Abstract
A sequence 30 bp downstream from the start site of the Drosophila melanogaster retrotransposon mdg1 is shown to be responsible for correct and precise initiation of mdg1 RNA synthesis in combination with the RNA start-site sequence TCAGTT. A sequence-specific DNA binding protein is demonstrated to interact with the +30 sequence, and the efficient binding of this factor is necessary for in vivo transcriptional activity of the plasmid constructs containing mdg1 promoter fragments. The nucleotides -8/+34 of mdg1 represent a minimal promoter which is able to provide correct initiation of transcription by RNA polymerase II at basal levels. A comparison with properties of some other retrotransposable elements and several developmentally regulated cellular genes allows us to conclude that together they form a specific class of RNA polymerase II promoter. This promoter class characteristically lacks upstream sequences necessary for transcription initiation, such as TATA boxes, but requires a specific downstream promoter element within 40 bp downstream of the RNA start site. The level of transcription can, however, be modulated by upstream regulatory elements. The identified sequence-specific downstream initiation factor may be responsible for transcription initiation on promoters of some genes which belong to this class.
Full text
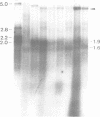
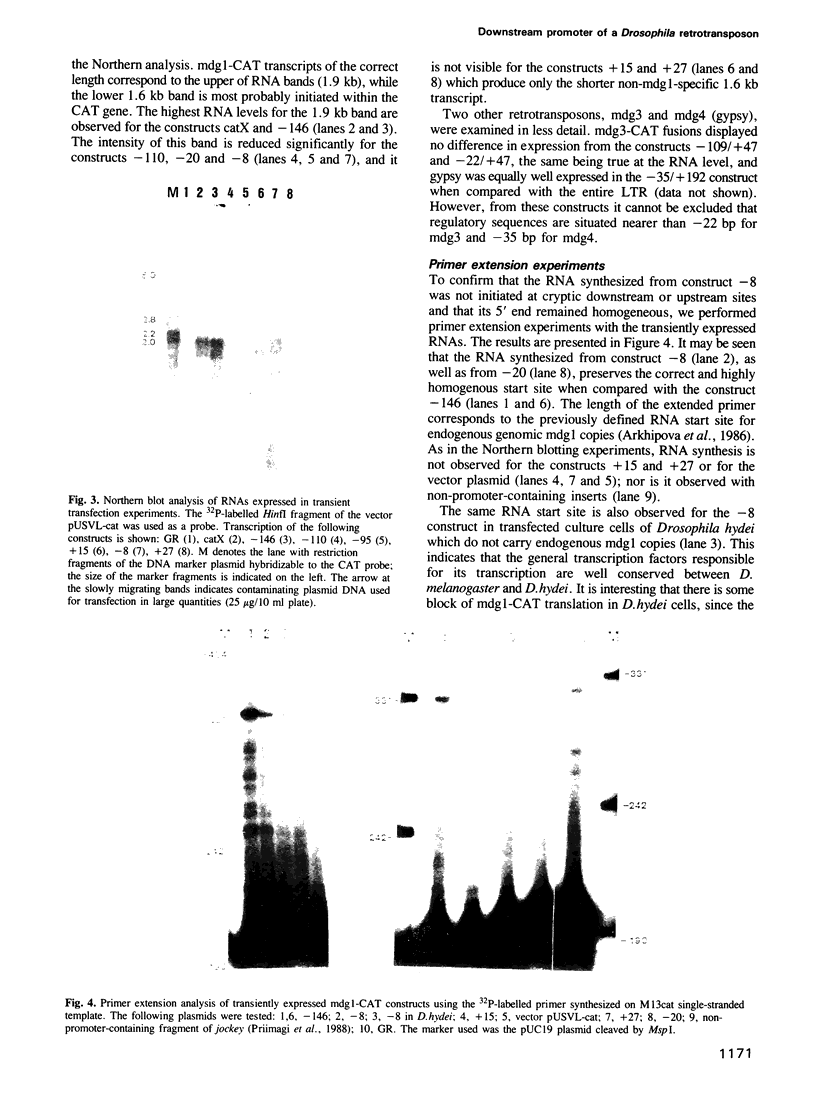
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arkhipova I. R., Gorelova T. V., Ilyin Y. V., Schuppe N. G. Reverse transcription of Drosophila mobile dispersed genetic element RNAs: detection of intermediate forms. Nucleic Acids Res. 1984 Oct 11;12(19):7533–7548. doi: 10.1093/nar/12.19.7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkhipova I. R., Mazo A. M., Cherkasova V. A., Gorelova T. V., Schuppe N. G., Llyin Y. V. The steps of reverse transcription of Drosophila mobile dispersed genetic elements and U3-R-U5 structure of their LTRs. Cell. 1986 Feb 28;44(4):555–563. doi: 10.1016/0092-8674(86)90265-5. [DOI] [PubMed] [Google Scholar]
- Ayer D. E., Dynan W. S. A downstream-element-binding factor facilitates assembly of a functional preinitiation complex at the simian virus 40 major late promoter. Mol Cell Biol. 1990 Jul;10(7):3635–3645. doi: 10.1128/mcb.10.7.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer D. E., Dynan W. S. Simian virus 40 major late promoter: a novel tripartite structure that includes intragenic sequences. Mol Cell Biol. 1988 May;8(5):2021–2033. doi: 10.1128/mcb.8.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988 Jun 3;53(5):699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Corces V. G. Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol. 1989;43:403–434. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Bray S. J., Hirsh J. The Drosophila virilis dopa decarboxylase gene is developmentally regulated when integrated into Drosophila melanogaster. EMBO J. 1986 Sep;5(9):2305–2311. doi: 10.1002/j.1460-2075.1986.tb04498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S., Hahn S., Guarente L., Sharp P. A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989 Feb 24;56(4):549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- Carcamo J., Maldonado E., Cortes P., Ahn M. H., Ha I., Kasai Y., Flint J., Reinberg D. A TATA-like sequence located downstream of the transcription initiation site is required for expression of an RNA polymerase II transcribed gene. Genes Dev. 1990 Sep;4(9):1611–1622. doi: 10.1101/gad.4.9.1611. [DOI] [PubMed] [Google Scholar]
- Cherbas L., Schulz R. A., Koehler M. M., Savakis C., Cherbas P. Structure of the Eip28/29 gene, an ecdysone-inducible gene from Drosophila. J Mol Biol. 1986 Jun 20;189(4):617–631. doi: 10.1016/0022-2836(86)90492-4. [DOI] [PubMed] [Google Scholar]
- Di Nocera P. P. Close relationship between non-viral retroposons in Drosophila melanogaster. Nucleic Acids Res. 1988 May 11;16(9):4041–4052. doi: 10.1093/nar/16.9.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera P. P., Digan M. E., Dawid I. B. A family of oligo-adenylate-terminated transposable sequences in Drosophila melanogaster. J Mol Biol. 1983 Aug 25;168(4):715–727. doi: 10.1016/s0022-2836(83)80071-0. [DOI] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Driver A., Lacey S. F., Cullingford T. E., Mitchelson A., O'Hare K. Structural analysis of Doc transposable elements associated with mutations at the white and suppressor of forked loci of Drosophila melanogaster. Mol Gen Genet. 1989 Dec;220(1):49–52. doi: 10.1007/BF00260854. [DOI] [PubMed] [Google Scholar]
- Dynan W. S. Modularity in promoters and enhancers. Cell. 1989 Jul 14;58(1):1–4. doi: 10.1016/0092-8674(89)90393-0. [DOI] [PubMed] [Google Scholar]
- Emori Y., Shiba T., Kanaya S., Inouye S., Yuki S., Saigo K. The nucleotide sequences of copia and copia-related RNA in Drosophila virus-like particles. 1985 Jun 27-Jul 3Nature. 315(6022):773–776. doi: 10.1038/315773a0. [DOI] [PubMed] [Google Scholar]
- Fawcett D. H., Lister C. K., Kellett E., Finnegan D. J. Transposable elements controlling I-R hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell. 1986 Dec 26;47(6):1007–1015. doi: 10.1016/0092-8674(86)90815-9. [DOI] [PubMed] [Google Scholar]
- Finnegan D. J., Fawcett D. H. Transposable elements in Drosophila melanogaster. Oxf Surv Eukaryot Genes. 1986;3:1–62. [PubMed] [Google Scholar]
- Friesen P. D., Rice W. C., Miller D. W., Miller L. K. Bidirectional transcription from a solo long terminal repeat of the retrotransposon TED: symmetrical RNA start sites. Mol Cell Biol. 1986 May;6(5):1599–1607. doi: 10.1128/mcb.6.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano A. V., Robb S. M., Robb F. T. The structure of the regulatory region of the rat L1 (L1Rn, long interspersed repeated) DNA family of transposable elements. Nucleic Acids Res. 1988 Oct 11;16(19):9215–9231. doi: 10.1093/nar/16.19.9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev G. P. Mobile genetic elements in animal cells and their biological significance. Eur J Biochem. 1984 Dec 3;145(2):203–220. doi: 10.1111/j.1432-1033.1984.tb08541.x. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh J., Morgan B. A., Scholnick S. B. Delimiting regulatory sequences of the Drosophila melanogaster Ddc gene. Mol Cell Biol. 1986 Dec;6(12):4548–4557. doi: 10.1128/mcb.6.12.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultmark D., Klemenz R., Gehring W. J. Translational and transcriptional control elements in the untranslated leader of the heat-shock gene hsp22. Cell. 1986 Feb 14;44(3):429–438. doi: 10.1016/0092-8674(86)90464-2. [DOI] [PubMed] [Google Scholar]
- Ilyin Y. V., Chmeliauskaite V. G., Georgiev G. P. Double-stranded sequences in RNA of Drosophila melanogaster: relation to mobile dispersed genes. Nucleic Acids Res. 1980 Aug 11;8(15):3439–3457. doi: 10.1093/nar/8.15.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Hattori K., Yuki S., Saigo K. Structural variations in the Drosophila retrotransposon, 17.6. Nucleic Acids Res. 1986 Jun 25;14(12):4765–4778. doi: 10.1093/nar/14.12.4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis J. A., Desplan C., Wright D. K., O'Farrell P. H. Evolutionary conservation of homeodomain-binding sites and other sequences upstream and within the major transcription unit of the Drosophila segmentation gene engrailed. Mol Cell Biol. 1989 Oct;9(10):4304–4311. doi: 10.1128/mcb.9.10.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990 Jun 29;61(7):1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- Mathis D. J., Chambon P. The SV40 early region TATA box is required for accurate in vitro initiation of transcription. Nature. 1981 Mar 26;290(5804):310–315. doi: 10.1038/290310a0. [DOI] [PubMed] [Google Scholar]
- Matsuo Y., Kugimiya W., Kadokami Y., Saigo K. Nucleotide sequence of a retrotransposon 297 isolated from Drosophila simulans. Nucleic Acids Res. 1986 Dec 9;14(23):9521–9522. doi: 10.1093/nar/14.23.9521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo A. M., Mizrokhi L. J., Karavanov A. A., Sedkov Y. A., Krichevskaja A. A., Ilyin Y. V. Suppression in Drosophila: su(Hw) and su(f) gene products interact with a region of gypsy (mdg4) regulating its transcriptional activity. EMBO J. 1989 Mar;8(3):903–911. doi: 10.1002/j.1460-2075.1989.tb03451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrokhi L. J., Georgieva S. G., Ilyin Y. V. jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell. 1988 Aug 26;54(5):685–691. doi: 10.1016/s0092-8674(88)80013-8. [DOI] [PubMed] [Google Scholar]
- Moura-Neto R., Dudov K. P., Perry R. P. An element downstream of the cap site is required for transcription of the gene encoding mouse ribosomal protein L32. Proc Natl Acad Sci U S A. 1989 Jun;86(11):3997–4001. doi: 10.1073/pnas.86.11.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy N. B., Pays A., Tebabi P., Coquelet H., Guyaux M., Steinert M., Pays E. Trypanosoma brucei repeated element with unusual structural and transcriptional properties. J Mol Biol. 1987 Jun 20;195(4):855–871. doi: 10.1016/0022-2836(87)90490-6. [DOI] [PubMed] [Google Scholar]
- O'Hare K., Murphy C., Levis R., Rubin G. M. DNA sequence of the white locus of Drosophila melanogaster. J Mol Biol. 1984 Dec 15;180(3):437–455. doi: 10.1016/0022-2836(84)90021-4. [DOI] [PubMed] [Google Scholar]
- Perkins K. K., Dailey G. M., Tjian R. In vitro analysis of the Antennapedia P2 promoter: identification of a new Drosophila transcription factor. Genes Dev. 1988 Dec;2(12A):1615–1626. doi: 10.1101/gad.2.12a.1615. [DOI] [PubMed] [Google Scholar]
- Pirrotta V., Manet E., Hardon E., Bickel S. E., Benson M. Structure and sequence of the Drosophila zeste gene. EMBO J. 1987 Mar;6(3):791–799. doi: 10.1002/j.1460-2075.1987.tb04821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priimägi A. F., Mizrokhi L. J., Ilyin Y. V. The Drosophila mobile element jockey belongs to LINEs and contains coding sequences homologous to some retroviral proteins. Gene. 1988 Oct 30;70(2):253–262. doi: 10.1016/0378-1119(88)90197-7. [DOI] [PubMed] [Google Scholar]
- Regulski M., McGinnis N., Chadwick R., McGinnis W. Developmental and molecular analysis of Deformed; a homeotic gene controlling Drosophila head development. EMBO J. 1987 Mar;6(3):767–777. doi: 10.1002/j.1460-2075.1987.tb04819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenk T. Transcriptional control regions: nucleotide sequence requirements for initiation by RNA polymerase II and III. Curr Top Microbiol Immunol. 1981;93:25–46. doi: 10.1007/978-3-642-68123-3_3. [DOI] [PubMed] [Google Scholar]
- Smale S. T., Baltimore D. The "initiator" as a transcription control element. Cell. 1989 Apr 7;57(1):103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Smale S. T., Schmidt M. C., Berk A. J., Baltimore D. Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneddon A., Flavell A. J. The transcriptional control regions of the copia retrotransposon. Nucleic Acids Res. 1989 Jun 12;17(11):4025–4035. doi: 10.1093/nar/17.11.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeller W. C., Poole S. J., Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 1988 Jan;2(1):68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- Spana C., Harrison D. A., Corces V. G. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 1988 Nov;2(11):1414–1423. doi: 10.1101/gad.2.11.1414. [DOI] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Thummel C. S. The Drosophila E74 promoter contains essential sequences downstream from the start site of transcription. Genes Dev. 1989 Jun;3(6):782–792. doi: 10.1101/gad.3.6.782. [DOI] [PubMed] [Google Scholar]
- Wasylyk B. Enhancers and transcription factors in the control of gene expression. Biochim Biophys Acta. 1988 Nov 10;951(1):17–35. doi: 10.1016/0167-4781(88)90021-8. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Wilde C. D., Akam M. Conserved sequence elements in the 5' region of the Ultrabithorax transcription unit. EMBO J. 1987 May;6(5):1393–1401. doi: 10.1002/j.1460-2075.1987.tb02380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. The site-specific ribosomal DNA insertion element R1Bm belongs to a class of non-long-terminal-repeat retrotransposons. Mol Cell Biol. 1988 Jan;8(1):114–123. doi: 10.1128/mcb.8.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Elder R. T. A region internal to the coding sequences is essential for transcription of the yeast Ty-D15 element. Mol Cell Biol. 1989 Sep;9(9):3667–3678. doi: 10.1128/mcb.9.9.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki S., Inouye S., Ishimaru S., Saigo K. Nucleotide sequence characterization of a Drosophila retrotransposon, 412. Eur J Biochem. 1986 Jul 15;158(2):403–410. doi: 10.1111/j.1432-1033.1986.tb09767.x. [DOI] [PubMed] [Google Scholar]