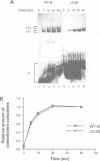
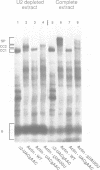
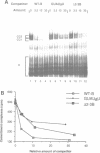
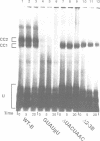
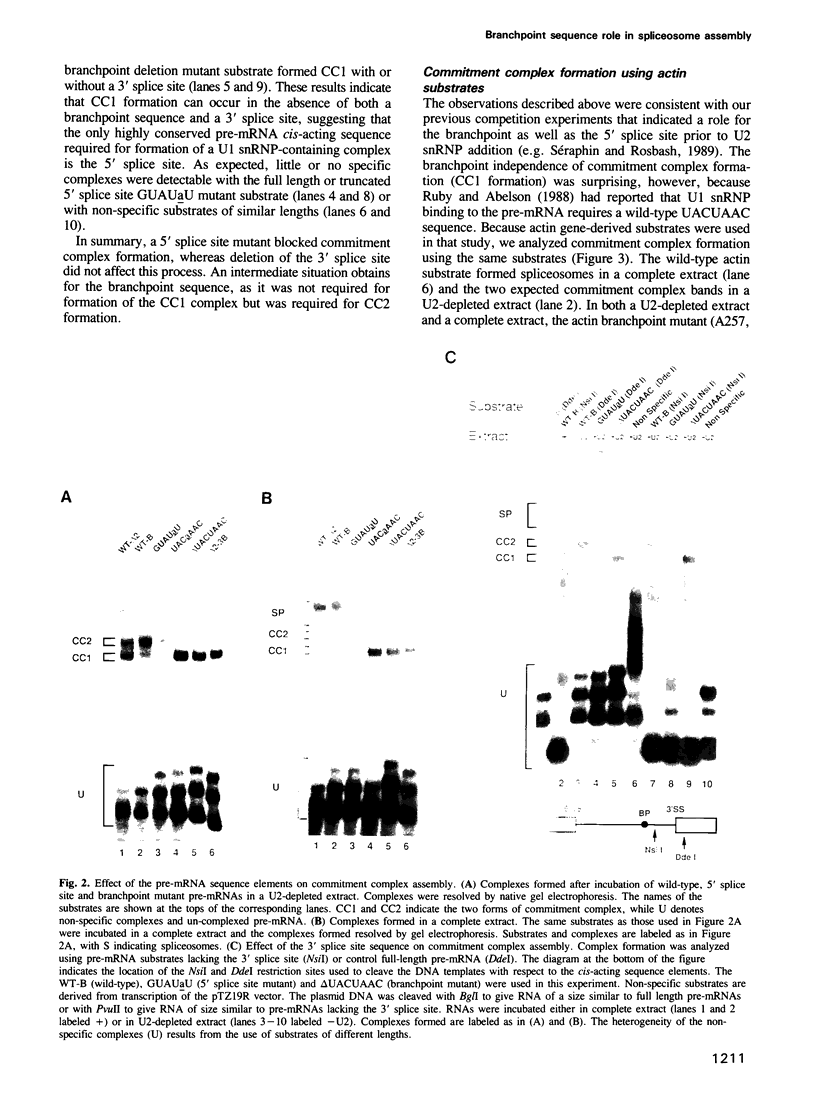
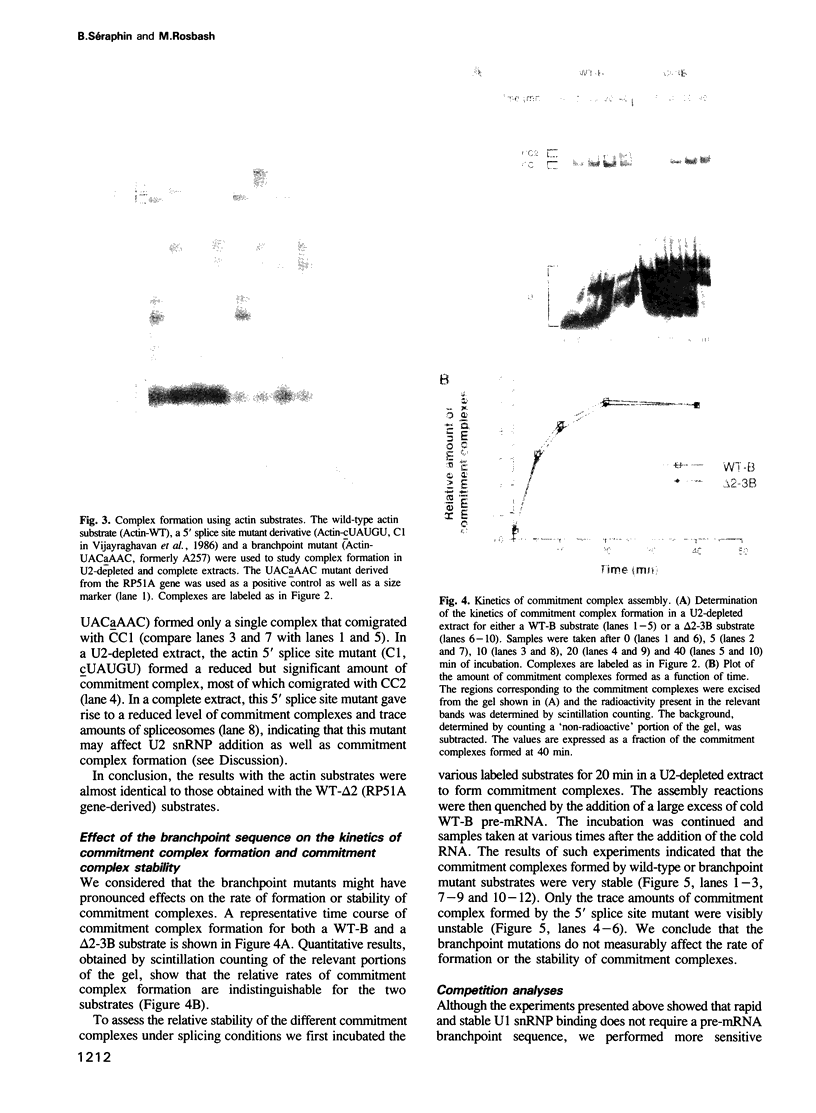
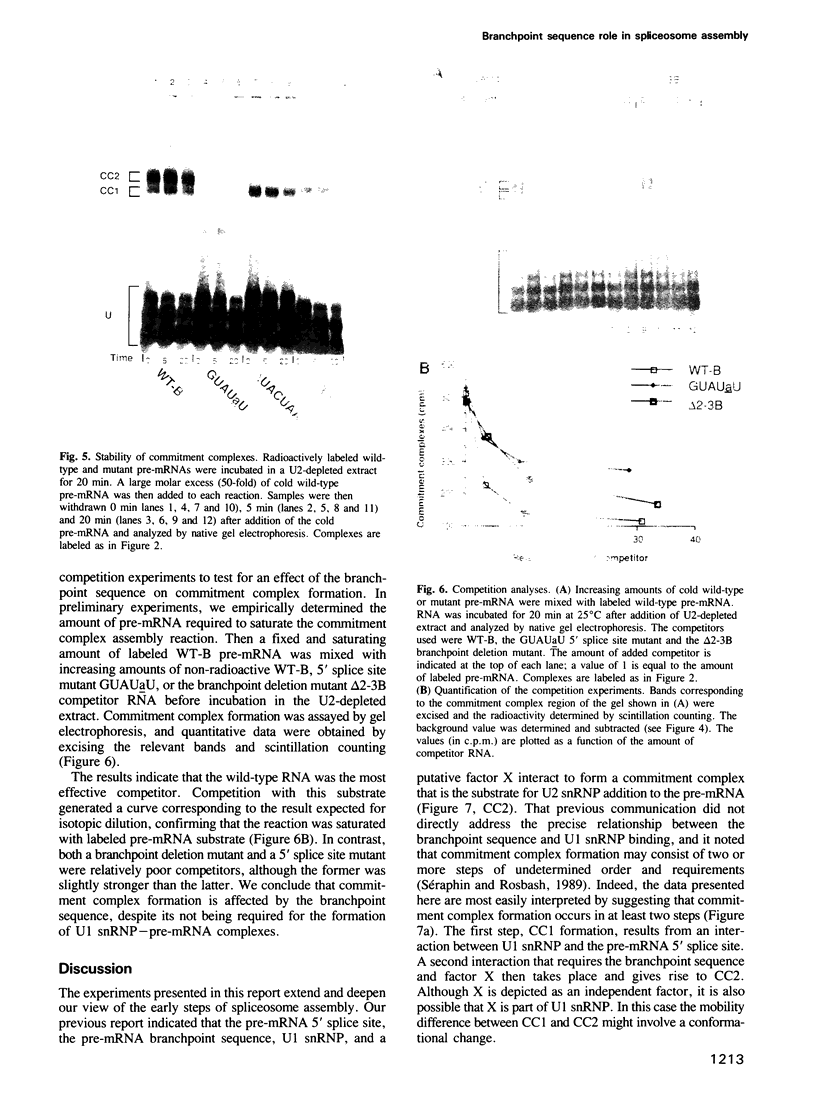
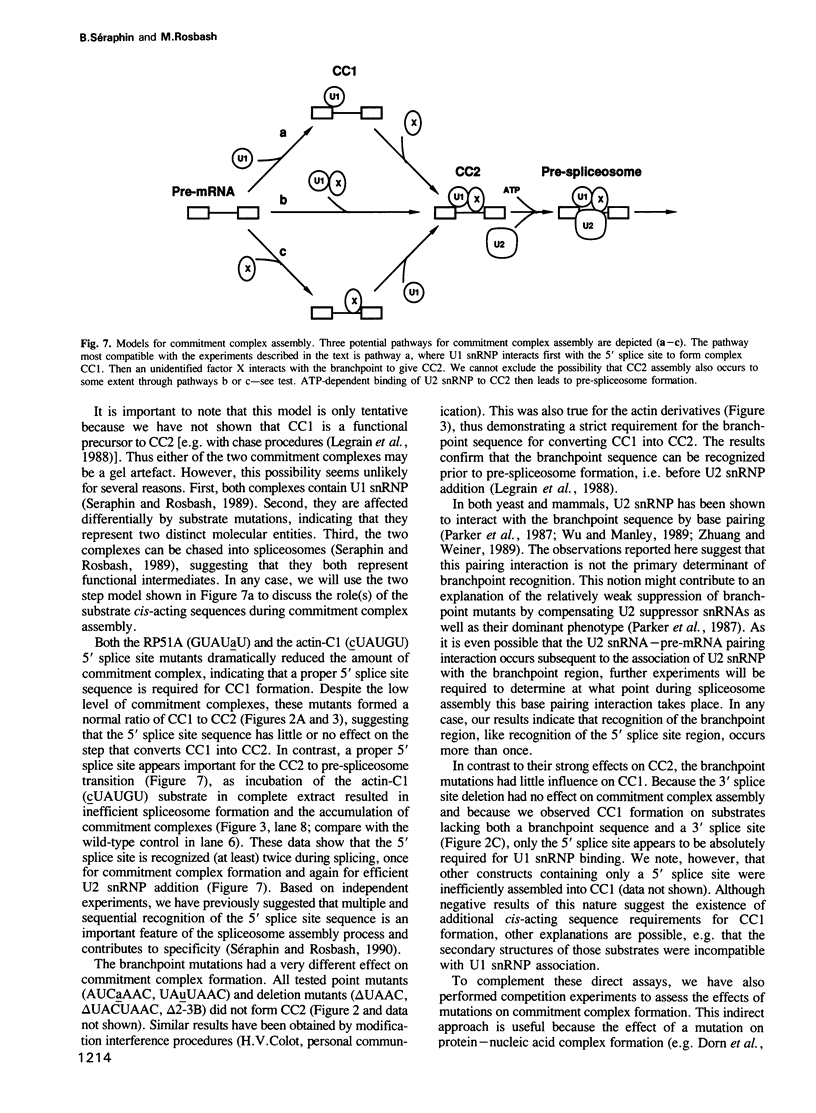
Abstract
Commitment complexes contain U1 snRNP as well as pre-mRNA and are the earliest functional complexes that have been described during in vitro spliceosome assembly. We have used a gel retardation assay to analyze the role of the yeast pre-mRNA cis-acting sequences in commitment complex formation. The results suggest that only a proper 5' splice site sequence is required for efficient U1 snRNA-pre-mRNA complex formation. A role for the highly conserved UACUAAC branchpoint sequence is indicated, however, by competition experiments and by the direct analysis of branchpoint mutant substrates, which cannot form one of the two commitment complex species observed with wild-type substrates. The results suggest that the formation of a U1 snRNP-pre-mRNA complex is not dependent upon the presence of a branchpoint sequence but that the branchpoint sequence is recognized prior to U2 snRNP addition during in vitro spliceosome assembly.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Legrain P., Rosbash M. The yeast PRP6 gene encodes a U4/U6 small nuclear ribonucleoprotein particle (snRNP) protein, and the PRP9 gene encodes a protein required for U2 snRNP binding. Mol Cell Biol. 1990 Dec;10(12):6417–6425. doi: 10.1128/mcb.10.12.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabino S. M., Blencowe B. J., Ryder U., Sproat B. S., Lamond A. I. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell. 1990 Oct 19;63(2):293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- Bindereif A., Green M. R. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. EMBO J. 1987 Aug;6(8):2415–2424. doi: 10.1002/j.1460-2075.1987.tb02520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B. J., Sproat B. S., Ryder U., Barabino S., Lamond A. I. Antisense probing of the human U4/U6 snRNP with biotinylated 2'-OMe RNA oligonucleotides. Cell. 1989 Nov 3;59(3):531–539. doi: 10.1016/0092-8674(89)90036-6. [DOI] [PubMed] [Google Scholar]
- Cheng S. C., Abelson J. Spliceosome assembly in yeast. Genes Dev. 1987 Nov;1(9):1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Dorn A., Bollekens J., Staub A., Benoist C., Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987 Sep 11;50(6):863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Krämer A., Keller W. Different small nuclear ribonucleoprotein particles are involved in different steps of splicing complex formation. Cold Spring Harb Symp Quant Biol. 1987;52:287–298. doi: 10.1101/sqb.1987.052.01.034. [DOI] [PubMed] [Google Scholar]
- Hamm J., Dathan N. A., Mattaj I. W. Functional analysis of mutant Xenopus U2 snRNAs. Cell. 1989 Oct 6;59(1):159–169. doi: 10.1016/0092-8674(89)90878-7. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Rodriguez J. R., Rosbash M. A quantitative analysis of the effects of 5' junction and TACTAAC box mutants and mutant combinations on yeast mRNA splicing. Cell. 1985 Dec;43(2 Pt 1):423–430. doi: 10.1016/0092-8674(85)90172-2. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986 Sep 12;46(6):845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987 Jun 19;49(6):763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Krämer A. Analysis of RNase-A-resistant regions of adenovirus 2 major late precursor-mRNA in splicing extracts reveals an ordered interaction of nuclear components with the substrate RNA. J Mol Biol. 1987 Aug 5;196(3):559–573. doi: 10.1016/0022-2836(87)90032-5. [DOI] [PubMed] [Google Scholar]
- Krämer A., Keller W., Appel B., Lührmann R. The 5' terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984 Aug;38(1):299–307. doi: 10.1016/0092-8674(84)90551-8. [DOI] [PubMed] [Google Scholar]
- Krämer A. Presplicing complex formation requires two proteins and U2 snRNP. Genes Dev. 1988 Sep;2(9):1155–1167. doi: 10.1101/gad.2.9.1155. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A. I., Konarska M. M., Grabowski P. J., Sharp P. A. Spliceosome assembly involves the binding and release of U4 small nuclear ribonucleoprotein. Proc Natl Acad Sci U S A. 1988 Jan;85(2):411–415. doi: 10.1073/pnas.85.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A. I., Konarska M. M., Sharp P. A. A mutational analysis of spliceosome assembly: evidence for splice site collaboration during spliceosome formation. Genes Dev. 1987 Aug;1(6):532–543. doi: 10.1101/gad.1.6.532. [DOI] [PubMed] [Google Scholar]
- Legrain P., Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989 May 19;57(4):573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- Legrain P., Seraphin B., Rosbash M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol. 1988 Sep;8(9):3755–3760. doi: 10.1128/mcb.8.9.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Lin R. J., Lustig A. J., Abelson J. Splicing of yeast nuclear pre-mRNA in vitro requires a functional 40S spliceosome and several extrinsic factors. Genes Dev. 1987 Mar;1(1):7–18. doi: 10.1101/gad.1.1.7. [DOI] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Rosbash M. Specific small nuclear RNAs are associated with yeast spliceosomes. Cell. 1986 Jun 20;45(6):869–877. doi: 10.1016/0092-8674(86)90561-1. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Rymond B. C., Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. 1986 Nov 27-Dec 3Nature. 324(6095):341–345. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Teem J. L., Rosbash M. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: implications for U1 RNA and metazoan mRNA splicing. Cell. 1983 Sep;34(2):395–403. doi: 10.1016/0092-8674(83)90373-2. [DOI] [PubMed] [Google Scholar]
- Reed R. Protein composition of mammalian spliceosomes assembled in vitro. Proc Natl Acad Sci U S A. 1990 Oct;87(20):8031–8035. doi: 10.1073/pnas.87.20.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revzin A. Gel electrophoresis assays for DNA-protein interactions. Biotechniques. 1989 Apr;7(4):346–355. [PubMed] [Google Scholar]
- Rogers J., Wall R. A mechanism for RNA splicing. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1877–1879. doi: 10.1073/pnas.77.4.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988 Nov 18;242(4881):1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Rosbash M. A chemical modification/interference study of yeast pre-mRNA spliceosome assembly and splicing. Genes Dev. 1988 Apr;2(4):428–439. doi: 10.1101/gad.2.4.428. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Rosbash M. Cleavage of 5' splice site and lariat formation are independent of 3' splice site in yeast mRNA splicing. Nature. 1985 Oct 24;317(6039):735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Torrey D. D., Rosbash M. A novel role for the 3' region of introns in pre-mRNA splicing of Saccharomyces cerevisiae. Genes Dev. 1987 May;1(3):238–246. doi: 10.1101/gad.1.3.238. [DOI] [PubMed] [Google Scholar]
- Seraphin B., Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989 Oct 20;59(2):349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Rosbash M. Exon mutations uncouple 5' splice site selection from U1 snRNA pairing. Cell. 1990 Nov 2;63(3):619–629. doi: 10.1016/0092-8674(90)90457-p. [DOI] [PubMed] [Google Scholar]
- Tatei K., Takemura K., Mayeda A., Fujiwara Y., Tanaka H., Ishihama A., Ohshima Y. U1 RNA-protein complex preferentially binds to both 5' and 3' splice junction sequences in RNA or single-stranded DNA. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6281–6285. doi: 10.1073/pnas.81.20.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatei K., Takemura K., Tanaka H., Masaki T., Ohshima Y. Recognition of 5' and 3' splice site sequences in pre-mRNA studied with a filter binding technique. J Biol Chem. 1987 Aug 25;262(24):11667–11674. [PubMed] [Google Scholar]
- Vijayraghavan U., Parker R., Tamm J., Iimura Y., Rossi J., Abelson J., Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986 Jul;5(7):1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Zamore P. D., Green M. R. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- Zillmann M., Rose S. D., Berget S. M. U1 small nuclear ribonucleoproteins are required early during spliceosome assembly. Mol Cell Biol. 1987 Aug;7(8):2877–2883. doi: 10.1128/mcb.7.8.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]