Abstract
Escherichia albertii occasionally causes food-borne outbreaks of gastroenteritis in humans; however, little is known about the vehicle of transmission. To screen retail chicken products for the presence of E. albertii, 104 retail chicken products were investigated. Portions of enrichment cultures that were PCR-positive for E. albertii (n=3) were sub-cultured on agar medium. Only 2 strains obtained from 2 chicken giblet samples were identified as E. albertii by multi locus sequence typing. Antimicrobial susceptibility testing showed that 1 strain was resistant to streptomycin and sulfisoxazole. Both strains harbored the virulence genes cdt and eae. This study is the first description of E. albertii isolation from retail food, suggesting that chicken products are a potential vehicle of E. albertii transmission.
Keywords: chicken giblet, chicken meat, chicken products, Escherichia albertii, infection source
Escherichia albertii can infect humans and birds, and was first described in 2003 [5]. E. albertii occasionally causes outbreaks of gastroenteritis in humans [1, 9, 15], yet the vehicle of this presumably foodborne pathogen remains unknown. Oaks et al. [12] reported that chickens harbor E. albertii; therefore, we suspected that chicken meat may be a vehicle for transmission of this enteropathogen to humans. It is important to identify the sources of contamination to help develop effective food hygiene protocols. The aim of the present study was to screen retail chicken meat and giblet samples for the presence of E. albertii.
A total of 104 retail chicken meat (n=54: 15 thigh, 8 skin, 6 inner fillet, 5 mix of thigh and breast, 5 neck meat, 4 mince, 3 breast and 8 uncertain meat) and giblet (n=50: 42 liver, 4 crop, 2 heart and 2 reproductive tract) samples were collected from 10 supermarkets in Fukuoka Prefecture, Japan, between June and October 2014. Samples were incubated in 9 volumes of buffered peptone water (BPW) (Oxoid, Basingstoke, U.K.) at 35°C for 18 hr. Aliquots of each BPW culture were then centrifuged at 13,800 × g for 5 min, and the supernatant was discarded. The pellet was resuspended in normal saline (1 ml) and centrifuged at 13,800 × g for 5 min. The supernatant was discarded, and the pellet was resuspended in 1 ml of 0.1× TE buffer with 5% Chelex 100 (Bio-Rad Laboratories, Hercules, CA, U.S.A.) before being boiled for 10 min, and then centrifuged at 13,800 × g for 10 min. The supernatant was used as a template for a PCR assay to detect E. albertii using primer pair lysP107F/lysP358R, as described previously [6]. To isolate the pathogen, the remainder of the BPW cultures of the PCR-positive samples were streaked onto deoxycholate-hydrogen sulfide-lactose agar (Eiken Chemical Co., Tokyo, Japan) and incubated at 37°C overnight. Presumptive E. albertii colonies (50–100/sample) were isolated on nutrient agar plates (Eiken Chemical Co.) and incubated at 37°C overnight. The isolates were then re-tested using the PCR method described above. The confirmed PCR-positive isolates were then examined for fermentation of glucose, motility and hydrogen sulfide production on triple-sugar iron agar (Eiken Chemical Co.) and sulfide indole motility medium agar (Eiken Chemical Co.). Isolates with positive glucose fermentation, negative motility and negative hydrogen sulfide production test results were then confirmed as E. albertii using multi locus sequence typing (MLST) as previously described [9]. Sequences of isolates in the present study were submitted to MLST databases run by the University of Warwick (http://mlst.warwick.ac.uk/mlst/, accessed 5th October, 2014), and isolates were assigned sequence types (ST) defined by the database (http://mlst.warwick.ac.uk/mlst/dbs/Senterica/GetTableInfo_html). Allele sequences for each isolate were then concatenated in the order adk − fumC − gyrB − icd − mdh − purA − recA, for a final composite length of 3,423 bp. The concatenated sequences were aligned using ClustalW [21], and a phylogenetic tree was constructed using the neighbor-joining method in MEGA5 software [20] to compare the results with other published STs.
Antimicrobial susceptibility testing was performed using the Kirby-Bauer disc-diffusion method, in accordance with the guidelines of the Clinical and Laboratory Standards Institute [2, 3]. Ampicillin, chloramphenicol, ciprofloxacin, kanamycin, nalidixic acid, streptomycin, sulfisoxazole, gentamicin, tetracycline and cefixime discs used for the test were purchased from Becton Dickinson (Franklin Lakes, NJ, U.S.A.). The presence of the virulence-related genes stx2f (Shiga toxin 2f gene) [10], cdt (cytolethal distending toxin gene) [18] and eae (intimin gene) [16] was also examined by PCR as described previously.
PCR results showed that 2 enrichment cultures of chicken giblets (liver) and 1 chicken meat sample were positive for E. albertii. However, only 2 E. albertii strains, from the 2 chicken giblet samples, were subsequently isolated from the 104 chicken product samples tested. These samples were from different manufacturers and were obtained from different supermarkets. Both strains were identified as ST4633, and the phylogenetic tree confirmed that this ST is an E. albertii lineage (Fig. 1). The 2 E. albertii strains showed different resistance patterns, although both strains harbored the virulence-related genes eae and cdt, but not stx2f (Table 1).
Fig. 1.
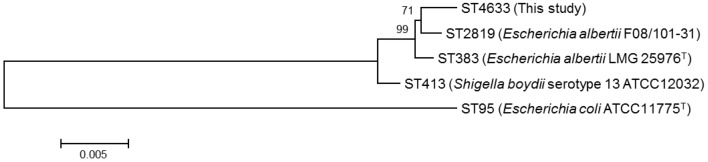
Phylogenetic tree showing nucleotide sequence clusters of the tested Escherichia albertii strains obtained using multi-locus sequence typing. ST383, ST413, ST95 and ST2819 were included as outgroups. Reference sequences were tested in a previous study [9]. Shigella boydii serotype 13 ATCC 12032 has been reclassified into the Escherichia albertii lineage. The scale bar indicates the number of nucleotide substitutions per site.
Table 1. Features of Escherichia albertii isolates in this study and in previous studies.
|
E. albertii isolates |
Origins | Number of strains tested |
Virulence-related genesa) | Antimicrobial resistance (ampicillin, chloramphenicol, ciprofloxacin, kanamycin, nalidixic acid, streptomycin, sulfisoxazole, gentamicin, tetracycline and cefixime) |
References | |||
|---|---|---|---|---|---|---|---|---|
| eae | cdt | stx2f | ||||||
| This study | ||||||||
| ID 3428 | Chicken giblet | +b) | + | –c) | Susceptible to all antimicrobials tested | This study | ||
| ID 3483 | Chicken giblet | + | + | – | Streptomycin and sulfisoxazole | This study | ||
| Other studies | ||||||||
| Birds | 29 | 100% | 100% | NT | NTd) | [12] | ||
| Birds | 9 | 100%e) | 100% | NT | NT | [13] | ||
| Birds | 11 | 100%e) | 100% | 9.1% | NT | [14] | ||
| Feline | 1 | 100%e) | 0% | 0% | NT | [14] | ||
| Swine | 1 | 100% | 100%f) | NT | NT | [4] | ||
| Humans | 4 | NT | NT | NT | Ampicillin (0%), chloramphenicol (0%), kanamycin (0%) and tetracycline (100%)g) | [5] | ||
| Humans | 48 | 100% | 85% | 4.2% | NT | [11] | ||
| Humans | 14 | 100%e) | 100% | 7.1% | NT | [14] | ||
| Humans | 21 | NT | NT | NT | Ampicillin (14%), chloramphenicol (62%), ciprofloxacin (0%), kanamycin (0%), streptomycin (76%), gentamicin (0%), tetracycline (100%) and cefixime (0%)h) | [19] | ||
| Humans | 1 | 100%e) | 100% | 100% | NT | [9] | ||
| Environmental water | 18 | 0% | 83% | 0% | NT | [8] | ||
| Uncertain | 3 | NT | NT | NT | Ampicillin (0%), chloramphenicol (0%), gentamicin (0%) and tetracycline (100%)i) | [17] | ||
a) eae: Intimin gene, cdt: Cytolethal distending toxin gene, stx2f: Shiga toxin 2f gene. b) +: Detected. c) –: Not detected. d) NT: Not tested. e) Only eae-positive isolates were used in this study. f) Only cdt-positive isolates were used in this study. g) Did not test with ciprofloxacin, nalidixic acid, streptomycin, sulfisoxazole, gentamicin or cefixime. h) Did not test with nalidixic acid or sulfisoxazole. i) Did not test with ciprofloxacin, kanamycin, nalidixic acid, streptomycin, sulfisoxazole or cefixime.
These results show that retail chicken can be contaminated by E. albertii. The inability to isolate E. albertii from 1 of the 3 PCR-positive samples might be caused by a low density of the bacterium in the sample and/or non-specific annealing of the primer pair lysP107F/lysP358R, as described previously [7]. The virulence-related gene patterns of the 2 strains, showing positive for eae and cdt, and negative for stx2f, were similar to those of previous reports that showed that most reported E. albertii strains carry eae (114/132) [4, 8, 9, 11,12,13,14] and cdt (121/132) [4, 8, 9, 11,12,13,14], and several contain stx2f (5/93) [8, 9, 11, 14], including bird isolates (Table 1). Interestingly, both E. albertii strains from the current study were susceptible to tetracycline, although only one of the 2 strains was resistant to streptomycin and sulfisoxazole. This is in contrast to 3 previous studies [5, 17, 19], which reported that E. albertii was resistant to tetracycline (Table 1). The results of the current study show that E. albertii contamination of chicken giblets might pose a potential health risk to humans because of the presence of virulence-related and antimicrobial resistance factors.
To the best of our knowledge, the present study is the first description of E. albertii isolation from retail food, although E. albertii has previously been isolated from birds, including chickens [12], feline [14] and swine [4]. Because only 104 samples collected from a small area were tested in the present study, further analyses are needed to determine the prevalence of E. albertii in chicken products. However, this study supports the hypothesis that chicken products might be a potential vehicle of E. albertii transmission.
Sequences of strains in the present study were submitted to MLST databases run by the University of Warwick (http://mlst.warwick.ac.uk/mlst/) as ST4633.
Acknowledgments
This work was supported by the Japanese Society for the Promotion of Science KAKENHI, Grant No. 24590846, and was also supported in part by a grant from the Ministry of Health Labour and Welfare of Japan (H24-Shinko-Ippan-005) and the Daido Life Welfare Foundation, Osaka, Japan. We are grateful to Dr. Hirata, Dr. Chijiwa, Mr. Kimoto, Ms. Uemura and Dr. Saeki, of the Fukuoka Institute of Health and Environmental Sciences, for their invaluable advice. The MLST data are publicly available at http://www2.warwick.ac.uk/mlst, which is currently supported by a grant from the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Asoshima N., Matsuda M., Shigemura K., Honda M., Yoshida H., Hiwaki H., Ogata K., Oda T.2014. Identification of Escherichia albertii as a causative agent of a food-borne outbreak occurred in 2003. Jpn. J. Infect. Dis. 67: 139–140. doi: 10.7883/yoken.67.139 [DOI] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial disk and dilution susceptibility tests; approved standard, tenth edition. M02-A11, CLSI, Wayne, PA. [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement. M100-S24, CLSI, Wayne, PA. [Google Scholar]
- 4.Hinenoya A., Shima K., Asakura M., Nishimura K., Tsukamoto T., Ooka T., Hayashi T., Ramamurthy T., Faruque S. M., Yamasaki S.2014. Molecular characterization of cytolethal distending toxin gene-positive Escherichia coli from healthy cattle and swine in Nara, Japan. BMC Microbiol. 14: 97. doi: 10.1186/1471-2180-14-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huys G., Cnockaert M., Janda J. M., Swings J.2003. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int. J. Syst. Evol. Microbiol. 53: 807–810. doi: 10.1099/ijs.0.02475-0 [DOI] [PubMed] [Google Scholar]
- 6.Hyma K. E., Lacher D. W., Nelson A. M., Bumbaugh A. C., Janda J. M., Strockbine N. A., Young V. B., Whittam T. S.2005. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J. Bacteriol. 187: 619–628. doi: 10.1128/JB.187.2.619-628.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maeda E., Murakami K., Okamoto F., Etoh Y., Sera N., Ito K., Fujimoto S.2014. Nonspecificity of primers for Escherichia albertii detection. Jpn. J. Infect. Dis. 67: 503–505. doi: 10.7883/yoken.67.503 [DOI] [PubMed] [Google Scholar]
- 8.Maheux A. F., Boudreau D. K., Bergeron M. G., Rodriguez M. J.2014. Characterization of Escherichia fergusonii and Escherichia albertii isolated from water. J. Appl. Microbiol. 117: 597–609. doi: 10.1111/jam.12551 [DOI] [PubMed] [Google Scholar]
- 9.Murakami K., Etoh Y., Tanaka E., Ichihara S., Horikawa K., Kawano K., Ooka T., Kawamura Y., Ito K.2014. Shiga toxin 2f-producing Escherichia albertii from a symptomatic human. Jpn. J. Infect. Dis. 67: 204–208. [DOI] [PubMed] [Google Scholar]
- 10.Nakao H., Kimura K., Murakami H., Maruyama T., Takeda T.2002. Subtyping of Shiga toxin 2 variants in human-derived Shiga toxin-producing Escherichia coli strains isolated in Japan. FEMS Immunol. Med. Microbiol. 34: 289–297. doi: 10.1111/j.1574-695X.2002.tb00636.x [DOI] [PubMed] [Google Scholar]
- 11.Nimri L. F.2013. Escherichia albertii, a newly emerging enteric pathogen with poorly defined properties. Diagn. Microbiol. Infect. Dis. 77: 91–95. doi: 10.1016/j.diagmicrobio.2013.06.028 [DOI] [PubMed] [Google Scholar]
- 12.Oaks J. L., Besser T. E., Walk S. T., Gordon D. M., Beckmen K. B., Burek K. A., Haldorson G. J., Bradway D. S., Ouellette L., Rurangirwa F. R., Davis M. A., Dobbin G., Whittam T. S.2010. Escherichia albertii in wild and domestic birds. Emerg. Infect. Dis. 16: 638–646. doi: 10.3201/eid1604.090695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh J. Y., Kang M. S., Hwang H. T., An B. K., Kwon J. H., Kwon Y. K.2011. Epidemiological investigation of eaeA-positive Escherichia coli and Escherichia albertii strains isolated from healthy wild birds. J. Microbiol. 49: 747–752. doi: 10.1007/s12275-011-1133-y [DOI] [PubMed] [Google Scholar]
- 14.Ooka T., Seto K., Kawano K., Kobayashi H., Etoh Y., Ichihara S., Kaneko A., Isobe J., Yamaguchi K., Horikawa K., Gomes T. A., Linden A., Bardiau M., Mainil J. G., Beutin L., Ogura Y., Hayashi T.2012. Clinical significance of Escherichia albertii. Emerg. Infect. Dis. 18: 488–492. doi: 10.3201/eid1803.111401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ooka T., Tokuoka E., Furukawa M., Nagamura T., Ogura Y., Arisawa K., Harada S., Hayashi T.2013. Human gastroenteritis outbreak associated with Escherichia albertii, Japan. Emerg. Infect. Dis. 19: 144–146. doi: 10.3201/eid1901.120646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paton A. W., Paton J. C.1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36: 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez K. L., Alam M. J., Castillo A., Taylor T. M.2013. Antibiotic resistance and growth of the emergent pathogen Escherichia albertii on raw ground beef stored under refrigeration, abuse, and physiological temperature. J. Food Prot. 76: 124–128. doi: 10.4315/0362-028X.JFP-12-277 [DOI] [PubMed] [Google Scholar]
- 18.Pickett C. L., Pesci E. C., Cottle D. L., Russell G., Erdem A. N., Zeytin H.1996. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB gene. Infect. Immun. 64: 2070–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stock I., Rahman M., Sherwood K. J., Wiedemann B.2005. Natural antimicrobial susceptibility patterns and biochemical identification of Escherichia albertii and Hafnia alvei strains. Diagn. Microbiol. Infect. Dis. 51: 151–163. doi: 10.1016/j.diagmicrobio.2004.10.008 [DOI] [PubMed] [Google Scholar]
- 20.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson J. D., Higgins D. G., Gibson T. J.1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. doi: 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]


