Highlight
This study presents new members of the IDA/IDL and PIP/PIPL families of peptide ligands in Arabidopsis, and highlights that family members are linked to stress responses as well as development.
Key words: Arabidopsis, evolution, gene expression, INFLORESCENCE DEFICIENT IN ABSCISSION, peptide ligand.
Abstract
Peptide ligands play crucial roles in the life cycle of plants by modulating the innate immunity against pathogens and regulating growth and developmental processes. One well-studied example is INFLORESCENCE DEFICIENT IN ABSCISSION (IDA), which controls floral organ abscission and lateral root emergence in Arabidopsis thaliana. IDA belongs to a family of five additional IDA-LIKE (IDL) members that have all been suggested to be involved in regulation of Arabidopsis development. Here we present three novel members of the IDL subfamily and show that two of them are strongly and rapidly induced by different biotic and abiotic stresses. Furthermore, we provide data that the recently identified PAMP-INDUCED SECRETED PEPTIDE (PIP) and PIP-LIKE (PIPL) peptides, which show similarity to the IDL and C-TERMINALLY ENCODED PEPTIDE (CEP) peptides, are not only involved in innate immune response in Arabidopsis but are also induced by abiotic stress. Expression patterns of the IDA/IDL and PIP/PIPL genes were analysed using in silico data, qRT-PCR and GUS promoter lines. Transcriptomic responses to PIPL3 peptide treatment suggested a role in regulation of biotic stress responses and cell wall modification.
Introduction
Plants, like all other multicellular organisms, are dependent on cell-to-cell communication for growth and development, as well as for managing and surviving in a challenging and unpredictable environment. Plant cells are linked together by a cellulose wall, and signals between cells passes through plasmodesmata (Gallagher and Benfey, 2005; Kim, 2005) or through ligand-receptor interactions on the cell surface (Shiu and Bleecker, 2001a). Recent evidence indicates that there could be a connection between these two (Stahl and Simon, 2013). For many years, research focused on the classical phytohormones and their abilities to mediate physiological responses, but during the last decade, peptide ligands have emerged as important mediators of cell-to-cell communication in both development and defence (Butenko et al., 2009; Matsubayashi, 2014). Most peptide ligands are translated as prepropeptides and shuttled into the secretory pathway through their N-terminal signal peptide (SP). The SP is removed, followed by further structural modifications to yield the mature peptide ligand (Murphy et al., 2012). Peptide ligands can be divided into two main groups based on these modifications. Cysteine-rich peptides are characterized by an even number of cysteine residues that form intramolecular disulfide bonds upon maturation (Matsubayashi, 2014). Small peptides may on the other hand be generated from the C-terminus of propeptides with a general absence of cysteine residues, and the active peptides may contain post-translational modifications of key amino acids, like tyrosine sulfation, proline hydroxylation and hydroxyproline arabinosylation (Matsubayashi, 2014; Tabata and Sawa, 2014). A large number of genes encoding putative RECEPTOR-LIKE KINASEs (RLKs) and peptide ligands have been identified in the Arabidopsis genome (Shiu and Bleecker, 2001b; Lease and Walker, 2006); still, only a few ligands have been characterized and linked to a receptor and a cellular response (Butenko et al., 2009, 2014).
A well-studied peptide of the second category is INFLORESCENCE DEFICIENT IN ABSCISSION (IDA), known to regulate cell separation processes in A. thaliana (Aalen et al., 2013). The ida mutant fails to undergo floral organ abscission (Butenko et al., 2003), and overexpression of IDA leads to premature and ectopic abscission (Stenvik et al., 2006). Twenty amino acids in the C-terminal region, termed EPIP, were shown in genetic experiments to be sufficient to rescue the ida phenotype, thus suggesting that EPIP encompasses the active ligand motif of the peptide (Stenvik et al., 2008). IDA mediates its effect through the two LEUCINE-RICH REPEAT RLKs (LRR-RLKs) HAESA (HAE) and HAESA-LIKE 2 (HSL2), as the double knockout hae hsl2 is phenotypically similar to the ida mutant and overexpression of IDA is not able to rescue this phenotype (Cho et al., 2008; Stenvik et al., 2008). Moreover, a dodeca hydroxyprolinated peptide within the EPIP domain can activate and bind HSL2, and also activate HAE, although at substantially higher concentration (Butenko et al., 2014). So far, five genes encoding peptides with similarity to IDA, named IDA-LIKE 1 to 5 (IDL1 to 5), have been identified in the Arabidopsis genome (Butenko et al., 2003). It has previously been suggested that the IDL genes may share a common role in regulating cell separation events, as they are expressed at sites where cell separation occurs, such as during vascular development, stomata formation, root cap sloughing, lateral root emergence and seed shedding (Stenvik et al., 2008; Kumpf et al., 2013).
Bioinformatic tools have been used to identify ~1000 putative peptides in Arabidopsis, based on their general features and similarities to known peptides (Lease and Walker, 2010). Currently, only a small fraction of these have been assigned a function. In this paper we present three new members of the IDA-LIKE family named IDA-LIKE 6 (IDL6), IDA-LIKE 7 (IDL7) and IDA-LIKE 8 (IDL8), encoding putative proteins with a ligand motif similar to IDA. In addition, we have in parallel with Hou et al. (2014) identified a new family of 11 genes termed PAMP-INDUCED SECRETED PEPTIDES (PIPs) and PIP-LIKE (PIPLs) (Hou et al., 2014) encoding peptides with similarity to IDA/IDLs and the C-TERMINALLY ENCODED PEPTIDEs (CEPs) (Ohyama et al., 2008; Delay et al., 2013; Imin et al., 2013; Roberts et al., 2013). These families can be recognized by the presence of one or both of two C-terminal, conserved core motifs: SGPS, a motif present in the functional peptide of IDA (Butenko et al., 2014), is found in IDA/IDLs and PIP/PIPLs, whereas the GxGH motif located at the extreme C-terminal is common for PIP/PIPLs and CEPs. Interestingly, while the IDA/IDL and CEP members characterized so far are involved in developmental processes, we show that the PIP/PIPL peptides are involved in stress responses.
Materials and methods
Identification of IDA/IDL and PIP/PIPL family genes and phylogenetic analyses
Full-length protein sequences and the conserved C-terminal domain of IDA and IDL1-5 were used in TBLASTN searches against expressed sequence tag (EST), genomic and non-redundant nucleotide databases at NCBI (Altschul et al., 1997). In order to further investigate the presence of IDA/IDL family members in other plant species, similar BLAST searches were performed on the Phytozome v9.1 genome (Goodstein et al., 2012) and OneKP EST (https://sites.google.com/a/ualberta.ca/onekp/ accessed 28 May 2015) databases. Protein alignments were made using the ClustalX programme (Larkin et al., 2007) and later manually refined with GeneDoc (Nicholas et al., 1997). Neighbour-joining (N-J) trees were produced from the protein alignments using the N-J method (Saitou and Nei, 1987) and Kimura’s correction for multiple substitutions as implemented in the ClustalX programme. In total 1000 bootstrap trials were run on the N-J tree. Maximum-likelihood (ML) analysis of the full-length IDA/IDL and PIP/PIPL protein alignments were performed using the RAxML programme (Stamatakis, 2006) with the PROTGAMMABLOSUM62 substitution model and running 1000 bootstrap replicas. BLOSUM substitution matrices were used in both ML and N-J analyses. Trees were visualized using TreeView 1.6.6 (Page, 2002) and refined in Adobe Illustrator CS6. SP sequences were identified through SignalP 4.0 (Petersen et al., 2011) (http://www.cbs.dtu.dk/services/SignalP/ accessed 28 May 2015). Protein sequence motif visualization was done using WebLogo (Crooks et al., 2004; Schneider and Stephens, 1990). Analysis of gene duplication events and identification of syntenic regions was done by screening the 40 nearest protein coding genes flanking each of the IDA/IDL-PIP/PIPL gene loci for other closely related genes located next to the IDA/IDL and PIP/PIPL genes. Each region was analysed by BLASTP searches, and a custom-made Perl script was used to parse BLAST tables and identify high scoring proteins (included in the top 5 score list) that had corresponding genes mapping to IDA/IDL-PIP/PIPL genomic regions.
Plant material
Seeds of the Arabidopsis thaliana ecotype Col-0 (N1092) were obtained from the European Arabidopsis Stock Centre (NASC, Nottingham, UK).
The five pIDL:GUS constructs were made using Gateway technology. The promoters included 1555, 1864, 1908, 1980 and 2020bp upstream of the ATG start codon of IDL1 to IDL5, respectively (Stenvik et al. 2008).
Plant growth conditions and plant tissue collection for expression analysis during development
Seeds of Col-0 ecotype were surface sterilized and sown on half-strength MS plates supplemented by 2% (w/v) sucrose at a density of 44 seeds per Petri dish (14cm diameter) and stratified for 3 d at 4°C before being transferred to a controlled in vitro growth room under a 16h light (70 µmol m-2 sec-1): 8h dark photoperiod at 22°C. At stage 1.10 (Boyes et al., 2001), plants were transferred to soil and grown further in a controlled growth chamber (VB1514, Vötsch Industrietechnik, Balingen, Germany) under the same light conditions at 22°C until the end of the experiment.
Tissue was harvested at different growth stages as defined by Boyes et al. (2001). For stages 1.0, 1.06 and 1.10, whole plantlets were harvested from in vitro cultivation medium. At the later stages roots, rosette leaves, cauline leaves, stem, inflorescences and siliques were harvested separately. All material was immediately flash frozen in liquid nitrogen upon harvesting and stored at −80°C until further processing. Three biological replicates were harvested, where each replicate consisted of plant material pooled from eight Petri dishes (stage 1.0), four Petri dishes (stages 1.06 and 1.10) and five plants (stages 6.00 and 8.00), respectively.
Stress treatments
All treatments were conducted on 2-week-old wild-type seedlings corresponding to growth stage 1.06 (Boyes et al., 2001) unless otherwise stated. Seeds of Col-0 ecotype were surface sterilized and sown out on half-strength MS plates supplemented by 2% (w/v) sucrose at a density of 20 seeds per Petri dish (14cm diameter). For chitin, cycloheximide (CHX) and anisomycin treatments, seedlings were sprayed with 10 µg/ml chitin, 10 µg/ml CHX or 15 µg/ml anisomycin in MilliQ (MQ) water added 0.02% Silwet L-77 (Lehle Seeds) and vacuum infiltrated at 20 inches Hg for 1min. As control, plants were treated with MQ water added 0.02% Silwet L-77 and vacuum infiltrated at 20 inches Hg for 1min. Seedlings were incubated 1h (chitin) and 6h (CHX and anisomycin) after treatment under normal growth conditions before harvesting. Salt treatment was conducted in 24 well plates (1 seed per well) containing 1ml liquid half-strength MS and 2% (w/v) sucrose. At stage 1.06 (Boyes et al., 2001), the medium was replaced with liquid half-strength MS supplemented with 2% sucrose (w/v) and NaCl (150mM). Control plants were placed in fresh half-strength MS medium [2% sucrose (w/v)] without NaCl. The seedlings were treated for 6h before harvesting. For all stress experiments, three biological replicates were harvested, each replicate consisting of plant material pooled from three Petri dishes. Brevicoryne brassicae treatments were conducted as described in Kuśnierczyk et al. (2011).
Peptide treatments for microarray analyses
Peptides of the putative ligand motif of PIPL3 [LSSAGERMHTMASG(HYP)SRRGAGH, where HYP is hydroxyproline] and a mock peptide (LSPGKNLSAPGRVGSNPFTKLRGS) were synthesized with a purity of >95% by Biomatik (Cambridge, Canada). Seeds of Col-0 ecotype were surface-sterilized and sown out on half-strength MS plates at a density of 20 seeds per Petri dish (14cm diameter), and stratified for 3 d at 4°C. Plates were grown under a 16h photoperiod (70 µmol m-2 s-1) at 22°C for 2 weeks. Seedlings were sprayed with an aqueous peptide solution (100nM) supplemented with 0.02% silwet L-77 (Lehle Seeds, UK). Whole rosettes were collected 3h after treatment, snap-frozen in liquid nitrogen, and stored at −80°C.
RNA extraction and cDNA synthesis
100mg frozen plant tissue each from four biological replicas were homogenized using TissueLyser II (Qiagen, Hilden, Germany) for 2×2min at 25 Hz. Total RNA was extracted with the Spectrum Plant Total RNA kit (Sigma-Aldrich, Saint Louis, USA) as described by the supplier, but with lysis solution being added to the plant tissue between the two disruption cycles. An on-column DNase digestion was performed using the RNase-Free DNase Set (Qiagen, Hilden, Germany). Total RNA was quantified using NanoDrop ND-1000 (Nanodrop, Delaware, USA) and RNA quality was verified by formaldehyde gel electrophoresis. RNA was stored at −80°C until used.
cDNA synthesis was performed on 1 µg total RNA using the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany), following the supplier’s instructions. cDNA samples were diluted 10-fold before use in qRT-PCR reactions.
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed on a LightCycler 480 using the LightCycler 480 SYBR Green I Master kit (Roche Applied Science, Mannheim, Germany), with PCR parameters as recommended by the supplier: pre-incubation was performed at 95°C for 5min, followed by 50 amplification cycles, each consisting of 10 s denaturation at 95°C, 10 s annealing at 55°C and 10 s elongation at 72°C. TIP41-LIKE (At4g34270) was used as reference gene (Czechowski et al., 2005) for the stress and developmental analyses, and CYP71A13 (At2g30770) was used as negative RT control. PCR efficiencies and Ct values were calculated by linear regression using the LinRegPCR software (Ramakers et al., 2003; Ruijter et al., 2009), and mean PCR efficiency was calculated for each pair of primers. Ct-values and PCR efficiencies were then imported into the REST 2008 software (Pfaffl et al., 2002) to calculate the statistical significance of differences in expression levels upon various treatments. Primers used are listed in Supplementary Table S1.
Microarray and statistical analysis
Genome-wide expression analysis was performed using the Arabidopsis (V4) Gene Expression Microarray 4×44K (Agilent Technology, USA) as described by the supplier’s manual: total RNA (~0.2 µg) was reverse transcribed, amplified and labelled using the Low Input Quick Amp Labeling Kit, two-colour protocol, (Agilent p/n 5190-2306) (Agilent Technologies, USA). Hybridization was performed with the Gene Expression Hybridization Kit (Agilent p/n 5188–5242). 825ng cRNA from both mock-treated plants and PIPL3 peptide-treated plants were used. The cRNA mixture was fragmented and hybridized on Arabidopsis (V4) Gene Expression Microarray 4×44K arrays in a rotary oven at 65°C for ~15h. cRNA from the mock- and PIPL3-treated plants were alternately labelled with Cy3 or Cy5, which makes it possible to assess dye bias effects during the statistical analysis. The slides were washed with Gene Expression Wash Buffer 1 (Agilent p/n 5188–5325), Gene Expression Wash Buffer 2 (Agilent p/n 5188–5326), acetonitrile (VWR International) and Stabilization and Drying Solution (Agilent Technologies) according to the manufacturer’s instructions). The slides were scanned at 5 µm resolution on an Agilent DNA microarray scanner (Agilent Technologies). The image files were analysed with the Agilent Feature Extraction Software.
Prior to the statistical analysis spots from control spikes, landmarks and genes with low expression (absent) were filtered out. The data were analysed using the limma package (Smyth, 2005) and the R statistical data analysis programme package (R 2.10.1). No background subtraction was performed, and data were normalized using the Global Loess Normalization method. Benjamini and Hochberg’s method to control the false discovery rate (FDR) was used to identify differentially regulated genes (Benjamini and Hochberg, 1995). Genes with dye bias effects were removed and genes with an adjusted P-value of less than 0.05 were regarded as significantly differentially expressed. The study is MIAME compliant. Raw data has been deposited in GEO (accession GSE66201).
GO analysis
Gene ontology (GO) annotation analysis was performed using the Cytoscape 3.1.0 (Smoot et al., 2011) plug-in Bingo 3.0.2 (Maere et al., 2005). Over-represented categories were identified using a hypergeometric test with a significance threshold of 0.05 after Benjamini and Hochberg’s FDR correction (Benjamini and Yekutieli, 2001) using the whole annotated genome as the reference set.
Histochemical GUS assays
Histochemical GUS assays were performed as described by Butenko et al. (2003).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_105550.1 (IDA), NM_113464.2 (IDL1), NM_001085327.2 (IDL2), NM_001085091.1 (IDL3), NM_001084711.1 (IDL4), AY642386.1 (IDL5), NM_120612.1 (IDL6), AK118348.1 (IDL7), AK221754.1 (IDL8), NM_118988.2 (PIP1), NM_119892.1 (PIP2), NM_127891.1 (PIP3), NM_103867.2 (PIPL1), NM_111484.1 (PIPL2), NM_119893.1 (PIPL3), NM_001125892.1 (PIPL4), CB254609.1 (PIPL5), EF183199.1 (PIPL6), NC_003075.7 (PIPL7), NC_003076.8 (PIPL8), NM_103641 (CEP1), NM_148611.1 (CEP2), NM_127908.1 (CEP3), NM_201876 (CEP4), NM_126080.1 (CEP5), NM_114921.1 (CEP6), NC_003076.8 (CEP7 and CEP8), NM_114921.1 (CEP9), NC_003070.9 (CEP11), NC_003071.7 (CEP12), NM_101556.3 (CEP13), NM_102669.3 (CEP14) and NM_129615.3 (CEP15).
Results
Identification of IDL and PIP/PIPL genes in Arabidopsis
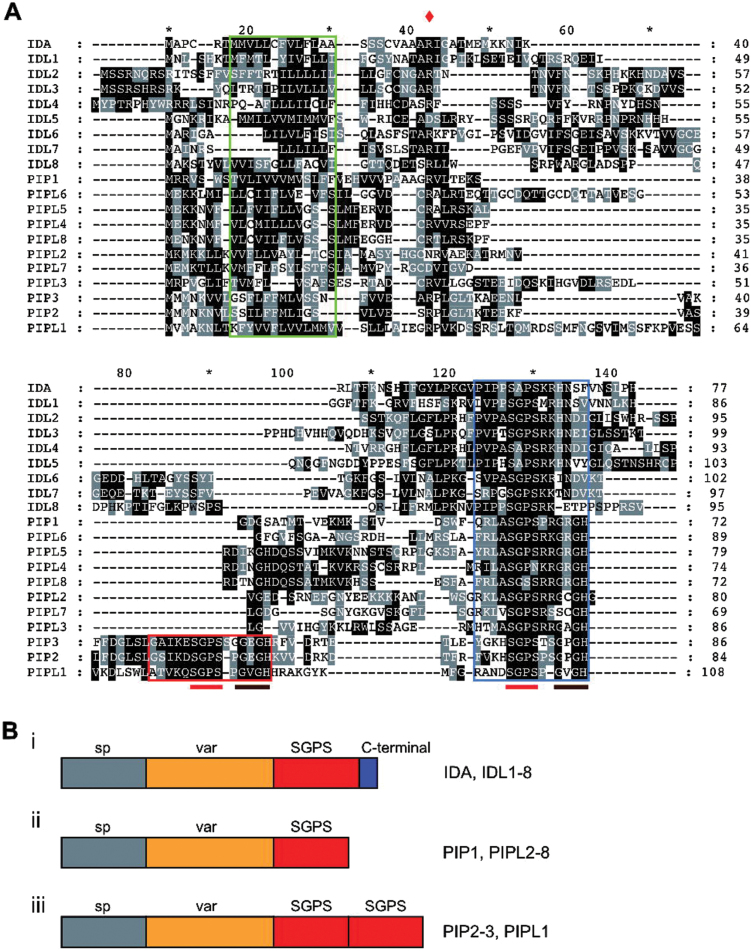
Six members of the IDA gene family have been identified in Arabidopsis to date: IDA and IDL1 to IDL5. All members of the IDA gene family are intronless and encode small proteins (<110 amino acids) characterized by an N-terminal secretory SP, a variable region and a C-terminal, conserved region (Butenko et al., 2003). Through database searches, three new IDL genes were found (Table 1; Fig. 1A; Supplementary Dataset S1). In addition, we identified 11 genes with similarity to the IDLs. During preparation of this paper an article was published presenting these genes as a family encoding secreted PAMP-INDUCED PEPTIDES (PIPs) and PIP-LIKE (PIPL) peptides (Hou et al., 2014) (Table 1; Fig. 1A; Supplementary Dataset S1). For all members the N-terminal SP contains a stretch of aliphatic residues typical of secreted proteins (Fig. 1A, green box). This motif is followed by a conspicuously conserved arginine residue (Fig. 1A, red diamond). The C-terminal is characterized by the conserved core motif S(G,A,V)PS (hereafter called the SGPS motif) conserved in both IDLs and PIP/PIPLs (Fig. 1A, blue box). The SGPS motif in IDL proteins is followed by four highly conserved residues [(R/K)(R/K)HN] followed by up to 13 additional less conserved residues (Fig. 1A, Bi). The PIP/PIPL proteins lack the variable region C-terminal to the SGPS motif that is found for the IDL proteins (Fig. 1A, Bii, iii). Three of the PIP/PIPLs (PIP2, PIP3 and PIPL1) contain two SPGS motifs in a tandem orientation at the C-terminal (Fig. 1A, Biii), as identified by Hou et al. (2014).
Table 1.
The IDA/IDL and PIP/PIPL gene families in Arabidopsis
| Gene name | Locus | Accession number | Signal peptide aa | Protein aa |
|---|---|---|---|---|
| IDA | At1g68765 | NM_105550.1 | 26 | 77 |
| IDL1 | At3g25655 | NM_113464.2 | 27 | 86 |
| IDL2 | At5g64667 | NM_001085327.2 | 36 | 95 |
| IDL3 | At5g09805 | NM_001085091.1 | 32 | 99 |
| IDL4 | At3g18715 | NM_001084711.1 | 36 | 93 |
| IDL5 | At1g76952 | AY642386.1 | 27 | 103 |
| IDL6 | At5g05300 | NM_120612.1 | 24 | 102 |
| IDL7 | At3g10930 | AK118348.1 | 21 | 97 |
| IDL8 | At5g02591a | AK221754.1 | 22 | 95 |
| PIP1 | At4g28460 | NM_118988.2 | 30 | 72 |
| PIP2 | At4g37290 | NM_119892.1 | 24 | 84 |
| PIP3 | At2g23270 | NM_127891.1 | 19 | 86 |
| PIPL1 | At1g49800 | NM_103867.2 | 27 | 108 |
| PIPL2 | At3g06090 | NM_111484.1 | 22 | 79 |
| PIPL3 | At4g37295 | NM_119893.1 | 22 | 86 |
| PIPL4 | At5g43066 | NM_001125892.1 | 21 | 74 |
| PIPL5 | At5g43068a | CB254609.1 | 21 | 79 |
| PIPL6 | At1g47178a | EF183199.1 | 22 | 88 |
| PIPL7 | At4g11402a | NC_003075.7 (w/6941763-6941972)b | 23 | 69 |
| PIPL8 | At5g43064a | NC_003076.8 (w/17282272-17282490)b | 21 | 72 |
a Preliminary AtID from TAIR
b Chromosomal coordinates of cds
Fig. 1.
The IDA/IDL and PIP/PIPL peptide families. (A) Protein alignment based on full-length sequences of the IDA/IDL/PIP/PIPL proteins. The green box indicates the SP, the blue box indicates the C-terminal putative peptide ligand motif and the red box indicates the second ligand motif identified in PIP2, PIP3 and PIPL1. The conserved arginine following the predicted SP is marked by a red diamond. The SGPS and GxGH motifs are indicated by red and brown bars below the alignment, respectively. (B) Schematic representation of the proteins in the IDA/IDL and PIP/PIPL peptide families. Grey boxes (marked with ‘sp’) represent the SP sequences identified by SignalP 4.1, orange boxes (marked with ‘var’) indicates a variable region with little homology, and red boxes represents the conserved, C-terminal EPIP domain known to be the active part of the IDA peptide indicated by the conserved core motif SGPS. The IDA and IDL proteins possess a C-terminal, variable sequence (i). This non-conserved sequence is not found among the PIP/PIPLs (ii). PIP2, PIP3 and PIPL1 contain two tandem SPGS motifs (iii).
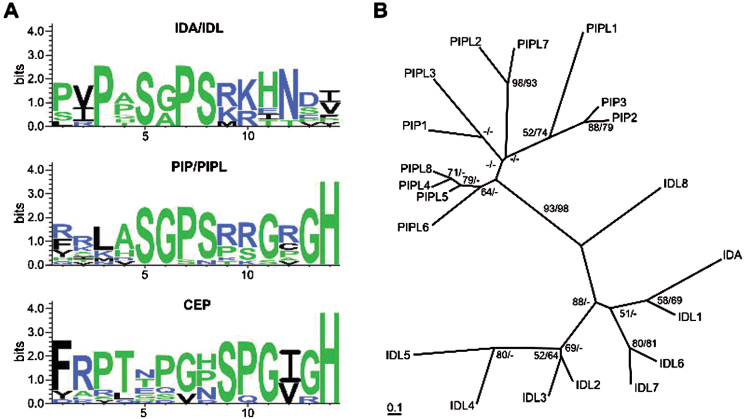
Two studies of the CEP family of small peptides, consisting of 15 members, have recently been published (Delay et al., 2013; Roberts et al., 2013). Two of the CEP family members, CEP13 and CEP14, show similarity to the PIP/PIPLs. A sequence consensus logo of the SGPS motif and surrounding residues was made for the IDL and PIP/PIPL families as well as for the CEP family (Fig. 2A). The SGPS motif (position 4–7) is conserved in IDLs and PIP/PIPLs. A second conserved motif (GxGH, where x is any amino acid) was seen at the C-terminal end of all PIP/PIPLs. The SGPS motif was not found in the CEPs; however, the family was characterized by the C-terminal SPG(I/V)GH sequence, which resembles the C-terminal end of the PIP/PIPLs. Thus, the putative ligand domain of the PIP/PIPLs shares features with both IDLs and CEPs.
Fig. 2.
Phylogenetic relationship between the IDA/IDL and PIP/PIPL peptides. (A) Sequence logo representation of the conserved C-terminal of IDA/IDL, PIP/PIPL and CEP peptides. (B) N-J and ML trees were constructed based on the protein alignment of the IDA/IDL and PIP/PIPL families shown in Fig. 1A. The N-J trees are shown. The overall topologies for the N-J and ML trees are the same. Bootstrap confidence values above 50% for N-J (first value) and ML (second value) are shown in the tree.
Phylogenetic analysis of the IDA/IDL and PIP/PIPL families
In order to examine the relationship of the IDA/IDL and PIP/PIPL gene families in Arabidopsis, a phylogenetic analysis on the full-length protein sequences of all IDL and PIP/PIPL members was performed. Two methods were used: a distance-matrix method combined with the N-J algorithm as implemented in the ClustalX programme, and a ML method using a gamma model and the RAxML programme (Fig. 2B). The resulting tree topologies from the two analyses were highly similar (data not shown), although bootstrap confidence values were a bit lower for the ML analysis. The analysis shows that the IDA/IDL and PIP/PIPL families split into separate branches with a high level of bootstrap confidence, even though the highly variable central part of the proteins is included in the analysis. Due to the high sequence divergence of these genes, not all branches in the tree are supported with high confidence levels. However, the IDA/IDL cluster can be divided into two subgroups: one containing IDL2, IDL3, IDL4 and IDL5 and the other containing IDA, IDL1, IDL6 and IDL7. Identical ML and N-J tree topologies support this division. Of the IDL proteins, IDL8 can be regarded as an outlier, and does not cluster well with any of the other IDLs. The PIP/PIPL proteins can be broadly divided into two groups: those with a single SGPS motif (PIP1 and PIPL2–PIPL8) and those with two SGPS motifs (PIP2, PIP3 and PIPL1).
A phylogenetic analysis (using the ML method) was also performed on a full-length protein alignment of the IDA/IDL and PIP/PIPL families as well as the CEP family (Supplementary Fig. S1). CEP9 was not included in the analysis due to its aberrant length and number of peptide motifs (five). The IDA/IDL family was clearly separated from the PIP/PIPL and CEP families with a high bootstrap confidence value (100%). Furthermore, CEP13, CEP14 and CEP15 [defined as group II CEPs by Delay et al. (2013) and Roberts et al. (2013) formed a clade with significant bootstrap values.
To further study the evolutionary relationship between the IDA/IDL and PIP/PIPL genes, the region surrounding the IDA/IDL-PIP/PIPL gene loci were analysed for ancient chromosomal or gene duplications. The results are summarized in Supplementary Table S2. The chromosomal localization of the genes are shown in Supplementary Fig. S2. PIPL5, PIPL4 and PIPL8 are organized in tandem repeats, as are PIP2 and PIPL3. IDA and IDL1 are likely the result of a recent duplication event, as seven of the 40 genes flanking IDA and IDL1 are closely related to each other. This is the case for several other genes in the IDA/IDL family as well; IDL2 and IDL3 share 13–15 common neighbouring genes, as previously noted by Stenvik et al. (2008), while IDL6 and IDL7 shares six. PIPL6 have five neighbouring genes with corresponding homologues surrounding the PIPL5-PIPL4-PIPL8 loci and PIP3 shares four to eight genes with PIPL3-PIP2, depending on the region used for BLAST search.
The regions flanking IDL5 do not share any closely related genes located within the other IDA-PIPL regions, while IDL8 and PIPL6 only share a few. This lack of synteny suggests that these genes have evolved through mechanisms other than tandem gene duplication, such as movement via RNA intermediates like retrotransposable elements.
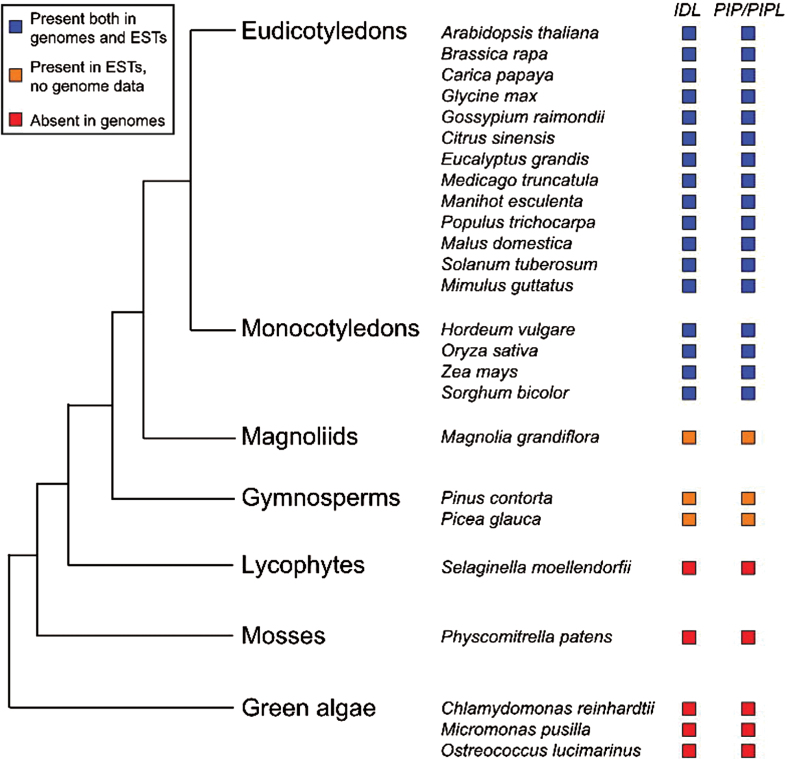
BLAST searches for IDA/IDL and PIP/PIPL family genes within the Viridiplantae were performed using full-length protein sequences as well as mature peptide sequences of Arabidopsis proteins. The searches showed that genes encoding both peptide families are present in seed plants, but absent in lycophytes (Selaginella), mosses (Physcomitrella) and green algae (Fig. 3). This distribution is similar to the one previously reported for CEPs (Delay et al., 2013; Roberts et al., 2013).
Fig. 3.
Distribution of IDA/IDL and PIP/PIPL genes within Viridiplantae. The conserved C-terminal domain of IDA/IDL and PIP/PIPL proteins were used in TBLASTN searches against the NCBI (genomes and ESTs), Phytozome v9.1 (genomes) and OneKP (ESTs) databases. The tree was adapted from Phytozome (Goodstein et al., 2012) and Delay et al. (2013).
Expression patterns of IDA/IDL and PIP/PIPL genes during development
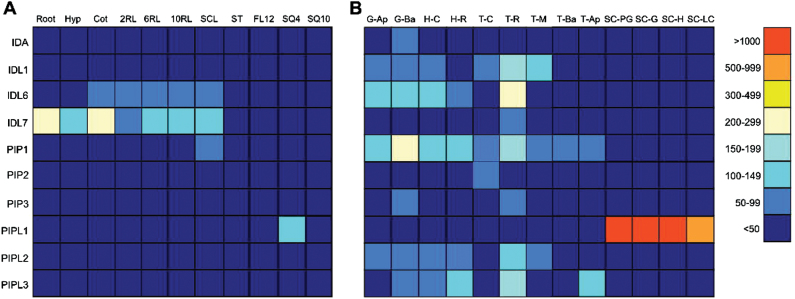
Since the expression pattern of a gene may provide an indication of its function, we conducted an in silico analysis of the transcription levels of the genes included on the Affymetrix ATH1 microarrays (IDA, IDL1, IDL6, IDL7, PIP1-PIP3 and PIPL1-PIPL3) during plant development (Schmid et al., 2005), obtaining data from the eFP Browser (Winter et al., 2007). These genes are expressed at low levels both at post-germination stages and during embryogenesis (Fig. 4). However, IDL1, IDL6, PIP1 and PIPL3 are expressed in several embryonic tissues [data obtained from Casson et al. (2005)], with the highest expression found in root primordia during the torpedo stage. PIP1 is also expressed in basal tissue during the globular stage. PIPL1 is the most strongly expressed gene during seed development. In contrast to the rest of the genes, PIPL1 is not expressed in the embryo, but shows high expression levels in the seed coat during early stages of seed development.
Fig. 4.
Developmental expression patterns of IDA/IDL and PIP/PIPL genes based on in silico data. (A) Expression in different vegetative tissues during development. (B) Expression during embryo and seed development (Casson et al., 2005). All data was obtained from the Arabidopsis eFP browser at the Bio-Array Resource database (Winter et al., 2007). The arithmetic expression values are given next to the colour scale. Hyp, hypocotyl; Cot, cotyledon; 2RL, second rosette leaf; 6RL, sixth rosette leaf; 10RL, tenth rosette leaf; SCL, senescing leaves; ST, stem; FL12, flower stage 12; SQ4, silique position 4; SQ10, silique position 10; G-Ap, globular stage apical; G-Ba, globular stage basal; H-C, heart stage cotyledon; H-R, heart stage root; T-C, torpedo stage cotyledon; T-R, torpedo stage root; T-M, torpedo stage meristem; T-Ap, torpedo stage apical; T-Ba, torpedo stage basal; SC-PG, seed coat preglobular stage; SC-G, seed coat globular stage; SC-H, seed coat heart stage; SC-LC, seed coat linear cotyledon.
Although the publicly available gene expression databases provide valuable information about the IDA/IDL and PIP/PIPL genes, it is very incomplete. Ten of the genes (IDL2–5, IDL8, PIPL4–8) are not included on the Affymetrix ATH1 microarrays used to generate these data. To fully assess the expression pattern of all the genes, qRT-PCR was performed on RNA isolated from Col-0 ecotype tissue harvested at various growth stages during the plant life cycle, as described by Boyes et al. (2001). Since none of the genes in the family contain introns, a control for genomic contamination was included in the analysis, using primers for CYP71A13 spanning the third intron in this gene.
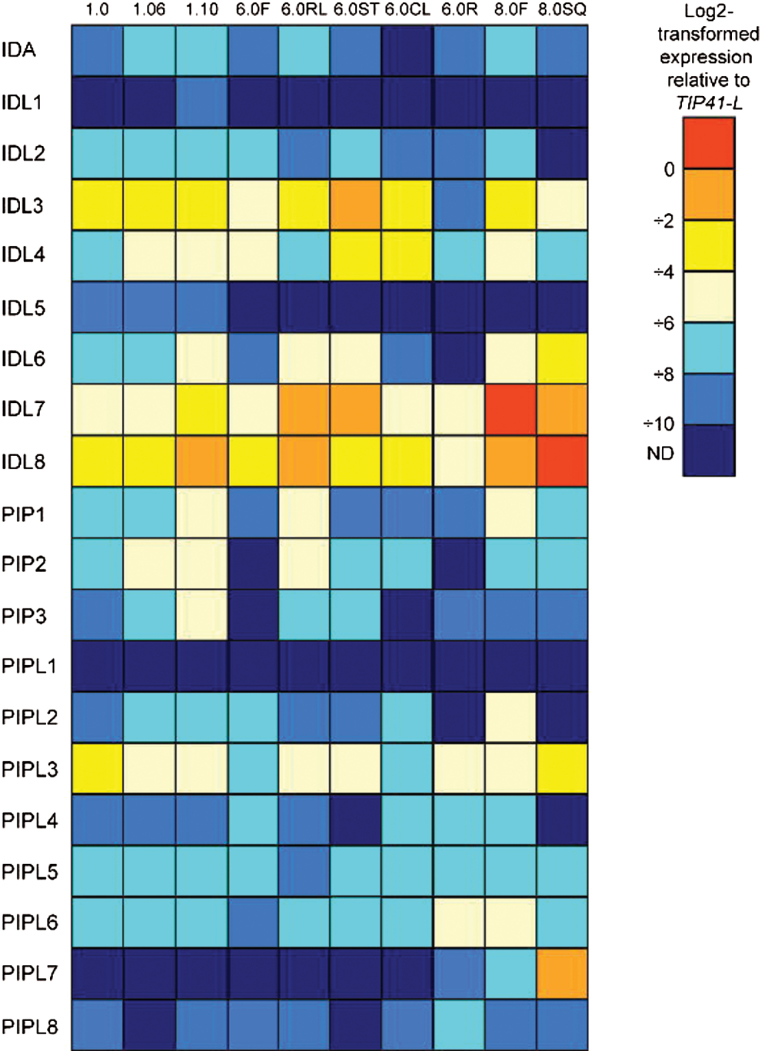
The expression levels of the IDA/IDL and PIP/PIPL genes under normal growth conditions were in general very low (Fig. 5); for some of the genes (IDL1, IDL5 and PIPL1) hardly any transcripts were detected. The highest transcript levels were found at the later stages of Arabidopsis development (i.e. IDL6, IDL7 and IDL8), indicating a possible role for these genes during seed development or senescence. Other genes, like IDL3 and PIPL3, were weakly expressed during all stages.
Fig. 5.
Developmental expression patterns of IDA/IDL and PIP/PIPL genes based on quantitative real-time PCR. The different developmental stages are annotated according to Boyes et al. (2001). F, flower; RL, rosette leaf; ST, stem; CL, cauline leaf; R, root; SQ, silique; ND, not detected. The expression levels (log2-transformed) relative to TIP41-LIKE are given next to the colour scale. n=3.
GUS expression analyses of IDL genes
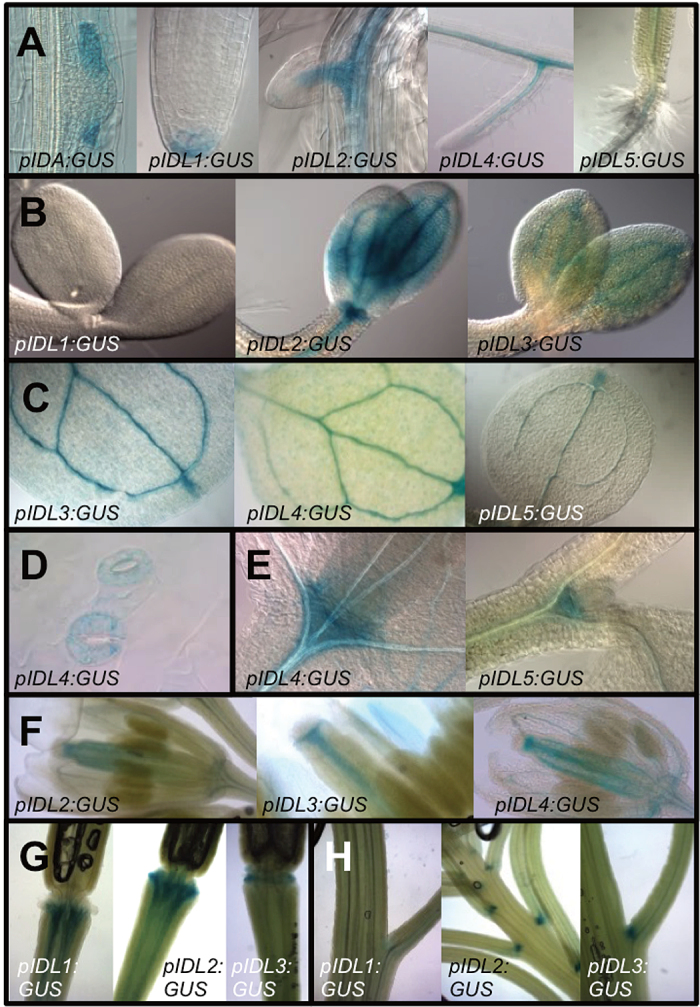
To further investigate the expression pattern of the different IDL genes during early Arabidopsis development, plants expressing promoter:GUS reporter constructs for IDA and IDL genes (pIDA/pIDL:GUS) were investigated from germination up until 14 d after germination (Supplementary Table S3). pIDA:GUS expression was observed in the cortex and epidermal cells overlaying the lateral root primordia in accordance with the recently reported function in cell separation allowing lateral root emergence (Fig. 6A; Kumpf et al., 2013). pIDL1:GUS had a specific pattern of expression in the columella root cap cells of the primary root (Fig. 6A), where cells undergo cell separation during root cap sloughing, which allows the primary root to penetrate the soil (del Campillo et al., 2004). After germination, pIDL2:GUS, pIDL4:GUS and pIDL5:GUS were expressed in the vascular tissue of the primary and lateral roots (Fig. 6A). Consistent with the expression in leaves identified by qRT-PCR (Fig. 5), the pIDL:GUS constructs were all expressed in the vascular tissue of the expanding cotyledons and/or primary leaves (Fig. 6B, C), and IDL4 expression was also observed in the guard cells (Fig. 6D) (Stenvik et al., 2008). The IDL promoter:GUS constructs were all expressed in the shoot apical meristem region, here represented by IDL4 and IDL5 in Fig. 6E. Expression was then investigated in various tissues at later developmental stages, as described by Boyes et al. (2001). Previous studies have shown that IDA, IDL2, IDL3 and IDL4 are expressed in the floral abscission zone (Stenvik et al., 2008). None of the genes were expressed in the abscission zone region at the time points investigated by qRT-PCR, although expression was detected in the vasculature of the developing floral organs (Fig. 6F). However, IDL1, IDL2 and IDL3 showed expression in the abscission zone at later stages, when the floral organs are already abscised (Fig. 6G). In addition, the same genes showed expression in the vestigial abscission zones of the pedicel region (Fig. 6H).
Fig. 6.
Histochemical analysis of promoter:GUS expression using the promoters of IDA and IDL1 to IDL5 genes. (A) pIDA:GUS is expressed during lateral root emergence in endodermis (63× magnification), cortex and epidermis cells; pIDL1:GUS in columella root cap cells (40×); pIDL3:GUS and pIDL4:GUS in the vasculature and pIDL5:GUS next to the hypocotyl (20×). (B) pIDL2:GUS and pIDL3:GUS are, in contrast to pIDL1:GUS, expressed in the cotyledons 3 d after germination (40×). (C) pIDL3:GUS, pIDL4:GUS and pIDL5:GUS are expressed in the vasculature of the first true leaves, including the hydathodes (20×). (D) pIDL4:GUS is expressed in the guard cells (40×); (E) pIDL4:GUS and IDL5:GUS are expressed in the shoot apical meristem (40×), (F) pIDL2:GUS, pIDL3:GUS and pIDL4:GUS are expressed in the vasculature of flowers at developmental stage 6.0F (20×). (G) pIDL1:GUS, pIDL2:GUS and pIDL3:GUS are expressed in the vasculature in the abscission zone region after the floral organs have been shed (20×). (H) pIDL2:GUS and pIDL3:GUS are in contrast to pIDL1:GUS expressed in vestigial abscission zones at the base of pedicels and branches (20×).
A subset of IDA/IDL and PIP/PIPL genes are induced by biotic and abiotic stress
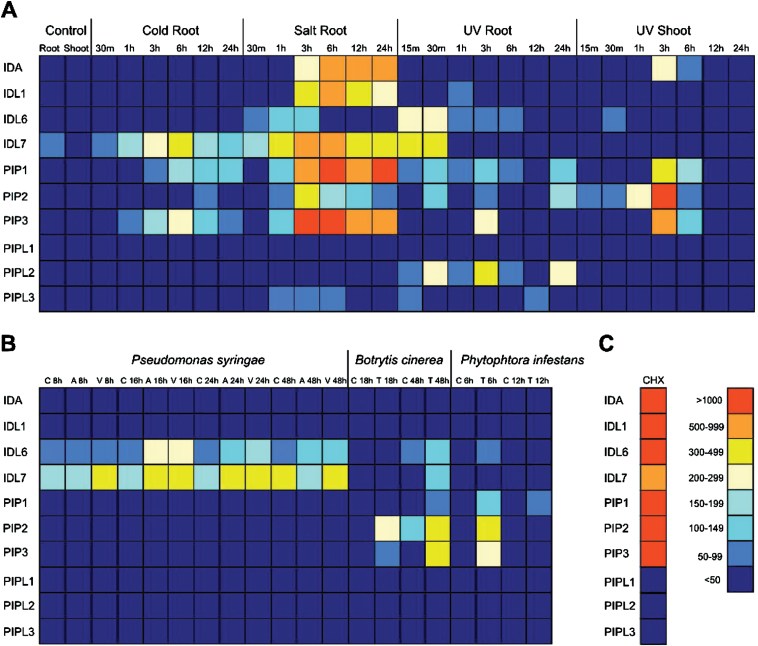
In general, the members of the IDA/IDL and PIP/PIPL families were found to be only weakly expressed under normal growth and development, so the response of IDA/IDL and PIP/PIPL genes to different abiotic and biotic stresses were therefore investigated (Fig. 7). Figure 7A summarizes the in silico analysis of abiotic stresses (Kilian et al., 2007). Cold stress induces expression of IDL7, PIP1 and PIP3 in roots, whereas UV induces eight out of 11 of the IDA/IDL/PIP/PIPL genes present on the Affymetrix ATH1 microarrays. The highest expression is observed in roots during salt stress. IDA, IDL1, IDL7, PIP1 and PIP3 are especially highly induced upon such stress, with expression levels up-regulated 500–1000 times compared to the control. PIPL1 is not expressed during any of the specified treatments. Biotic stress and treatments with elicitors (Fig. 7B; Supplementary Fig. S3) induces fewer genes than abiotic stress. Treatments with both virulent and avirulent strains of the biotrophic pathogen Pseudomonas syringae induce IDL6 and IDL7 expression. It should be noticed that expression of IDL6 is induced 1h after treatments with the pathogen-derived elicitors flg22, HrpZ and NPP1. PIP1 is up-regulated upon treatment with NPP1 and HrpZ (Supplementary Fig. S3). The necrotrophic pathogens Botrytis cinerea and Phytophtora infestans both induce expression of PIP2 and PIP3. This is in accordance with data recently published by Hou et al. (2014), showing that PIP1, PIP2 and PIP3 are involved in amplification of the immune response. IDL6 and IDL7 are in general induced at earlier time points than the rest of the genes (Fig. 7; Supplementary Fig. S3). Transcripts of IDL6 and IDL7 are detected as early as 15min post UV exposure, while PIPL2 and PIP3 are detected 30min (PIPL2) and 3h (PIP3) after exposure (Fig. 7A). Data obtained from Genevestigator (Hruz et al., 2008) confirmed these results (Supplementary Dataset S2).
Fig. 7.
Stress-induced expression of IDA/IDL and PIP/PIPL genes based on in silico data. (A) Abiotic stress. Cold root, root tissue collected from cold-treated seedlings (continuous 4°C); salt root, root tissue collected from salt-treated seedlings (150mM NaCl); UV root/shoot, root and shoot tissue collected from UV-treated seedlings (15min treatment in UV-B field). (B) Biotic stress. Pseudomonas syringae half-leaf infiltration: C, control (10mM MgCl2); A, avirulent P. syringae ES4326 avrRPt2; V, virulent P. syringae ES4326. Botrytis cinerea treatments: C, control (potato dextrose broth); T, treated (B. cinerea 5×105 spores/ml). Phytophthora infestans treatments: C, control (water); T, treated (Phytophthora infestans 106 spores/ml). (C) Cycloheximide (CHX) treatment (10 μM CHX, 3h). All data was obtained from the Arabidopsis eFP browser at the Bio-Array Resource database (Winter et al., 2007). The arithmetic expression values are given next to the colour scale.
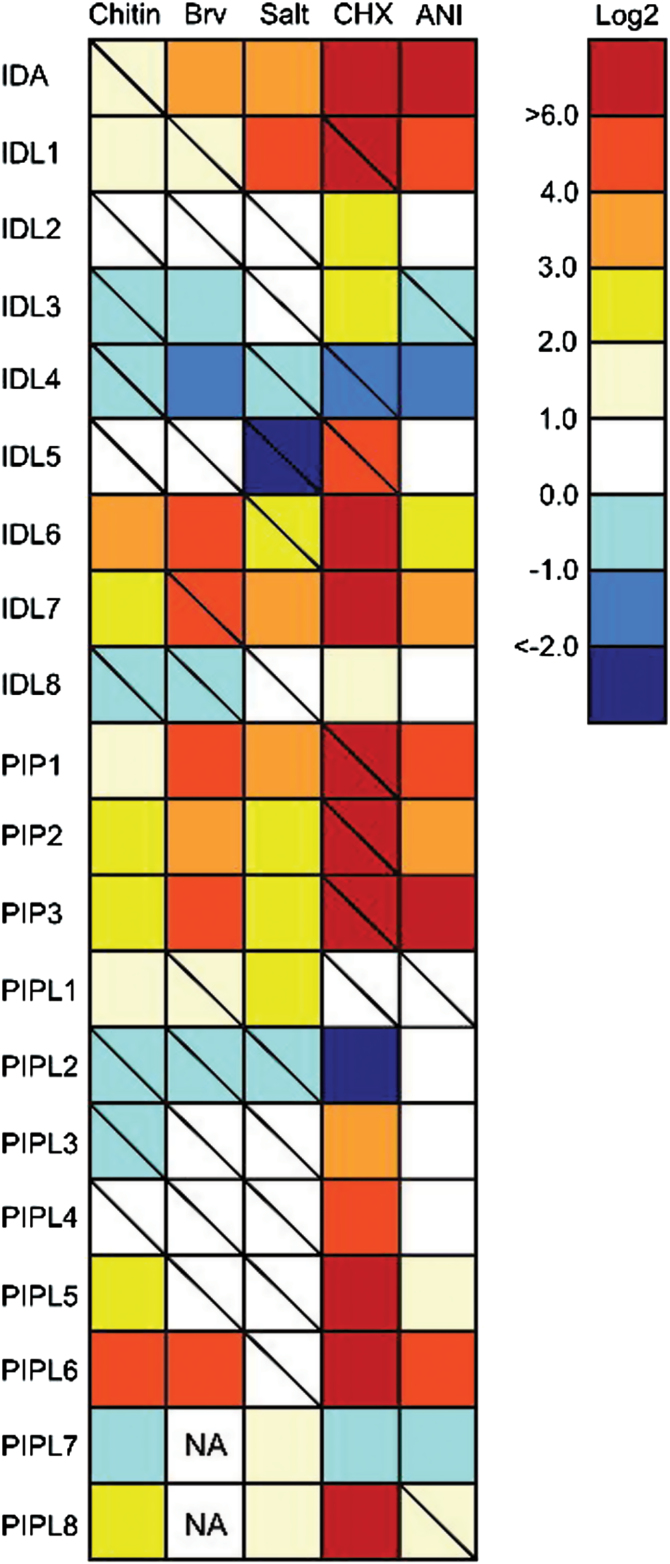
IDA/IDL/PIP/PIPL responses to hormone treatments were also studied using in silico data (Supplementary Fig. S4). Treatments with the ethylene precursor 1-aminocyclopropane-l-carboxylic acid (ACC), zeatin, abscisic acid (ABA), gibberellin A3 (GA-3) or brassinolide (BL) does not lead to any change in expression. IDL7 is weakly induced by methyl jasmonate, whereas PIPL3 and to a lesser extent PIP2 are induced by indole-3-acetic acid (IAA).
In order to complement the in silico data, Arabidopsis Col-0 seedlings were subjected to various biotic and abiotic treatments and a qRT-PCR analysis was performed to obtain information on all IDA/IDL and PIP/PIPL members (Fig. 8). Treatments with the elicitor chitin induced several of the genes, with the highest log2 ratio observed for IDL6 and PIPL6. The aphid Brevicoryne brassicae, which is a specialist on Brassicaceae species, was used as biotic treatment, and induced the expression of a large subset of the genes. Ten genes were up-regulated with log2>1; among these were IDA, PIP2, PIP3 and PIPL1. Salt stress induced eight of the genes log2>1, confirming the in silico data.
Fig. 8.
Stress-induced expression of IDA/IDL and PIP/PIPL genes based on qRT-PCR. Two-week-old plants were subjected to the following treatments for 6h unless otherwise specified: 10 μg/ml chitin (1h), 150mM NaCl, 10 μg/ml CHX and 15 μg/ml anisomycin. Infestation with Brevicoryne brassicae was done on 23-day-old plants, and rosette leaves were harvested after 72h (34). n=3. The relative expression (log2 ratios) values between treated and mock-treated samples are given next to the colour scale. Statistical differences between treated and mock-treated plants are indicated, where a diagonal line indicates no significance (REST analysis; P>0.05), and no diagonal line indicates significant difference (REST analysis; P<0.05). Brv, Brevicoryne brassicae; Salt, NaCl; CHX, cycloheximide; ANI, anisomycin.
CHX is a known inhibitor of protein synthesis (Schneider-Poetsch et al., 2010). Interestingly, all the IDL genes present on the ATH1 array, as well as PIP1, PIP2 and PIP3, are strongly induced upon CHX treatment (Fig. 7C). Treatment of seedlings with CHX led to an increase in the expression level of most of the IDA/IDL/PIP/PIPL genes 2-fold or more (Fig. 8). A subset of 12 genes showed an astounding response upon CHX treatment, with >1000-fold increase in expression levels. Generally, the genes found to be most inducible, either by abiotic or biotic stress, were strongly induced upon CHX treatment as well. These results were confirmed using another known protein synthesis inhibitor, anisomycin (Grollman, 1967). Similar to CHX, treatments with anisomycin highly induced the expression of ten of the IDA/IDL/PIP/PIPL genes (Fig. 8).
Transcriptomic responses to PIPL3 peptide treatment
In order to investigate possible roles of IDA/IDL and PIP/PIPL peptides, the transcriptomic response of Arabidopsis seedlings to treatment with PIPL3 peptide was analysed. PIPL3 was chosen, as no functional data was available for this peptide; furthermore, PIPL3 was expressed in leaf tissue during seedling stages.
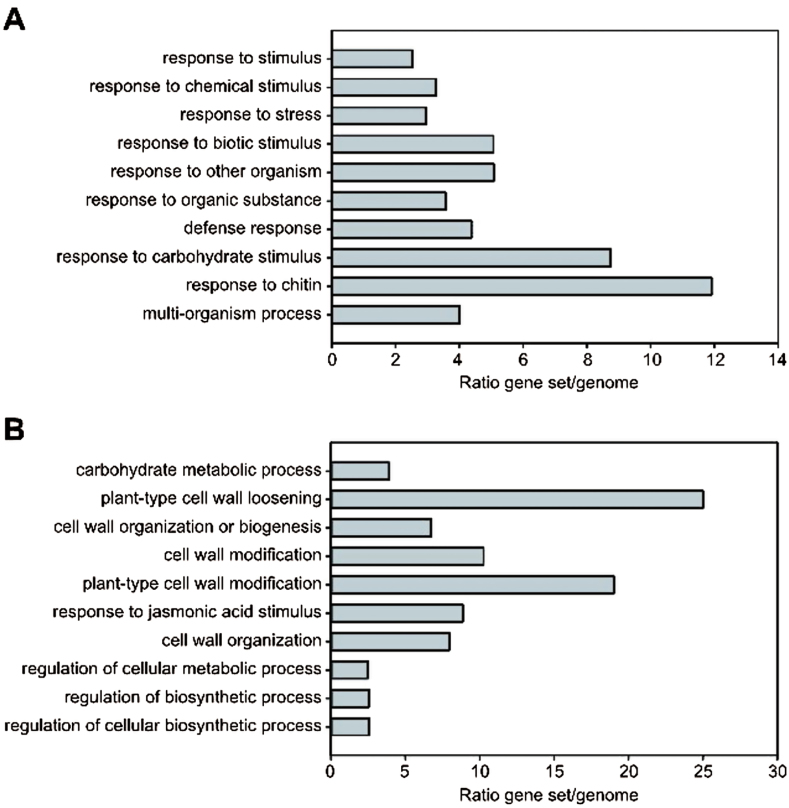
Treatment with 100nM PIPL3 peptide (containing a hydroxyproline in position 15) for 3h led to a widespread response compared with mock peptide-treated seedlings: 1599 genes were significantly (P<0.05) induced, whereas 1133 genes were significantly repressed. Genes showing low expression ratios (>log2 1.0, <log2 −0.7) were removed, and the filtered dataset (291 induced and 129 repressed genes, respectively, Supplementary Dataset S3) was subjected to a Gene Ontology (GO) enrichment analysis (Ashburner et al., 2000). The ten GO biological process categories most overrepresented in the up-regulated gene set indicated that the peptide treatment induced processes related to biotic stress and responses to chemical substances (possibly derived from other organisms) (Fig. 9A). The GO categories most enriched among the down-regulated genes were related to cell wall modification and loosening (Fig. 9B).
Fig. 9.
Transcriptional responses to PIPL3 peptide treatment. GO enrichment analysis of significantly regulated (P<0.05) genes in 2-week-old seedlings 3h after treatment with 100nM PIPL3 peptide. (A) Up-regulated genes; (B) Down-regulated genes. Control seedlings were treated with 100nM mock peptide. The dataset was filtered for expression ratio ratios (>log2 1.0, <log2 −0.7). The 10 most significantly enriched terms are listed from top to bottom. The bars show the frequency ratios of each GO term in the PIPL3-responsive gene set versus the genome.
A more detailed analysis of the significantly regulated genes revealed that a majority of the genes encoding enzymes of the camalexin biosynthetic pathway were strongly induced by PIPL3 peptide treatment (Supplementary Fig. S5; Supplementary Dataset S3). Camalexin, an indole phytoalexin, is synthesized from tryptophan via indole-3-acetaldoxime (IAOx) (Glawischnig et al., 2004; Ahuja et al., 2012). IAOx is a branching point between the biosynthetic pathways of camalexin and two other groups of compounds: indole glucosinolates and IAA. Whereas the first biosynthetic components of the indole glucosinolate pathway were not transcriptionally responsive, the last steps, from indol-3-ylmethyl glucosinolate (I3M) to 4-methoxy-indol-3-ylmethyl glucosinolate (4MO-I3M), were induced (Supplementary Fig. S5). Indole glucosinolate and camalexin biosynthetic genes have been reported to be positively regulated by HIG1, WRKY33 and ANAC042, respectively (Gigolashvili et al., 2007; Qiu et al., 2008; Saga et al., 2012). The expression of all three transcription factors was induced by PIPL3 treatment (Supplementary Dataset S3). In contrast, no genes encoding enzymes of the IAA biosynthetic pathway showed any response to PIPL3 treatment.
Discussion
For the last decade, peptide ligands have been found to act as important regulatory factors in plants as well as in animal systems. A well-studied peptide is IDA, found to regulate floral organ abscission in Arabidopsis (Butenko et al., 2003). In this study, we searched the Arabidopsis genome for genes encoding peptide ligands related to IDA, using both in vivo and in silico expression data to investigate these genes. In addition to three novel IDL genes, this search identified the recently described PIP/PIPL gene family. Of these genes, IDL8 has not previously been annotated.
Our phylogenetic analyses indicate that the CEPs (Delay et al., 2013; Roberts et al., 2013) constitute a related peptide family, as they share the C-terminal GxGH motif with the PIP/PIPLs (Fig. 2A; Supplementary Fig. S1). CEP13, CEP14 and CEP15 might be considered to be a subclade of the CEP family, as suggested by Delay et al. (2013) and Roberts et al. (2013), or as a separate family. IDA/IDLs, PIP/PIPLs and CEPs all first appear in seed plants; the founder of these peptide families is therefore difficult to predict. A phylogenetic tree including IDA/IDL, PIP/PIPL and CEP members presented by Hou et al. (2014) differs topologically somewhat from ours (Supplementary Fig. S1). The tree produced by Hou et al. (2014) also includes members of the CLE and PEP families. Furthermore, it is based on an alignment of the C-terminal peptide motif, in contrast to the tree in Supplementary Fig. S1, which was generated from a full-length protein alignment. Thus, a direct comparison of these trees is difficult.
Families of post-translationally modified peptides are characterized by multiple paralogous genes encoding small, cysteine-poor peptides with high sequence diversity outside of the C-terminal domain, which contains the mature peptide (Matsubayashi, 2014). All IDA/IDL and PIP/PIPL family genes fulfil these criteria; they encode putative prepropeptides with an N-terminal SP followed by a variable sequence and a C-terminal, conserved motif related to the EPIP motif of IDA (Butenko et al., 2003; Stenvik et al., 2008). The highly conserved SGPS core motif contains a proline that is an attractive candidate for post-translational modification. Several characterized plant peptides, such as systemin (Pearce et al., 1991), TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF) (Ito et al., 2006) and CEP1 (Ohyama et al., 2008) have been shown to contain hydroxyproline. Furthermore, hydroxyprolines of PLANT PEPTIDE CONTAINING SULFATED TYROSINE 1 (PSY1) (Amano et al., 2007), CLAVATA 3 (CLV3) and CLV3/EMBRYO SURROUNDING REGION-RELATED 2 (CLE2) (Ohyama et al., 2009) are modified by the addition of an O-linked L-arabinose chain. These post-translational modifications increase the activity and/or specificity of the peptides (Shinohara and Matsubayashi, 2010, 2013).
We have previously noted that IDA/IDL peptides and several CLE peptides share a similar core with (hydroxy)prolines and small amino acids (Ala, Gly, Ser) (Stenvik et al., 2008). These amino acids are also found in mature peptides of the PIP/PIPL and CEP families, and one may speculate that they are involved in receptor binding. Biochemical evidence has recently been provided for the binding of PIP1 to RLK7 (Hou et al., 2014) and CEP peptides to CEPR1 and CEPR2 (Tabata et al., 2014). Intriguingly, these three receptors are highly similar LRR-RLKs belonging to a subgroup of the LRR-RLK subclass XI that also includes HAE and HSL2 (Yamaguchi et al., 2006; Butenko et al., 2009). Sequence alignment (Supplementary Fig. S6) indicates that the LRR ectodomains of these three receptors are more similar to HAE and HSL2 than for instance to BAM1, which is documented to bind the CLE9 peptide (Shinohara et al., 2012). Other members of the HAE/HSL2 branch of LRR-RLKs should be promising candidate receptors for peptides belonging to the IDA/IDL, PIP/PIPL and CEP families.
Interestingly, PIP2, PIP3 and PIPL1 apparently possess two SGPS motifs (Fig. 1; Hou et al., 2014). The double peptide motif might be processed into two independent peptides that may bind different receptor complexes, thus activating different pathways. As a result, the response induced by the peptides may be wider and/or stronger. Alternatively, the double peptide acts as one functional unit that may interact with two different binding sites, or even to different partners in a receptor dimer. Multiple peptide motifs have also been identified in members of the CEP family (Delay et al., 2013; Roberts et al., 2013). A member of the CLE family, CLE18, encodes a precursor protein that contains two functional peptide motifs. The 13 aa CLE18 peptide located in the CLE18 variable region inhibits tracheary element differentiation and suppresses root growth (Ito et al., 2006), whereas the newly discovered C-terminal 12 aa CLE-LIKE peptide motif, promotes root growth (Meng et al., 2012), suggesting that one gene can encode two peptides with different roles. Crosstalk between different pathways is an important way to fine-tune the response to a given signal, and is normally mediated by common components in signalling pathways (Knight and Knight, 2001; Fujita et al., 2006). The double peptides motifs of PIP2, PIP3 and PIPL1 might provide plants with an extra dimension in crosstalk; one gene encodes two possible peptides that may modulate two different pathways.
Many of the IDA/IDL/PIP/PIPL genes have tissue-specific expression (Figs 4–6), suggesting that these genes may play roles during plant growth and development (Stenvik et al., 2008). We were hardly able to detect expression of IDA, IDL1, IDL2 and IDL5 in different organs during development (Fig. 5), but as shown in Fig. 6, expression of IDA and IDL1– IDL5 is restricted to very specific tissues or cell types, indicating a strict developmental regulation of transcription. This is further confirmed by the abiotic and biotic stress assays (Fig. 8), where IDL2– IDL5 appear to be non-responsive. IDA is strongly regulated by IAA in roots (Kumpf et al., 2013), but our analysis of in silico data suggests that this is not the case for green tissue (Supplementary Fig. S5; Supplementary Dataset S2). PIPL1 shows the highest expression levels of the 20 genes in our in silico analysis (Fig. 4), but as for the IDL genes, the expression is too specific to be detected in our qRT-PCR analysis (Fig. 5). Transcriptome studies of Arabidopsis seed development indicate that PIPL1 expression is restricted to seed coat tissue during early stages of seed development (Le et al., 2010; Belmonte et al., 2013).
Stress-induced genes (Figs 7, 8) include PIP1, PIP2, PIP3, PIPL5 and PIPL6, while IDA, IDL1, IDL6 and IDL7 are up-regulated both during development and stress. The stress-induced genes can be separated into two categories: those induced by abiotic stress (like IDA, IDL1, PIP1) and those induced by biotic stress (like IDL6). IDL7, PIP2 and PIP3 are induced by both abiotic and biotic stress. PIP1 and PIP2 have been implicated in immune responses and pathogen resistance (Hou et al., 2014). Our results suggest that IDA/IDL and PIP/PIPL peptides also may be involved in regulation of responses to abiotic stresses such as salt stress. The peptides could act in positive or negative feedback loops for temporal and/or spatial fine-tuning of stress signalling pathways.
A subset of the IDA/IDL and PIP/PIPL genes were strongly induced by treatment with the translational inhibitors CHX and anisomycin (Figs 7, 8). Such superinduction has previously been reported in plants, for cold-induced genes (Berberich and Kusano, 1997; Zarka et al., 2003) and immediate-early response genes (Horvath et al., 1998; Uquillas et al., 2004), amongst others. The mechanism behind CHX superinduction could be related to the presence of a labile transcriptional repressor or increased mRNA stability.
Treating seedlings with PIPL3 peptide led to the induction of genes involved in defence responses, including the camalexin and indole glucosinolate biosynthesis pathways (Supplementary Fig. S5). Neither in silico data (Fig. 7; Supplementary Fig. S3) nor our own experiments (Fig. 8) indicate that PIPL3 expression is activated by biotic or abiotic treatments; instead, expression appears to be auxin-induced (Supplementary Fig. S4; Supplementary Dataset S2). However, PIP1 and PIP2, the other family members for which functional data exist, have been strongly implicated as positive regulators of defence responses (Hou et al., 2014). Both camalexin and indole glucosinolates are important parts of the chemical defence system of crucifers (Sønderby et al., 2010; Ahuja et al., 2012). Whereas glucosinolates are stored after synthesis and are detected in most tissues (Brown et al., 2003), camalexin is produced upon biotic or abiotic stress (Glawischnig, 2007). Why are these pathways induced by a peptide that shows developmentally regulated expression? It is tempting to speculate that PIPL3 is involved in regulation of the trade-off between growth and defence during development. PIPL3 may activate a limited set of defence-related pathways to a basal expression level in tissue surrounding meristematic regions. The down-regulation of cell wall loosening genes by PIPL3 indicate that the target cells could be differentiated cells that have reached their final size and shape.
In this study, we have characterized the IDA/IDL and PIP/PIPL families of peptide ligands in Arabidopsis. The family is characterized by one or both of two C-terminal conserved peptide motifs, SGPS and GxGH. Three PIP/PIPL members contain two tandem peptide motifs. Members of the IDA/IDL and PIP/PIPL gene families are expressed during development or induced by stress, suggesting distinct biological roles. Transcriptome analysis of PIPL3 peptide-treated seedlings indicates that although PIPL3 expression appears to be developmentally regulated, it activates defence-related processes.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Figure S1. Phylogenetic relationship between IDA/IDL, PIP/PIPL and CEP peptides in Arabidopsis.
Supplementary Figure S2. Chromosomal localization of Arabidopsis IDA/IDL and PIP/PIPL genes.
Supplementary Figure S3. Elicitor-induced expression of IDA/IDL and PIP/PIPL genes based on in silico data.
Supplementary Figure S4. Transcriptional responses of IDA/IDL and PIP/PIPL genes to hormone treatments.
Supplementary Figure S5. Transcriptional activation of the camalexin and indole glucosinolate biosynthetic pathways by PIPL3 peptide treatment.
Supplementary Figure S6. IDA, PIP1 and CEP1 bind related proteins.
Supplementary Table S1. List of PCR primers used in this study.
Supplementary Table S2. Gene duplication events of the IDA/IDL and PIP/PIPL gene families.
Supplementary Table S3. Expression of IDA and the IDL1 to IDL5 genes fourteen days after germination.
Supplementary Dataset S1. Protein sequences of IDA/IDL and PIP/PIPL in Arabidopsis.
Supplementary Dataset S2. Transcriptional responses of IDA/IDL/PIP/PIPL genes to perturbations, based on in silico data from Genevestigator.
Supplementary Dataset S3. Transcriptome responses to PIPL3 peptide treatment.
Acknowledgements
We thank Torfinn Sparstad for excellent technical assistance. We also thank the reviewers for critical reading. This work was supported by the Research Council of Norway through the FUGE (grant 175238/S10 to AKV, BL, RBA and AMB) and FRIMEDBIO (grants 204756 to MAB, and 178049/V40 to RBA and AMB) programmes.
References
- Aalen RB, Wildhagen M, Stø IM, Butenko MA. 2013. IDA: a peptide ligand regulating cell separation processes in Arabidopsis . Journal of Experimental Botany 64, 5253–5261. [DOI] [PubMed] [Google Scholar]
- Ahuja I, Kissen R, Bones AM. 2012. Phytoalexins in defense against pathogens. Trends in Plant Science 17, 73–90. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y. 2007. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis . Proceedings of the National Academy of Sciences, USA 104, 18333–18338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. 2000. Gene Ontology: tool for the unification of biology. Nature Genetics 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, et al. 2013. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proceedings of the National Academy of Sciences, USA 110, E435–E444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300. [Google Scholar]
- Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics 29, 1165–1188. [Google Scholar]
- Berberich T, Kusano T. 1997. Cycloheximide induces a subset of low temperature-inducible genes in maize. Molecular and General Genetics 254, 275–283. [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J. 2001. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. The Plant Cell 13, 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J. 2003. Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana . Phytochemistry 62, 471–481. [DOI] [PubMed] [Google Scholar]
- Butenko MA, Patterson SE, Grini PE, Stenvik G-E, Amundsen SS, Mandal A, Aalen RB. 2003. INFLORESCENCE DEFICIENT IN ABSCISSION controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. The Plant Cell 15, 2296–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko MA, Vie AK, Brembu T, Aalen RB, Bones AM. 2009. Plant peptides in signalling: looking for new partners. Trends in Plant Science 14, 255–263. [DOI] [PubMed] [Google Scholar]
- Butenko MA, Wildhagen M, Albert M, Jehle A, Kalbacher H, Aalen RB, Felix G. 2014. Tools and strategies to match peptide-ligand receptor pairs. The Plant Cell 26, 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson S, Spencer M, Walker K, Lindsey K. 2005. Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis . The Plant Journal 42, 111–123. [DOI] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, Walker JC. 2008. Regulation of floral organ abscission in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 105, 15629–15634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Research 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis . Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo E, Abdel-Aziz A, Crawford D, Patterson SE. 2004. Root cap specific expression of an endo-beta-1,4-D-glucanase (cellulase): a new marker to study root development in Arabidopsis . Plant Molecular Biology 56, 309–323. [DOI] [PubMed] [Google Scholar]
- Delay C, Imin N, Djordjevic MA. 2013. CEP genes regulate root and shoot development in response to environmental cues and are specific to seed plants. Journal of Experimental Botany 64, 5383–5394. [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. 2006. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Current Opinion in Plant Biology 9, 436–442. [DOI] [PubMed] [Google Scholar]
- Gallagher KL, Benfey PN. 2005. Not just another hole in the wall: understanding intercellular protein trafficking. Genes & Development 19, 189–195. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Berger B, Mock HP, Müller C, Weisshaar B, Flügge UI. 2007. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana . The Plant Journal 50, 886–901. [DOI] [PubMed] [Google Scholar]
- Glawischnig E. 2007. Camalexin. Phytochemistry 68, 401–406. [DOI] [PubMed] [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, Halkier BA. 2004. Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis . Proceedings of the National Academy of Sciences, USA 101, 8245–8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman AP. 1967. Inhibitors of protein biosynthesis. Journal of Biological Chemistry 242, 3226–3233. [PubMed] [Google Scholar]
- Horvath DM, Huang DJ, Chua NH. 1998. Four classes of salicylate-induced tobacco genes. Molecular Plant-Microbe Interactions 11, 895–905. [DOI] [PubMed] [Google Scholar]
- Hou S, Wang X, Chen D, Yang X, Wang M, Turrà D, Pietro AD, Zhang W. 2014. The secreted peptide PIP1 amplifies immunity through RECEPTOR-LIKE KINASE 7. PLoS Pathogens 10, e1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. 2008. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N, Mohd-Radzman NA, Ogilvie HA, Djordjevic MA. 2013. The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula . Journal of Experimental Botany 64, 5395–5409. [DOI] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. 2006. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313, 842–845. [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. 2007. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal 50, 347–363. [DOI] [PubMed] [Google Scholar]
- Kim J-Y. 2005. Regulation of short-distance transport of RNA and protein. Current Opinion in Plant Biology 8, 45–52. [DOI] [PubMed] [Google Scholar]
- Knight H, Knight MR. 2001. Abiotic stress signalling pathways: specificity and cross-talk. Trends in Plant Science 6, 262–267. [DOI] [PubMed] [Google Scholar]
- Kumpf RP, Shi C-L, Larrieu A, Stø IM, Butenko MA, Péret B, Riiser ES, Bennett MJ, Aalen RB. 2013. Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proceedings of the National Academy of Sciences, USA 110, 5235–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuśnierczyk A, Tran D, Winge P, Jørstad T, Reese J, Troczynska J, Bones A. 2011. Testing the importance of jasmonate signalling in induction of plant defences upon cabbage aphid (Brevicoryne brassicae) attack. BMC Genomics 12, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, et al. 2010. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proceedings of the National Academy of Sciences, USA 107, 8063–8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease KA, Walker JC. 2006. The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiology 142, 831–838.16998087 [Google Scholar]
- Lease K, Walker J. 2010. Bioinformatic identification of plant peptides. In: Soloviev M, ed. Peptidomics: Methods and Protocols , vol. 615 Humana Press, 375–383. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 21, 3448–3449. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. 2014. Posttranslationally modified small-peptide signals in plants. Annual Review of Plant Biology 65, 385–413. [DOI] [PubMed] [Google Scholar]
- Meng L, Buchanan BB, Feldman LJ, Luan S. 2012. CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis . Proceedings of the National Academy of Sciences, USA 109, 1760–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E, Smith S, De Smet I. 2012. Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. The Plant Cell 24, 3198–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas K, Nicholas H, Deerfield D. 1997. GeneDoc: analysis and visualization of genetic variation. EMBnet.news 4, 1–4. [Google Scholar]
- Ohyama K, Ogawa M, Matsubayashi Y. 2008. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. The Plant Journal 55, 152–160. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. 2009. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nature Chemical Biology 5, 578–580. [DOI] [PubMed] [Google Scholar]
- Page RD. 2002. Visualizing phylogenetic trees using TreeView. Current Protocols in Bioinformatics Chapter 6, Unit 6.2. [DOI] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA. 1991. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253, 895–897. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 8, 785–786. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JL, Fiil BK, Petersen K, et al. 2008. Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO Journal 27, 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339, 62–66. [DOI] [PubMed] [Google Scholar]
- Roberts I, Smith S, De Rybel B, Van Den Broeke J, Smet W, De Cokere S, Mispelaere M, De Smet I, Beeckman T. 2013. The CEP family in land plants: evolutionary analyses, expression studies, and role in Arabidopsis shoot development. Journal of Experimental Botany 64, 5371–5381. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Research 37, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Saga H, Ogawa T, Kai K, Suzuki H, Ogata Y, Sakurai N, Shibata D, Ohta D. 2012. Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis . Molecular Plant Microbe Interactions 25, 684–696. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. 2005. A gene expression map of Arabidopsis thaliana development. Nature Genetics 37, 501–506. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Research 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. 2010. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nature Chemical Biology 6, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y. 2010. Arabinosylated glycopeptide hormones: new insights into CLAVATA3 structure. Current Opinion in Plant Biology 13, 515–519. [DOI] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y. 2013. Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant and Cell Physiology 54, 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Moriyama Y, Ohyama K, Matsubayashi Y. 2012. Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. The Plant Journal 70, 845–854. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. 2001. a Plant receptor-like kinase gene family: diversity, function, and signaling. Science Signaling 2001, re22. [DOI] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. 2001. b Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proceedings of the National Academy of Sciences, USA 98, 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2005. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, eds. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer, 397–420. [Google Scholar]
- Stahl Y, Simon R. 2013. Gated communities: apoplastic and symplastic signals converge at plasmodesmata to control cell fates. Journal of Experimental Botany 64, 5237–5241. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. [DOI] [PubMed] [Google Scholar]
- Stenvik GE, Butenko MA, Urbanowicz BR, Rose JK, Aalen RB. 2006. Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis . The Plant Cell 18, 1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA. 2008. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. The Plant Cell 20, 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby IE, Geu-Flores F, Halkier BA. 2010. Biosynthesis of glucosinolates - gene discovery and beyond. Trends in Plant Science 15, 283–290. [DOI] [PubMed] [Google Scholar]
- Tabata R, Sawa S. 2014. Maturation processes and structures of small secreted peptides in plants. Frontiers in Plant Science 5, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y. 2014. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346. [DOI] [PubMed] [Google Scholar]
- Uquillas C, Letelier I, Blanco F, Jordana X, Holuigue L. 2004. NPR1-independent activation of immediate early salicylic acid-responsive genes in Arabidopsis . Molecular Plant-Microbe Interactions 17, 34–42. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘electronic fluorescent pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Pearce G, Ryan CA. 2006. The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proceedings of the National Academy of Sciences, USA 103, 10104–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarka DG, Vogel JT, Cook D, Thomashow MF. 2003. Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiology 133, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.