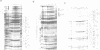Abstract
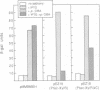
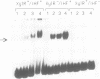
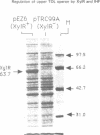
Transcription from promoter Pu of the upper catabolic operon of the Pseudomonas putida TOL plasmid which specifies conversion of toluene/xylenes to benzoate/toluates is activated by the TOL-encoded regulator XylR protein in the presence of substrates of the catabolic pathway and in conjunction with the sigma 54(NtrA)-containing form of RNA polymerase. This regulatory circuit was faithfully reproduced in Escherichia coli in single copy gene dosage by integrating the corresponding controlling determinants into the chromosomes of several K12 derivatives by means of specialized transposons. In vivo monitoring of the activity of a Pu-lacZ fusion in E. coli strains with different genetic backgrounds demonstrated that integration host factor (IHF) is involved in Pu regulation and that hyperproduction of the XylR protein leads to a decrease of Pu activity in a manner in which deletion of the putative DNA-binding domain of the XylR does not impair its inhibitory effect when hyperproduced. One discrete IHF binding site and two potential XylR sites (consensus sequence 5'-TTGANCAAATC-3'), bracketted by short stretches of DNase I-hypersensitive bonds, were detected upstream of the transcription initiation site. A model accounting for the features found is proposed which includes the IHF-promoted looping of upstream XylR-DNA complexes so that they contact the sigma 54(NtrA)-RNA polymerase bound at -12/-24 positions.
Full text
PDF


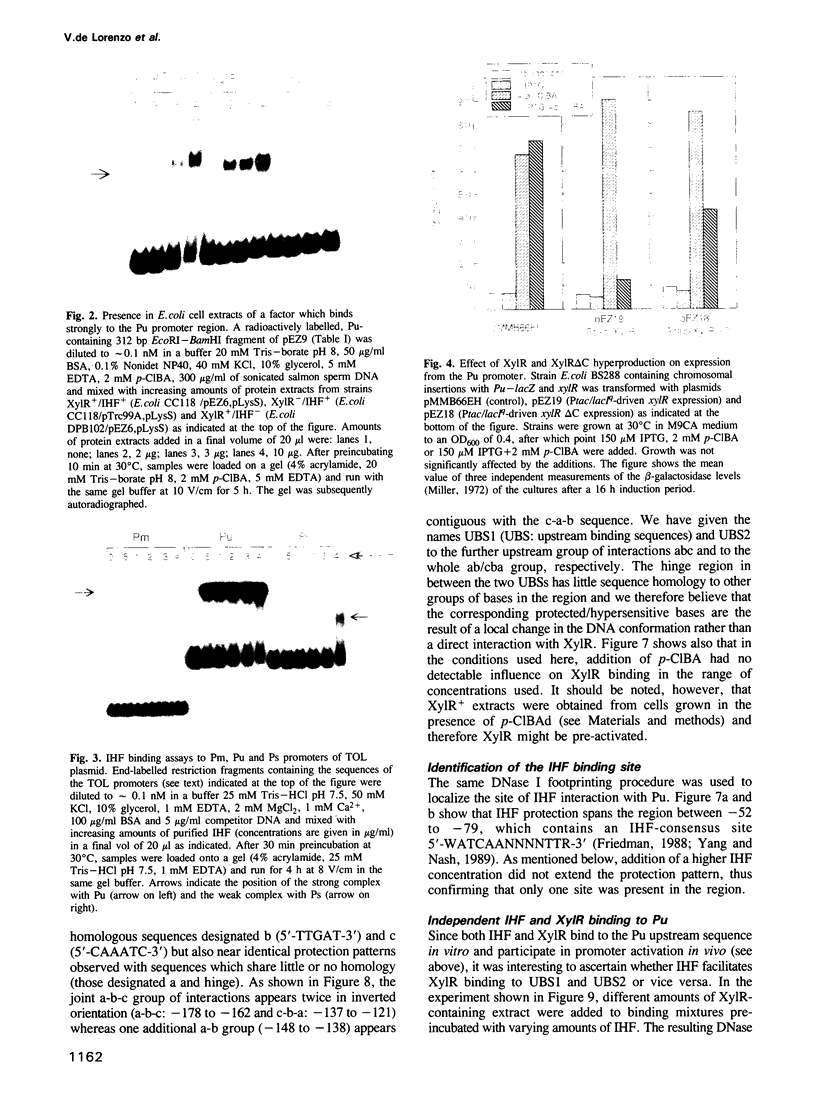
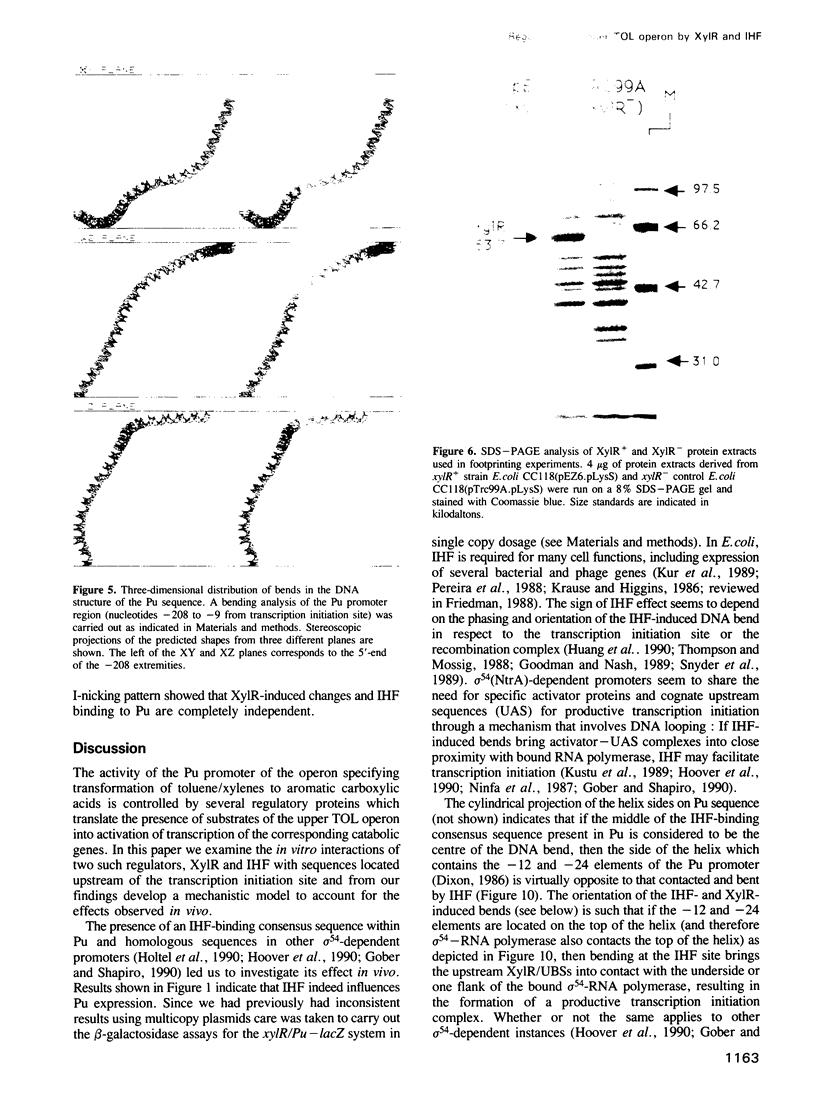
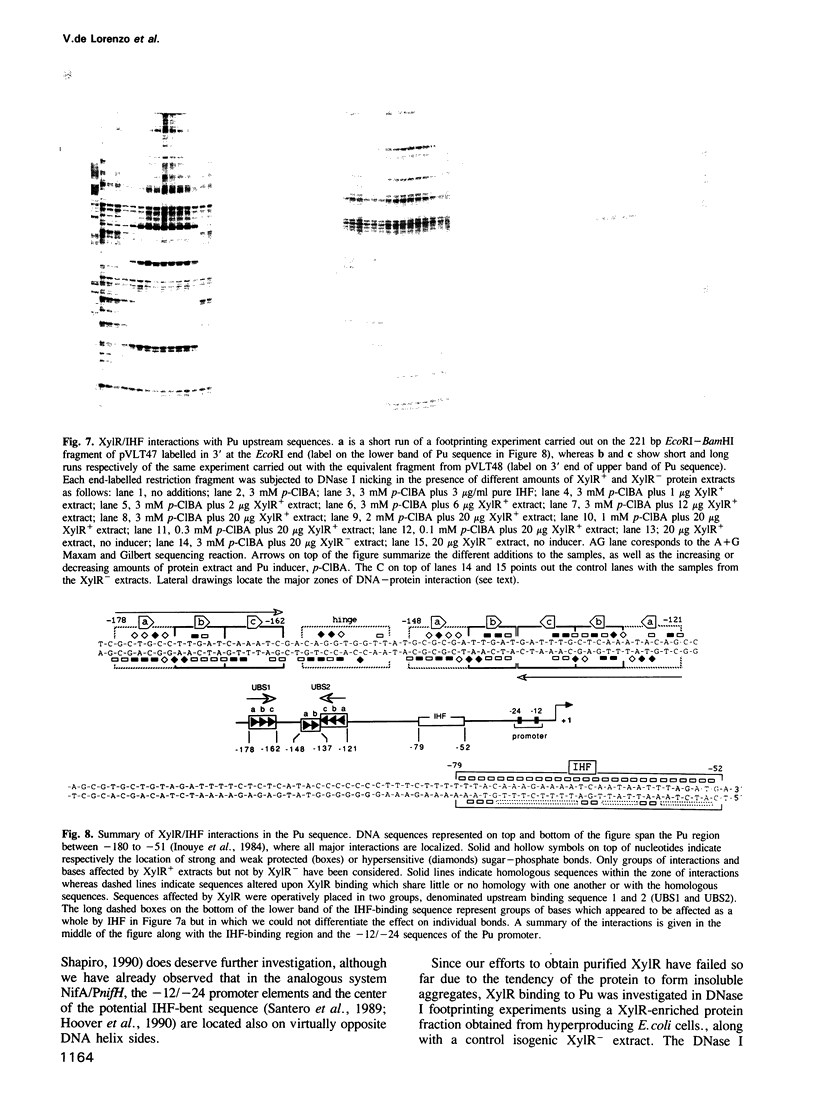



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abril M. A., Michan C., Timmis K. N., Ramos J. L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J Bacteriol. 1989 Dec;171(12):6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann E., Ochs B., Abel K. J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988 Sep 30;69(2):301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- Biek D. P., Cohen S. N. Involvement of integration host factor (IHF) in maintenance of plasmid pSC101 in Escherichia coli: characterization of pSC101 mutants that replicate in the absence of IHF. J Bacteriol. 1989 Apr;171(4):2056–2065. doi: 10.1128/jb.171.4.2056-2065.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracco L., Kotlarz D., Kolb A., Diekmann S., Buc H. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989 Dec 20;8(13):4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M., Cannon W. Mutations in the RNA polymerase recognition sequence of the Klebsiella pneumoniae nifH promoter permitting transcriptional activation in the absence of NifA binding to upstream activator sequences. Nucleic Acids Res. 1989 Apr 11;17(7):2597–2612. doi: 10.1093/nar/17.7.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M., Cannon W., Woodcock J. Transcriptional activation of the Klebsiella pneumoniae nitrogenase promoter may involve DNA loop formation. Mol Microbiol. 1987 Sep;1(2):243–249. doi: 10.1111/j.1365-2958.1987.tb00518.x. [DOI] [PubMed] [Google Scholar]
- Dixon R. The xylABC promoter from the Pseudomonas putida TOL plasmid is activated by nitrogen regulatory genes in Escherichia coli. Mol Gen Genet. 1986 Apr;203(1):129–136. doi: 10.1007/BF00330393. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Gober J. W., Shapiro L. Integration host factor is required for the activation of developmentally regulated genes in Caulobacter. Genes Dev. 1990 Sep;4(9):1494–1504. doi: 10.1101/gad.4.9.1494. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Nash H. A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989 Sep 21;341(6239):251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- Harayama S., Rekik M., Wubbolts M., Rose K., Leppik R. A., Timmis K. N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J Bacteriol. 1989 Sep;171(9):5048–5055. doi: 10.1128/jb.171.9.5048-5055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero M., de Lorenzo V., Timmis K. N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990 Nov;172(11):6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschman J., Wong P. K., Sei K., Keener J., Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Holtel A., Abril M. A., Marques S., Timmis K. N., Ramos J. L. Promoter-upstream activator sequences are required for expression of the xylS gene and upper-pathway operon on the Pseudomonas TOL plasmid. Mol Microbiol. 1990 Sep;4(9):1551–1556. doi: 10.1111/j.1365-2958.1990.tb02066.x. [DOI] [PubMed] [Google Scholar]
- Hoover T. R., Santero E., Porter S., Kustu S. The integration host factor stimulates interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990 Oct 5;63(1):11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- Huala E., Ausubel F. M. The central domain of Rhizobium meliloti NifA is sufficient to activate transcription from the R. meliloti nifH promoter. J Bacteriol. 1989 Jun;171(6):3354–3365. doi: 10.1128/jb.171.6.3354-3365.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Tsui P., Freundlich M. Integration host factor is a negative effector of in vivo and in vitro expression of ompC in Escherichia coli. J Bacteriol. 1990 Sep;172(9):5293–5298. doi: 10.1128/jb.172.9.5293-5298.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Ebina Y., Nakazawa A., Nakazawa T. Nucleotide sequence surrounding transcription initiation site of xylABC operon on TOL plasmid of Pseudomonas putida. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1688–1691. doi: 10.1073/pnas.81.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Gomada M., Sangodkar U. M., Nakazawa A., Nakazawa T. Upstream regulatory sequence for transcriptional activator XylR in the first operon of xylene metabolism on the TOL plasmid. J Mol Biol. 1990 Nov 20;216(2):251–260. doi: 10.1016/S0022-2836(05)80317-1. [DOI] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Expression of the regulatory gene xylS on the TOL plasmid is positively controlled by the xylR gene product. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5182–5186. doi: 10.1073/pnas.84.15.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Nucleotide sequence of the regulatory gene xylR of the TOL plasmid from Pseudomonas putida. Gene. 1988 Jun 30;66(2):301–306. doi: 10.1016/0378-1119(88)90366-6. [DOI] [PubMed] [Google Scholar]
- Inouye S., Yamada M., Nakazawa A., Nakazawa T. Cloning and sequence analysis of the ntrA (rpoN) gene of Pseudomonas putida. Gene. 1989 Dec 21;85(1):145–152. doi: 10.1016/0378-1119(89)90474-5. [DOI] [PubMed] [Google Scholar]
- Krause H. M., Higgins N. P. Positive and negative regulation of the Mu operator by Mu repressor and Escherichia coli integration host factor. J Biol Chem. 1986 Mar 15;261(8):3744–3752. [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kur J., Hasan N., Szybalski W. Physical and biological consequences of interactions between integration host factor (IHF) and coliphage lambda late p'R promoter and its mutants. Gene. 1989 Sep 1;81(1):1–15. doi: 10.1016/0378-1119(89)90331-4. [DOI] [PubMed] [Google Scholar]
- Kustu S., Santero E., Keener J., Popham D., Weiss D. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989 Sep;53(3):367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler T., Harayama S., Ramos J. L., Timmis K. N. Involvement of Pseudomonas putida RpoN sigma factor in regulation of various metabolic functions. J Bacteriol. 1989 Aug;171(8):4326–4333. doi: 10.1128/jb.171.8.4326-4333.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Manley J. L. Transcriptional repression of eukaryotic promoters. Cell. 1989 Nov 3;59(3):405–408. doi: 10.1016/0092-8674(89)90024-x. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mermod N., Lehrbach P. R., Reineke W., Timmis K. N. Transcription of the TOL plasmid toluate catabolic pathway operon of Pseudomonas putida is determined by a pair of co-ordinately and positively regulated overlapping promoters. EMBO J. 1984 Nov;3(11):2461–2466. doi: 10.1002/j.1460-2075.1984.tb02156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermod N., Ramos J. L., Bairoch A., Timmis K. N. The xylS gene positive regulator of TOL plasmid pWWO: identification, sequence analysis and overproduction leading to constitutive expression of meta cleavage operon. Mol Gen Genet. 1987 May;207(2-3):349–354. doi: 10.1007/BF00331600. [DOI] [PubMed] [Google Scholar]
- Morett E., Buck M. NifA-dependent in vivo protection demonstrates that the upstream activator sequence of nif promoters is a protein binding site. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9401–9405. doi: 10.1073/pnas.85.24.9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morett E., Cannon W., Buck M. The DNA-binding domain of the transcriptional activator protein NifA resides in its carboxy terminus, recognises the upstream activator sequences of nif promoters and can be separated from the positive control function of NifA. Nucleic Acids Res. 1988 Dec 23;16(24):11469–11488. doi: 10.1093/nar/16.24.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A. J., Reitzer L. J., Magasanik B. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell. 1987 Sep 25;50(7):1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- Pereira R. F., Ortuno M. J., Lawther R. P. Binding of integration host factor (IHF) to the ilvGp1 promoter of the ilvGMEDA operon of Escherichia coli K12. Nucleic Acids Res. 1988 Jul 11;16(13):5973–5989. doi: 10.1093/nar/16.13.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Vidal-Ingigliardi D., Richet E. A complex nucleoprotein structure involved in activation of transcription of two divergent Escherichia coli promoters. J Mol Biol. 1989 Feb 5;205(3):471–485. doi: 10.1016/0022-2836(89)90218-0. [DOI] [PubMed] [Google Scholar]
- Ramos J. L., Mermod N., Timmis K. N. Regulatory circuits controlling transcription of TOL plasmid operon encoding meta-cleavage pathway for degradation of alkylbenzoates by Pseudomonas. Mol Microbiol. 1987 Nov;1(3):293–300. doi: 10.1111/j.1365-2958.1987.tb01935.x. [DOI] [PubMed] [Google Scholar]
- Santero E., Hoover T., Keener J., Kustu S. In vitro activity of the nitrogen fixation regulatory protein NIFA. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7346–7350. doi: 10.1073/pnas.86.19.7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Role of eukaryotic-type functional domains found in the prokaryotic enhancer receptor factor sigma 54. Cell. 1990 Sep 7;62(5):945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- Snyder U. K., Thompson J. F., Landy A. Phasing of protein-induced DNA bends in a recombination complex. Nature. 1989 Sep 21;341(6239):255–257. doi: 10.1038/341255a0. [DOI] [PubMed] [Google Scholar]
- Thompson R. J., Mosig G. Integration host factor (IHF) represses a Chlamydomonas chloroplast promoter in E. coli. Nucleic Acids Res. 1988 Apr 25;16(8):3313–3326. doi: 10.1093/nar/16.8.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A. Why bend DNA? Cell. 1990 Jan 26;60(2):177–180. doi: 10.1016/0092-8674(90)90729-x. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Curved DNA. CRC Crit Rev Biochem. 1985;19(2):89–106. doi: 10.3109/10409238509082540. [DOI] [PubMed] [Google Scholar]
- Yang C. C., Nash H. A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989 Jun 2;57(5):869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]
- Zaballos A., Salas M., Mellado R. P. A set of expression plasmids for the synthesis of fused and unfused polypeptides in Escherichia coli. Gene. 1987;58(1):67–76. doi: 10.1016/0378-1119(87)90030-8. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V., Giovannini F., Herrero M., Neilands J. B. Metal ion regulation of gene expression. Fur repressor-operator interaction at the promoter region of the aerobactin system of pColV-K30. J Mol Biol. 1988 Oct 20;203(4):875–884. doi: 10.1016/0022-2836(88)90113-1. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V., Herrero M., Jakubzik U., Timmis K. N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990 Nov;172(11):6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]