al. (2013).
Measurement of ethylene production
For the measurement of ethylene accumulation, three leaf discs of 4–5-week-old plants were harvested using a 5mm cork borer and placed into a 6ml glass vial containing 0.5ml of ddH2O, then put back into the growth chamber and left overnight (~16h). Elicitor peptides (1 μM final concentration) and Spodoptera oral secretions (0.5% v/v final concentration) were added and vials were closed with air-tight rubber septa. After 4h of incubation at room temperature, ethylene accumulating in the free air space was measured by gas chromatography (GC-14A Shimadzu).
Results
The expression of PROPEP3 as well as both PEPRs is induced upon perception of Spodoptera littoralis oral secretions as well as herbivore feeding
In maize, individual PROPEPs and Peps were shown to have individual functions. Treatment with ZmPep1 led to an improved resistance against fungal pathogens whereas ZmPep3 application boosted plant defence against herbivores. Similarly, ZmPROPEP1 and ZmPROPEP3 transcription was induced upon treatment with a fungal pathogen or herbivore oral secretions, respectively (Huffaker et al., 2011, 2013).
The involvement of the Pep-PEPR system in fungal resistance has also been shown in Arabidopsis and tomato (Huffaker et al., 2006; Liu et al., 2013; Trivilin et al., 2014) but the biological relevance of the observation that a ZmPep3 pretreatment induces anti-herbivore resistance has not been shown due to the lack of PEPR mutants in maize. Thus a switch was made to the model plant Arabidopsis thaliana and the generalist herbivore Spodoptera littoralis to answer this question.
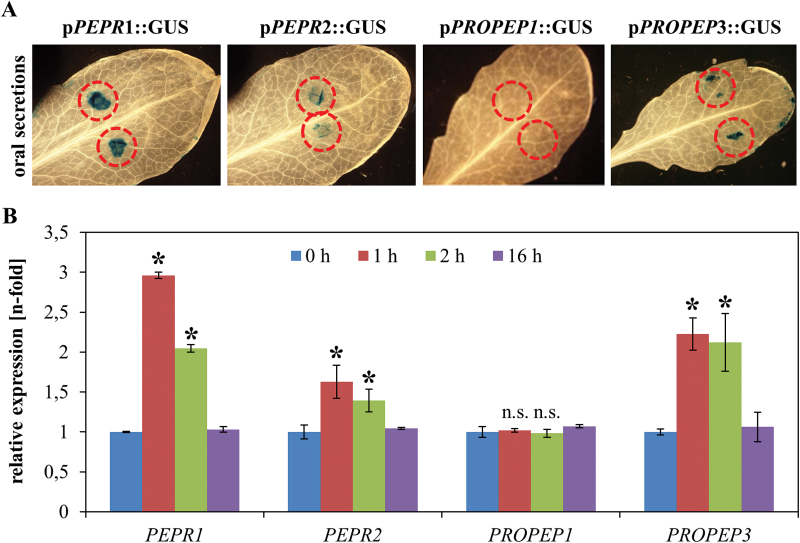
Transgenic Arabidopsis plant lines expressing a β-glucuronidase (GUS) reporter gene under the control of the promoter regions of either PROPEP1, PROPEP3, PEPR1, or PEPR2 were used as described by Bartels et al. (2013) and Spodoptera littoralis oral secretions (OS) were applied as two small droplets on to the upper leaf surface of unharmed leaves. In agreement with the up-regulation of ZmPROPEP3 upon OS detection (Huffaker et al., 2013) AtPROPEP3 is also induced locally at the site of OS application as detected by GUS staining (Fig. 1A) and via transcript quantification by real-time PCR (Fig. 1B). By contrast, AtPROPEP1 showed neither a detectable GUS-response (Fig. 1A) nor an increase in transcript abundance after OS application (Fig. 1B). The response of both PEPR promoters upon OS perception was also assessed. Similar to PROPEP3, both genes are induced upon OS application as shown by local GUS staining (Fig. 1A) as well as by real-time PCR (Fig. 1B). Notably, in contrast to the OS application procedure performed by Huffaker et al. which involved scratch-wounding, OS was just pipetted on to the leaf surface and so avoiding wounding and therefore any potential pleiotropic effects of the treatment procedure on our gene expression analysis (Huffaker et al., 2013).
Fig. 1.
Spodoptera oral secretions are sufficient to activate both PEPR and PROPEP3 promoters. (A) 1 μl of Spodoptera littoralis oral secretions were pipetted as two small droplets (red circles) onto the leaves of transgenic Arabidopsis plants expressing pPEPR::GUS, pPROPEP1::GUS, and pPROPEP3::GUS reporter constructs. After 12h, leaves were detached from the plant, fixed, and stained. For each construct, two independent lines were assessed with similar results. (B) Leaves of Arabidopsis Col-0 wild-type plants were treated with 1 μl of Spodoptera littoralis oral secretions as described above. After 0, 1, 2, and 16h they were detached from the plant and transcript levels of the respective genes were assessed by qRT PCR. Error bars show ±1 SE of three independent replicates, asterisks indicate significant differences in transcript accumulation compared with untreated plants (t test, P <0.05).
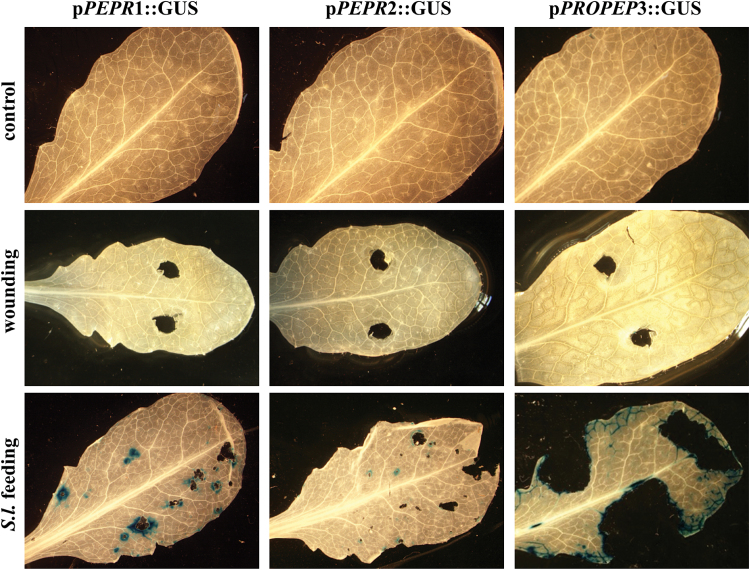
To assess directly PROPEP and PEPR gene expression upon herbivore attack, the response of PROPEP3 and both PEPRs to feeding S. littoralis larvae were analysed. Similar to the OS application, feeding of S. littoralis also strongly activated all three promoters (Fig. 2). The increased activity of the PEPR promoters is located directly around areas of herbivore attack and does not extend to unharmed parts of the leaves. In the case of the promoter of PROPEP3 the detected GUS staining was not limited to the actual feeding sites, but also spread into the leaf veins (Fig. 2). No GUS signal was detected upon wounding the plants by cutting out small leaf pieces using a sterile cork borer (Fig. 2).
Fig. 2.
Spodoptera feeding strongly induces the promoters of PEPR1, PEPR2, and PROPEP3. Leaves of transgenic Arabidopsis plants expressing pPEPR::GUS and pPROPEP3::GUS reporter constructs were either wounded using cork borers or exposed to feeding Spodoptera littoralis (S.l.) larvae. After 12h, the leaves were detached from the plant, fixed, and stained. For each construct, two independent lines were assessed with very similar results.
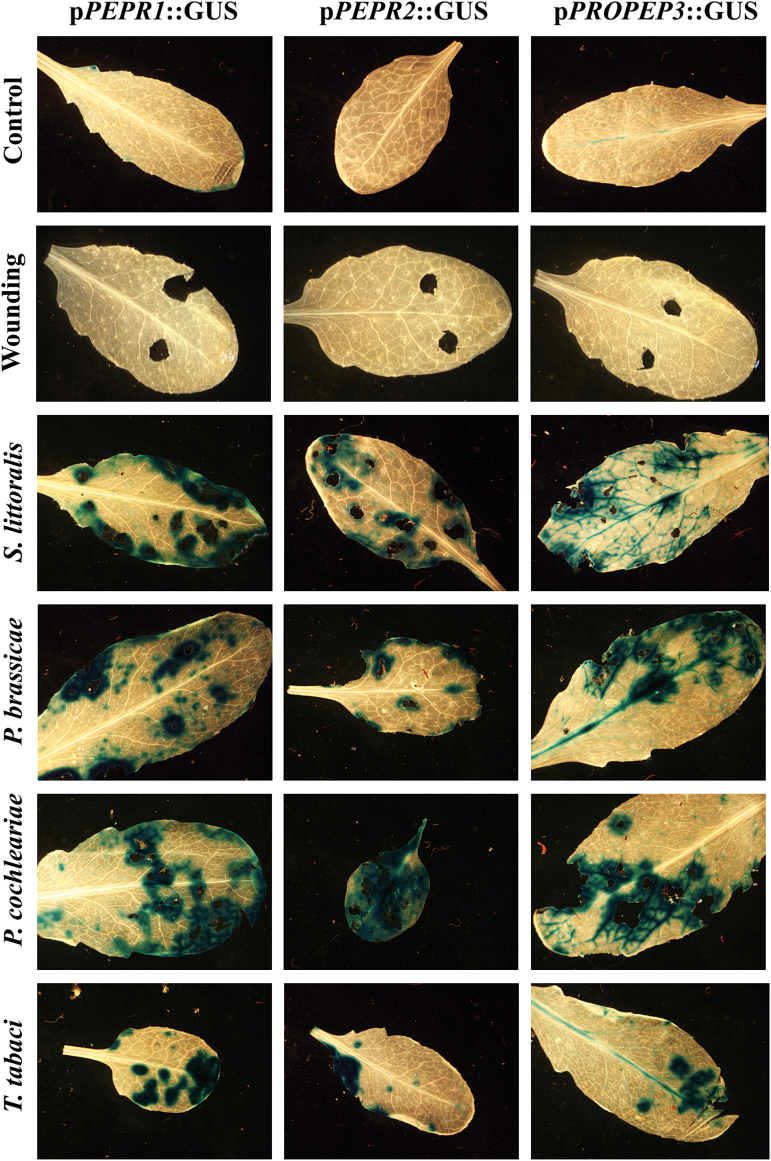
It was notable that the activation of PEPR promoters was not limited to feeding of S. littoralis. A variety of herbivores were tested on our promPEPR-GUS lines and GUS staining was found in all cases, whereas sterile wounding did not lead to detectable GUS staining (Fig 3). This was independent of the herbivores mode of attack as sucking herbivores such as thrips (T. tabaci) were also included, as well as whether the attackers were displaying a generalist (S. littoralis) or specialist feeding behaviour (e.g. P. cochleariae or P. brassicae). Overall, these findings further underline the importance of the Pep-PEPR system for herbivore resistance.
Fig. 3.
The promoters of PEPR1, PEPR2, and PROPEP3 are activated independently of feeding behaviour and specification of the feeding herbivore. Leaves of transgenic Arabidopsis plants expressing pPEPR::GUS and pPROPEP3::GUS reporter constructs were either wounded using cork borers or exposed to feeding insects. After 12h, they were detached from the plant, fixed, and stained. The following insects were assessed (from the top): Spodoptera littoralis (generalist, chewing), Pieris brassicae (specialist, chewing), Phaedon cochlearieae (specialist, chewing), and Thrips tabaci (generalist, sucking).
Spodoptera littoralis larvae perform better on pepr1 pepr2 double mutant plants
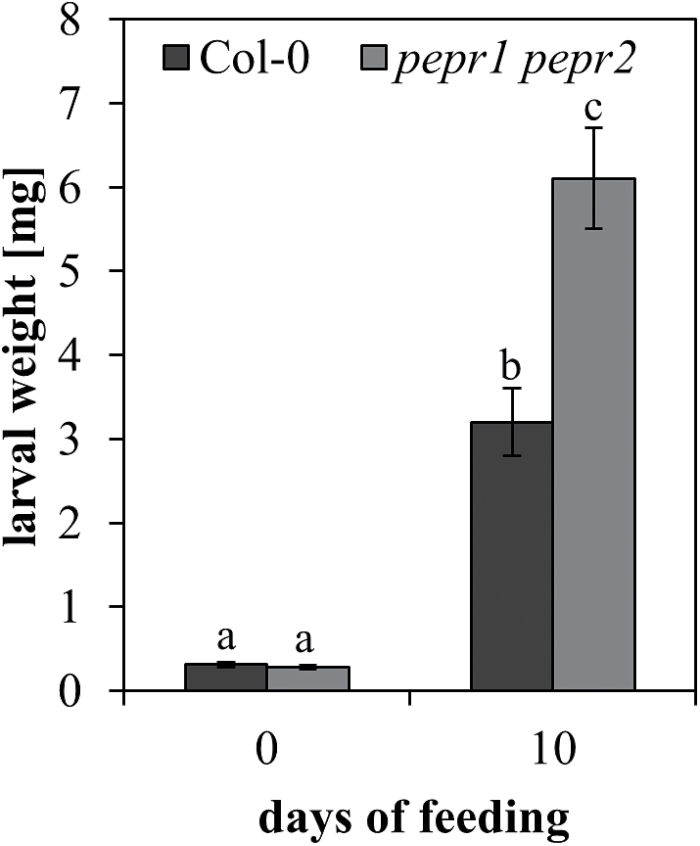
To asses further the indicated importance of the Pep-PEPR system during herbivore challenge, the feeding performance of Spodoptera littoralis on pepr1 pepr2 mutant plants, fully impaired in AtPep-signalling (Krol et al., 2010; Yamaguchi et al., 2010), was compared to Col-0 wild-type plants. Ten first instar larvae were placed on each plant for feeding. Ten days later, the larvae were removed and their performance was determined by weight gain. A remarkable difference was found in growth. Larvae feeding on Col-0 wild-type plants reached an average weight of 2.86mg whereas the ones feeding on pepr1 pepr2 plants showed an average weight of 5.37mg (Fig. 4). Comparing the performance, it was found that pepr1 pepr2 feeding larvae grew a significant 87% larger than their counterparts on Col-0 wild-type plants (F 1,13=4.82, P=0.047). Therefore, the biological relevance of the Pep-PEPR system for herbivore resistance could be proved.
Fig. 4.
Generalist herbivores perform better on plants impaired in AtPep-signalling. Mass of Spodoptera littoralis larvae (mean ±1 SE) at the beginning of the experiment (left) and after 10 d of feeding (right) on Arabidopsis Col-0 wild-type and pepr1 pepr2 mutant plants. Letters indicate significant differences between the means (α=0.05, one-way ANOVA, JMP9).
The response to AtPep perception and S. littoralis feeding overlaps in the induction of genes related to jasmonic acid signalling and herbivore resistance
Investigating the mechanism behind the contribution of an activated AtPep-signalling system to herbivore recognition and, potentially, deterrence, recently published gene expression data from Arabidopsis plants treated either with exogenously applied AtPep2 (Ross et al., 2014) or exposed to feeding Spodoptera littoralis larvae (Schweizer et al., 2013) were compared. This analysis revealed several genes which were similarly up-regulated under both circumstances (Table 1). The identified genes encode proteins potentially contributing to direct herbivore deterrence, such as proteinase inhibitors like LTP and TI1 and peroxidases (PRX52), transcription factors in defence signalling (FAD-binding proteins) as well as several genes involved in jasmonic acid (JA) biosynthesis and signalling pathways (JAZ10, LOX3, AOC1). Intriguingly, similar categories of genes were found to be induced upon the application of ZmPep3 in maize by Huffaker et al. (2013), namely proteinase inhibitors (WIP1, SerPIN) and genes involved in JA signalling (AOC, AOS).
Table 1.
Genes induced by both the exogenous application of AtPep2 and exposure to feeding Spodoptera littoralis larvae
Comparative analysis of data from two recent microarrays depicting gene expression patterns upon either 8 d Spodoptera littoralis feeding (Schweizer et al., 2013) or the exogenous application of 1 μM AtPep2 (Ross et al., 2014). The genes identified to be similarly induced are listed. Both studies reported P-values lower than 0.05 for all genes shown.
| Gene annotation | Description | Expression ratio (log2) Pep | Expression ratio (log2) Spodoptera |
|---|---|---|---|
| AT5G05340 | PRX52, peroxidase | 5.84 | 3.53 |
| AT5G13220 | JAZ 10 | 4.13 | 4.83 |
| AT3G44860 | FAMT, farnesoic acid methyl transferase | 4.11 | 3.79 |
| AT1G17420 | LOX3, lipoxygenase | 3.69 | 3.97 |
| AT5G05600 | Oxidereductase, 2OG-Fe(II) oxygenase | 2.97 | 4.57 |
| AT4G20860 | FAD-binding berberine family protein | 2.88 | 3.59 |
| AT4G12500 | Protease inhibitor (LTP) | 2.85 | 4.27 |
| AT2G38870 | Protease inhibitor | 2.53 | 3.55 |
| AT4G37990 | CAD8, cinnamyl- alcohol dehydrogenase | 2.18 | 3.86 |
| AT1G74010 | Strictosidine synthase | 2.09 | 3.64 |
| AT3G25760 | AOC1, allene oxide cyclase | 1.97 | 3.58 |
| AT2G43510 | TI1, trypsin inhibitor | 1.75 | 4.79 |
PEPR signalling contributes to JA signalling upon herbivore detection
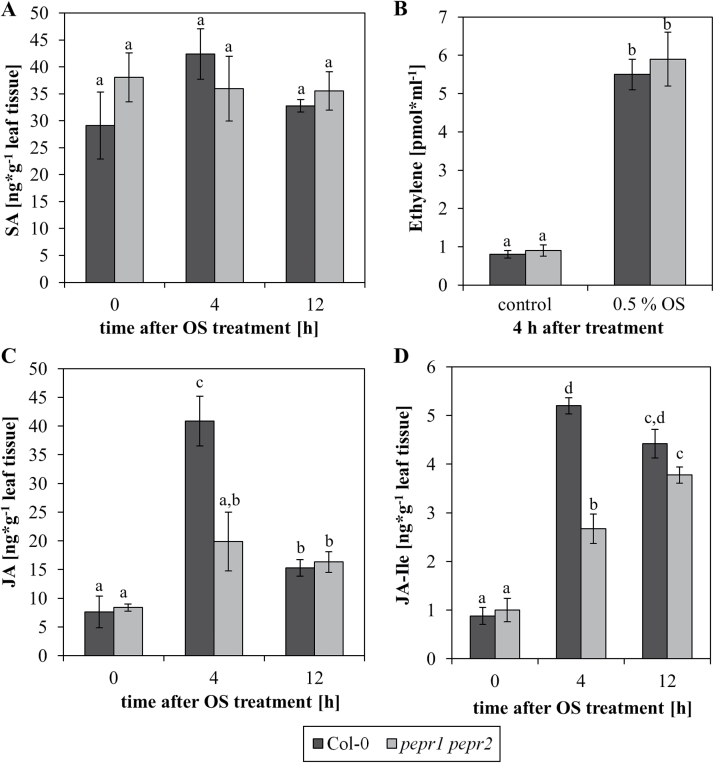
The induction of JA-related genes upon AtPep perception indicates a central role of JA to mediate the induction of herbivore resistance upon PEPR activation. However, the AtPep-PEPR system has been shown to interact positively with several hormonal pathways, enhancing defence responses against a variety of pathogens. These include the salicylic acid (SA) (Huffaker et al., 2006; Huffaker and Ryan, 2007; Ross et al., 2014), the ethylene (Huffaker and Ryan, 2007; Liu et al., 2013; Tintor et al., 2013; Ross et al., 2014), and the JA pathways (Huffaker et al., 2006; Huffaker and Ryan, 2007; Ross et al., 2014). To dissect this network further in the specific context of herbivory, the levels of the respective plant hormones were compared between Col-0 wild-type plants and the pepr1 pepr2 mutant plants before and after the application of herbivore OS (Fig. 5). Upon the perception of OS, the levels of SA did not increase at the time points assessed, with generally no difference being observed between wild-type and mutant plants (Fig. 5A). By contrast, the application of herbivore OS triggered the production of ethylene, with again no detectable difference between Col-0 wild-type and pepr1 pepr2 mutant plants (Fig. 5B). This, however, was different for the accumulation of JA and its active derivate JA-isoleucine (JA-Ile), where a greater increase of both JA and JA-Ile levels upon OS perception was observed in wild-type Col-0 plants compared with the pepr1 pepr2 mutant plants (Fig. 5C, D). This difference was most visible 4h after OS application and disappeared when JA levels flattened 12h after treatment, indicating an additional attenuation of the JA response in the mutant. Taken together, the lack of functional PEPR signalling during herbivore perception leads to reduced and/or slower production of JA which probably results in reduced herbivore resistance.
Fig. 5.
The detection of herbivore oral secretions induces JA biosynthesis in a PEPR-dependent manner. (A, C, D) Leaves of Col-0 and pepr1 pepr2 double mutant plants were treated by pipetting 1 μl of Spodoptera littoralis OS onto the upper leaf surface. After the time indicated, leaves were detached from the plant, flash frozen in liquid nitrogen and the levels of SA (A), JA (C), and JA-Ile (D) were determined by LC-MS. Bars show mean values of eight independent replicates with ±1 SE displayed as error bars. Letters indicate significant differences between the mean values (One-way ANOVA with α=0.05 and t test with P <0.05). (B) Leaf discs of Arabidopsis Col-0 and pepr1 pepr2 plants were either treated with 0.5% (v/v) OS or without any elicitor (control). Ethylene production was assessed in the headspace 4h after treatment using gas chromatography. Bars show mean values of eight independent replicates with ±1 SE displayed as error bars. Letters indicate significant differences between the mean values (one-way ANOVA with α=0.05 and t test with P <0.05).
Discussion
Recently, it was shown that the exogenous application of ZmPep3, an AtPep orthologue in maize, induced defence responses against herbivore feeding (Huffaker et al., 2013). Although these findings already suggest a role for Pep-signalling in the plant’s response against herbivores, the biological relevance remained unclear.
With this work, the biological relevance of the Pep-PEPR system in the context of herbivore resistance can now be ascertained by showing that, first, the Pep-PEPR system is induced upon herbivore recognition and, second, that plants lacking a functional Pep-PEPR system are indeed more susceptible to herbivore feeding.
Herbivore feeding activates the promoters of Arabidopsis PROPEP3, but also PEPR1 and PEPR2
In maize, the application of herbivore OS was shown to trigger transcript accumulation of ZmPROPEP3 (Huffaker et al., 2013). Using quantitative real-time PCR as well as transgenic plants expressing a GUS-reporter gene under the control of the AtPROPEP3 promoter sequence, these findings could now be confirmed for the model organism Arabidopsis thaliana. Intriguingly, a similar expression pattern was also observed for AtPROPEP1 and ZmPROPEP1, both of which are not induced by herbivore oral secretions (Huffaker et al. 2013), but respond to the detection of fungal pathogens (Huffaker et al., 2011; Liu et al., 2013). In addition to AtPROPEP3, the promoters of AtPEPR1 and AtPEPR2, the two receptors for AtPeps (Krol et al., 2010; Yamaguchi et al., 2010) also showed rapid activation upon exposure to herbivore OS. This activation was stronger for the promoter of AtPEPR1 than for AtPEPR2, supporting the assumption that AtPEPR1 is the more important Pep-receptor and reflecting the generally more pronounced expression of AtPEPR1 as well as the fact that AtPEPR1 is able to detect all AtPeps whereas AtPEPR2 can only detect AtPep1 and AtPep2 (Krol et al., 2010; Yamaguchi et al., 2010; Bartels et al., 2013). Moreover, based on the report that AtPEPR2 is important for the repression of Glutamine Dumper (GUD) genes and the inhibition of root growth, this receptor might play a more dominant role in the root (Ma et al., 2014). However, in addition to the already mentioned similarities between the regulation of maize PROPEPs and Arabidopsis PROPEPs, the activation of, specifically, the promoter of AtPROPEP3 seems to be in line with other recent expression studies, which have shown that, in particular, the expression patterns of AtPROPEP2 and AtPROPEP3 are linked to defence signalling (Logemann et al., 2013; Ross et al., 2014). However, despite the apparent central role of PROPEP3 regarding herbivore resistance other PROPEPs and Peps are likely to contribute as well. In Arabidopsis, PROPEP5 is constitutively expressed in leaves and Pep1 has been isolated from unharmed leaves indicating that these PROPEPs and Peps might be released upon damage due to herbivore feeding (Huffaker et al., 2006; Bartels et al., 2013). This would again activate PEPR-triggered defence responses probably contributing to herbivore resistance. Thus, an analysis of PROPEP knock-out mutants could help in understanding the specific contribution of each PROPEP to plant immunity in general and herbivore resistance in particular.
The application of herbivore OS alone, however, constitutes a slightly artificial system as it does not involve the mechanical damage generally occurring upon herbivore feeding (Howe and Jander, 2008). Wounding is known to induce JA accumulation rapidly which would activate anti-herbivore responses and so herbivores make use of elicitors of, for example, microbial origin present in their oral secretions to activate SA signalling and therewith counteract JA signalling (Glauser et al., 2009; Chung et al., 2013). This strategy was shown to be effective in suppressing the induction of wounding-responsive genes upon herbivore feeding using the same model system (Arabidopsis and S. littoralis) used here (Consales et al., 2012). It was found here that the Pep-PEPR system is induced upon OS perception. This is in line with the robustness of PROPEP3 induction upon microbial challenges which is not impaired by the dysfunction of either the JA, the ethylene or the SA signalling pathways (Ross et al., 2014). Thus the Pep-PEPR system seems to be immune to a potential perturbation of anti-herbivore signalling by OS elicitors.
Similar to OS application, Spodoptera feeding also led to a very strong and local induction of the promoters of both PEPRs and PROPEP3, whereas sterile wounding alone did not induce the promoters of both PEPRs nor PROPEP3. It is notable that, previously, activation of the PROPEP3 promoter was found upon mechanical damage applied with a forceps but this activation was limited to the damaged section of the central vasculature of the leaf and was not detectable in the areas which were treated in this study and where the Spodoptera larvae were feeding (Bartels et al., 2013). This indicates a distinct pattern of the Pep-PEPR system activation depending on the danger signal perceived.
Herbivores attack plants with different feeding strategies. Apart from chewing herbivores, such as S. littoralis, others, such as aphids and thrips, can nourish themselves from the plant tissue by using stylets either to attack single cells or to suck phloem juice from the plant’s vascular tissue (Howe and Jander, 2008). Most herbivore-derived elicitors have so far been identified in the regurgitant of chewing herbivores (Mithofer and Boland, 2008). However, the activation of the Pep-PEPR system upon feeding of thrips (lacking the production of regurgitant) indicates additional or different modes of herbivore detection, potentially through substances and/or microbes in the attacker’s saliva (Delphia et al., 2007; Chung et al., 2013).
Taken together, the observed local induction of the Pep-PEPR system is not a general response to mechanical damage but a specific and robust response to the perception of herbivore oral secretions and elicitors therein. Unfortunately, given this plethora of potential sources for the HAMP(s) triggering an activation of the Pep-PEPR system, it remains a challenge eventually to identify actual compound(s). Still, the combination of these findings with the fact that the Pep-PEPR signalling system seems to be abundant in all higher plants (Huffaker et al., 2013; Lori et al., 2015) suggests that the Pep-PEPR system is a conserved signalling mechanism for herbivore defence.
An intact AtPep-signalling system is required for full defence responses against herbivores
Several sources have proposed Peps to be considered as endogenous amplifiers of defence responses against a variety of biotic dangers, based on their ability to trigger defence responses and to interact positively with other defence signalling pathways (Boller and Felix, 2009; Yamaguchi and Huffaker, 2011; Macho and Zipfel, 2014). Our comparative analysis of recent transcription profile studies of plants either exposed to exogenously applied AtPep2 (Ross et al., 2014) or to feeding herbivores (Schweizer et al., 2013) has led to the identification of a set of similarly induced genes under both conditions, indicating that transcriptional changes upon Pep-signalling include a set of herbivory responsive genes. When combining these findings with the fact that the expression of PROPEP3 as well as both PEPRs is induced by feeding herbivores, it is tempting to expand the aforementioned amplifier theory for AtPep-signalling to herbivore deterrence. In agreement with this, feeding Spodoptera littoralis larvae perform significantly better on mutant plants lacking a functional AtPep-PEPR-signalling system.
The AtPep-system contributes to JA-mediated defence responses
AtPeps have been suggested to interact with several plant hormone pathways involved in responses to abiotic stress. These pathways include SA (Huffaker et al., 2006), ethylene (Liu et al., 2013; Tintor et al., 2013), and JA (Huffaker and Ryan, 2007; Flury et al., 2013), with ethylene and JA being particularly strongly and positively intertwined with Pep-signalling (Huffaker and Ryan, 2007; Flury et al., 2013; Ross et al., 2014). Furthermore, JA is particularly known to be a major mediator of plant defence responses upon herbivore attack (Howe and Jander, 2008). Aligned with this, our studies revealed that, upon OS detection, both ethylene as well as JA biosynthesis was strongly induced whereas SA levels did not increase.
Moreover, this JA and JA-Ile accumulation was significantly reduced in mutant plants lacking a functional Pep-signalling system. These findings are also aligned with the aforementioned transcriptome analysis, which led to the identification of several genes involved in JA biosynthesis pathways being induced upon both herbivore challenge and treatment with AtPep2 (Schweizer et al., 2013; Ross et al., 2014).
Interestingly, recent studies have shown a positive feedback loop between AtPep- and JA-signalling with the application of AtPeps leading to increased JA accumulation and a functional JA signalling system being required for full-strength Pep-signalling (Flury et al., 2013; Huffaker et al., 2013). In this context, our findings provide additional lines of evidence that support a potential role of the AtPep system as an amplifier of JA-mediated defence responses, as shown here in the case of herbivore deterrence.
Both the temporal as well as the spatial resolution of this positive interaction between AtPep- and JA-signalling in the context of herbivore deterrence remain at least partially elusive: First, since PROPEPs are induced by JA and Peps trigger JA accumulation, it needs to be investigated whether the detection of herbivory leads first to an activation of JA signalling, which then induces the Pep-system, or vice-versa. The use of a JA-insensitive mutant might give further insights here but will be complex to analyse due to the positive feedback between both, with not only JA signalling being impaired in JA mutants but also PEPR signalling being reduced which also feeds back on the induction of PROPEP and PEPR expression. Second, as the transcription of both AtPEPRs as well as AtPROPEP3 is induced locally around the site of herbivore detection, it is tempting to speculate that the AtPep-PEPR system is mainly involved in local defence responses. However, Ross et al. (2014) showed that, in addition to triggering local defence responses, Pep-signalling is also required for the full activation of systemic defence responses, although as yet only in the context of microbial pathogens. Therefore, apart from the temporal, the spatial resolution of Pep-signalling in the context of herbivore deterrence also requires further investigation and the assays and reporter lines described here could prove helpful tools to investigate these processes further.
Acknowledgements
We would like to thank Roland Reist and Oliver Kindler (Syngenta Crop Protection, Stein) for providing Spodoptera littoralis larvae for the feeding assays and the harvest of oral secretions, Ted Farmer (University of Lausanne) for critical input on JA signalling, and Rainer Böni for help with the promPEPR::GUS lines. We would also like to thank Yosra Chabaane and David Ermacora for their help in collecting the data for the herbivore performance assay.
GAD and TCJT are supported by a grant from the European Science Foundation in the context of Eurocore project EuroVol. TB is supported by the Swiss National Science Foundation [grant 31003A_127563].
References
- Albert M. 2013. Peptides as triggers of plant defence. Journal of Experimental Botany 64, 5269–5279. [DOI] [PubMed] [Google Scholar]
- Bartels S, Lori M, Mbengue M, van Verk M, Klauser D, Hander T, Boni R, Robatzek S, Boller T. 2013. The family of Peps and their precursors in Arabidopsis: differential expression and localization but similar induction of pattern-triggered immune responses. Journal of Experimental Botany 64, 5309–5321. [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G. 2009. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annual Review of Plant Biology 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Chung SH, Rosa C, Scully ED, Peiffer M, Tooker JF, Hoover K, Luthe DS, Felton GW. 2013. Herbivore exploits orally secreted bacteria to suppress plant defenses. Proceedings of the National Academy of Sciences, USA 110, 15728–15733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consales F, Schweizer F, Erb M, Gouhier-Darimont C, Bodenhausen N, Bruessow F, Sobhy I, Reymond P. 2012. Insect oral secretions suppress wound-induced responses in Arabidopsis . Journal of Experimental Botany 63, 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt DC, Refi-Hind S, Stratmann JW, Lincoln DE. 2010. Systemin and jasmonic acid regulate constitutive and herbivore-induced systemic volatile emissions in tomato, Solanum lycopersicum . Phytochemistry 71, 2024–2037. [DOI] [PubMed] [Google Scholar]
- Delphia CM, Mescher MC, De Moraes CM. 2007. Induction of plant volatiles by herbivores with different feeding habits and the effects of induced defenses on host–plant selection by thrips. Journal of Chemical Ecology 33, 997–1012. [DOI] [PubMed] [Google Scholar]
- D’Ovidio R, Mattei B, Roberti S, Bellincampi D. 2004. Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant–pathogen interactions. Biochimica et Biophysica Acta 1696, 237–244. [DOI] [PubMed] [Google Scholar]
- Flury P, Klauser D, Schulze B, Boller T, Bartels S. 2013. The anticipation of danger: MAMP perception enhances AtPep-triggered oxidative burst. Plant Physiology 161, 2023–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauser G, Dubugnon L, Mousavi SAR, Rudaz S, Wolfender JL, Farmer EE. 2009. Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis . Journal of Biological Chemistry 284, 34506–34513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauser G, Wolfender JL. 2013. A non-targeted approach for extended liquid chromatography–mass spectrometry profiling of free esterified jarsmonates after wounding. Methods in Molecular Biology 1011, 123–134. [DOI] [PubMed] [Google Scholar]
- Holton N, Harrison K, Yokota T, Bishop GJ. 2008. Tomato BRI1 and systemin wound signalling. Plant Signaling & Behavior 3, 54–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Jander G. 2008. Plant immunity to insect herbivores. Annual Review of Plant Biology 59, 41–66. [DOI] [PubMed] [Google Scholar]
- Huffaker A, Dafoe NJ, Schmelz EA. 2011. ZmPep1, an ortholog of Arabidopsis elicitor peptide 1, regulates maize innate immunity and enhances disease resistance. Plant Physiology 155, 1325–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Ryan CA. 2006. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proceedings of the National Academy of Sciences, USA 103, 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Pearce G, Veyrat N, et al. 2013. Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proceedings of the National Academy of Sciences, USA 110, 5707–5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A, Ryan CA. 2007. Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proceedings of the National Academy of Sciences, USA 104, 10732–10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauser D, Flury P, Boller T, Bartels S. 2013. Several MAMPs, including chitin fragments, enhance AtPep-triggered oxidative burst independently of wounding. Plant Signaling & Behavior 8, e25346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol E, Mentzel T, Chinchilla D, et al. 2010. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. Journal of Biological Chemistry 285, 13471–13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfermeijer FC, Staal M, Malinowski R, Stratmann JW, Elzenga JTM. 2008. Micro-electrode flux estimation confirms that the Solanum pimpinellifolium cu3 mutant still responds to systemin. Plant Physiology 146, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, Tian X, Zhou J-M. 2013. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proceedings of the National Academy of Sciences, USA 110, 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Birkenbihl RP, Rawat V, Schneeberger K, Schmelzer E, Somssich IE. 2013. Functional dissection of the PROPEP2 and PROPEP3 promoters reveals the importance of WRKY factors in mediating microbe-associated molecular pattern-induced expression. The New Phytologist 198, 1165–1177. [DOI] [PubMed] [Google Scholar]
- Lori M, van Verk M, Hander T, Schatowitz H, Klauser D, Flury P, Gehring C, Boller T, Bartels S. 2015. Evolutionary divergence of the plant elicitor peptides (Peps) and their receptors: interfamily incompatibility of perception but compatibility of downstream signalling. Journal of Experimental Botany 66, 5315–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Guo J, Kang Y, Doman K, Bryan AC, Tax FE, Yamaguchi Y, Qi Z. 2014. AtPEPTIDE RECEPTOR2 mediates the AtPEPTIDE1 induced cytosolic Ca2+ rise which is required for the suppression of Glutamine Dumper gene expression in Arabidopsis roots. Journal of Integrative Plant Biology 56, 684–694. [DOI] [PubMed] [Google Scholar]
- Macho AP, Zipfel C. 2014. Plant PRRs and the activation of innate immune signaling. Molecular Cell 54, 263–272. [DOI] [PubMed] [Google Scholar]
- Malinowski R, Higgins R, Luo Y, Piper L, Nazir A, Bajwa VS, Clouse SD, Thompson PR, Stratmann JW. 2009. The tomato brassinosteroid receptor BRI1 increases binding of systemin to tobacco plasma membranes, but is not involved in systemin signaling. Plant Molecular Biology 70, 603–616. [DOI] [PubMed] [Google Scholar]
- Mithofer A, Boland W. 2008. Recognition of herbivory-associated molecular patterns. Plant Physiology 146, 825–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PY, Janovjak H, Miserez AR, Dobbie Z. 2002. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32, 1372–1374. [PubMed] [Google Scholar]
- Ross A, Yamada K, Hiruma K, Yamashita-Yamada M, Lu X, Takano Y, Tsuda K, Saijo Y. 2014. The Arabidopsis PEPR pathway couples local and systemic plant immunity. The EMBO Journal 33, 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CA, Pearce G. 2003. Systemins: a functionally defined family of peptide signal that regulate defensive genes in Solanaceae species. Proceedings of the National Academy of Sciences, USA 100, 14577–14580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Alborn HT, Tumlinson JH, 3rd, Teal PE. 2009. Phytohormone-based activity mapping of insect herbivore-produced elicitors. Proceedings of the National Academy of Sciences, USA 106, 653–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F, Bodenhausen N, Lassueur S, Masclaux FG, Reymond P. 2013. Differential contribution of transcription factors to Arabidopsis thaliana defense against Spodoptera littoralis . Frontiers in Plant Science 4, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwessinger B, Ronald PC. 2012. Plant innate immunity: perception of conserved microbial signatures. Annual Review of Plant Biology 63, 451–482. [DOI] [PubMed] [Google Scholar]
- Sieber P, Schorderet M, Ryser U, Buchala A, Kolattukudy P, Metraux JP, Nawrath C. 2000. Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. The Plant Cell 12, 721–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JQ, Jiang HL, Li CY. 2011. Systemin/Jasmonate-mediated systemic defense signaling in tomato. Molecular Plant 4, 607–615. [DOI] [PubMed] [Google Scholar]
- Tintor N, Ross A, Kanehara K, Yamada K, Fan L, Kemmerling B, Nurnberger T, Tsuda K, Saijo Y. 2013. Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proceedings of the National Academy of Sciences, USA 110, 6211–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivilin AP, Hartke S, Moraes MG. 2014. Components of different signalling pathways regulated by a new orthologue of AtPROPEP1 in tomato following infection by pathogens. Plant Pathology 63, 1110–1118. [Google Scholar]
- Turlings TC, McCall PJ, Alborn HT, Tumlinson JH. 1993. An elicitor in caterpillar oral secretions that induces corn seedlings to emit chemical signals attractive to parasitic wasps. Journal of Chemical Ecology 19, 411–425. [DOI] [PubMed] [Google Scholar]
- Xu S, Zhou W, Pottinger S, Baldwin IT. 2015. Herbivore associated elicitor-induced defences are highly specific among closely related Nicotiana species. BMC Plant Biology 15, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A. 2011. Endogenous peptide elicitors in higher plants. Current Opinion in Plant Biology 14, 351–357. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA. 2010. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis . The Plant Cell 22, 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA. 2006. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proceedings of the National Academy of Sciences, USA 103, 10104–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]