Abstract
Aicardi–Goutières syndrome (AGS) is a severe childhood inflammatory disorder that shows clinical and genetic overlap with systemic lupus erythematosus (SLE). AGS is thought to arise from the accumulation of incompletely metabolized endogenous nucleic acid species owing to mutations in nucleic acid-degrading enzymes TREX1 (AGS1), RNase H2 (AGS2, 3 and 4), and SAMHD1 (AGS5). However, the identity and source of such immunogenic nucleic acid species remain undefined. Using genome-wide approaches, we show that fibroblasts from AGS patients with AGS1-5 mutations are burdened by excessive loads of RNA:DNA hybrids. Using MethylC-seq, we show that AGS fibroblasts display pronounced and global loss of DNA methylation and demonstrate that AGS-specific RNA:DNA hybrids often occur within DNA hypomethylated regions. Altogether, our data suggest that RNA:DNA hybrids may represent a common immunogenic form of nucleic acids in AGS and provide the first evidence of epigenetic perturbations in AGS, furthering the links between AGS and SLE.
DOI: http://dx.doi.org/10.7554/eLife.08007.001
Research organism: human
eLife digest
The immune system protects the body from attack by bacteria, viruses, and other microbes. A key feature of this system is the ability to discriminate between the body's own cells and potential foreign invaders. Occasionally, this process can go wrong and the immune system starts attacking its own tissues, which can lead to arthritis, diabetes, lupus, and other ‘autoimmune’ diseases.
Aicardi–Goutières syndrome (AGS) is an autoimmune disease that leads to severe mental and physical symptoms. Recent research has revealed that the disease is caused by mutations in genes that make enzymes called nucleases. In healthy people, these enzymes destroy DNA molecules and other nucleic acids. In AGS patients, the failure of the nucleases to act is thought to lead to the accumulation of unwanted DNA and RNA molecules. These molecules, in turn, are thought to be mistakenly identified by the immune system as ‘foreign’ and to cause an autoimmune response. However, it is not clear how this works.
Here, Lim et al. studied skin cells called fibroblasts from patients with Aicardi–Goutières syndrome. The experiments found that the patients' cells had excessive numbers of RNA molecules binding to sections of matching DNA. These unusual DNA–RNA ‘hybrids’ accumulated in regions of the genome that do not contain many genes, perhaps as a result of breaks in the DNA. It is possible that they may mimic nucleic acids from viruses and could trigger an autoimmune response.
In healthy individuals, small ‘methyl’ groups are often attached to DNA in a process known as DNA methylation. This serves to maintain the stability of the genome and controls the activity of genes. Unexpectedly, Lim et al. found that the DNA in AGS patients had far fewer methyl groups, especially in areas where the DNA–RNA hybrids had accumulated. This may lead to genome destabilization, alterations in gene activity, and may mean that the DNA in these regions may be mistaken for foreign DNA by the immune system.
Altogether, Lim et al.'s findings suggest that Aicardi–Goutières syndrome may be caused by immune responses triggered by the accumulation of RNA–DNA hybrids and lower levels of DNA methylation. These findings may aid the development of new therapies to treat Aicardi–Goutières syndrome, lupus, and other similar diseases.
Introduction
Aicardi–Goutières syndrome (AGS) is a severe inflammatory encephalopathy characterized by neurological dysfunction, psychomotor retardation, seizures, unexplained fevers, joint stiffness, basal ganglia calcification, and chilblain skin lesions (Rice et al., 2007b). Despite its rarity, AGS has captured the attention of the scientific community due to its clinical, molecular, and genetic overlap with another, common, systemic autoimmune disorder, systemic lupus erythematosus (SLE) (Rice et al., 2007a, 2007b; Ravenscroft et al., 2011) (Gunther et al., 2015). Both diseases are characterized, and to a large extent driven, by excessive expression of interferon alpha, a potent anti-viral cytokine associated with heightened innate immune response and inflammation (Crow, 2011). AGS is a monogenic disorder caused by mutations in any one of the six AGS genes. The AGS genes encode for the 3′ to 5′ single-stranded DNA exonuclease TREX1 (AGS1) (Crow et al., 2006a), the RNA:DNA hybrid-specific ribonuclease H2 subunits (RNase H2A, B and C, corresponding to AGS4, 2 and 3, respectively) (Crow et al., 2006b), the 3′ to 5′ exonuclease and dNTP hydrolase SAMHD1 (AGS5) (Rice et al., 2009; Goldstone et al., 2011), and the RNA adenosine deaminase ADAR1 (AGS6) (Rice et al., 2012). Recently, gain-of-function mutations in the cytosolic double-stranded RNA receptor gene IFIH1 have also been shown to be associated with AGS (Rice et al., 2014). Interestingly, mutations in TREX1 and the RNASEH2 genes have also been implicated in SLE, further illustrating the genetic link between AGS and SLE (Lee-Kirsch et al., 2007; Gunther et al., 2015). Given the role of AGS enzymes in DNA and RNA metabolism, the innate immune response in AGS is thought to be triggered by the accumulation of incompletely metabolized endogenous nucleic acid elements (Crow and Rehwinkel, 2009). However, the identity and source of such immunogenic nucleic acid elements remain undefined.
While genetically well defined, the potential contribution of epigenetic deregulation to AGS has not been addressed. Characterizing putative epigenetic deregulation in AGS is all the more justified given that DNA hypomethylation, either spontaneous or caused by drug exposure, triggers increased B cell autoreactivity and the development of drug-induced and idiopathic human lupus (Richardson, 1986; Quddus et al., 1993; Yung et al., 1995; Jeffries et al., 2011; Absher et al., 2013; Zhang et al., 2013). Furthermore, loss of DNA methylation, albeit modest and restricted to specific autoimmunity-related genes, was reported in SLE patients (Richardson et al., 1990; Ballestar et al., 2006; Javierre et al., 2010; Jeffries et al., 2011; Absher et al., 2013), raising the question of whether the same epigenetic deregulation is shared in AGS.
To uncover the mechanism driving inflammatory responses in AGS, we sought to identify transcriptional, genetic, and epigenetic perturbations shared among AGS subtypes. For this, we extensively profiled a series of primary fibroblast cells from AGS patients with mutations in AGS1, AGS2, and AGS4 and AGS5 using high-throughput sequencing technologies. Transcriptional profiling via RNA-seq showed that AGS fibroblasts are characterized by an activated antiviral, immune response with a characteristic interferon signature. Genome-wide profiling of RNA:DNA hybrid formation revealed that AGS cells accumulate excessive loads of RNA:DNA hybrids. Whole-genome DNA methylation profiling further indicated that AGS genomes experience global loss of DNA methylation. Our data therefore provide the first evidence that TREX1, RNASEH2, and SAMHD1 mutations share common molecular abnormalities including genome-wide DNA hypomethylation and accumulation of RNA:DNA hybrid species.
Results
Primary AGS fibroblasts show heightened immunomodulatory transcriptional responses
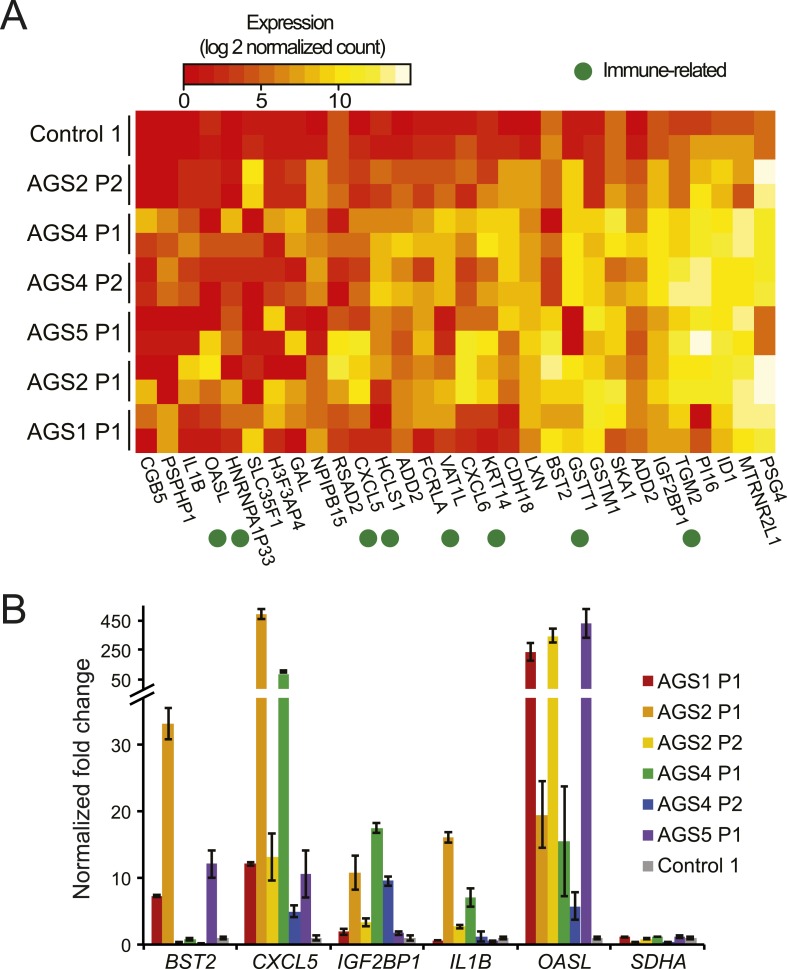
We first performed RNA-seq to ascertain transcriptional signatures of AGS using primary fibroblasts from four patients with mutations in AGS1, AGS2, AGS4, or AGS5, and an age-matched healthy control (see Supplementary file for detailed genotype information). We identified a total of 98 and 209 genes that were significantly up- and down-regulated, respectively, in at least one AGS sample (Figure 1A; Figure 1—figure supplement 1A; Supplementary file 2). AGS down-regulated genes significantly associated with ontologies linked to cell adhesion and the extracellular matrix (Supplementary file 2). These ontologies may be relevant to the skin lesions often observed in AGS and SLE patients (Rice et al., 2007b). Genes involved in inflammation, immune responses, chemokine signaling pathways, and sensing of viral nucleic acids were up-regulated in AGS patient cells. Genes for the major pro-inflammatory cytokine IL1β (Zhao et al., 2013b) and the CXCL5 and CXCL6 chemokines were up-regulated in several AGS patients (Figure 1A,B). Additional immune signaling genes (IL-33, CXCL3, CXCL1, IL-8, CCL11) were significantly up-regulated in at least one AGS sample (Supplementary file 2) and gene ontology analysis of AGS deregulated genes identified cytokine and chemokine signaling pathways as one of weakly enriched terms (Figure 1—figure supplement 1B). These observations are indicative of a heightened immunomodulatory and chemotactic response previously identified in multiple autoimmune conditions, including SLE (Tuller et al., 2013). Genes involved in anti-viral responses (RSAD2, OASL, IGF2BP1, BST2), in particular interferon-inducible genes, were also up-regulated in multiple AGS samples (Figure 1A,B). Additional interferon-inducible genes (IFI6, IF44L, ISG15) described previously as representing an interferon signature in SLE (Baechler et al., 2003) and AGS (Rice et al., 2013) showed up-regulation in at least one AGS sample (Supplementary file 2). AGS fibroblasts thus exhibit an activated immune, inflammatory, and anti-viral state, underscoring the systemic nature of the disease and indicating that fibroblasts are an appropriate cell type to study AGS.
Figure 1. AGS fibroblasts exhibit activated immune antiviral response.
(A) RNA-seq heatmap showing log2 expression level of genes up-regulated in at least two Aicardi–Goutières syndrome (AGS) samples. Two biological replicates are shown for each sample. Immune-related genes are marked with green dots. (B) Gene expression changes between AGS samples and control were measured for five immune-related genes by real time reverse-transcription PCR (RT-qPCR). Y-axis represents fold change in gene expression, normalized to GAPDH gene, relative to the control sample. SDHA is another housekeeping gene used as control. Each value is the average of at least two technical replicates. Error bars represent SEM.
Figure 1—figure supplement 1. RNA-seq of AGS and control fibroblasts.
Accumulation of ribonucleotides in genomic DNA is unique to RNASEH2-deficient AGS patients
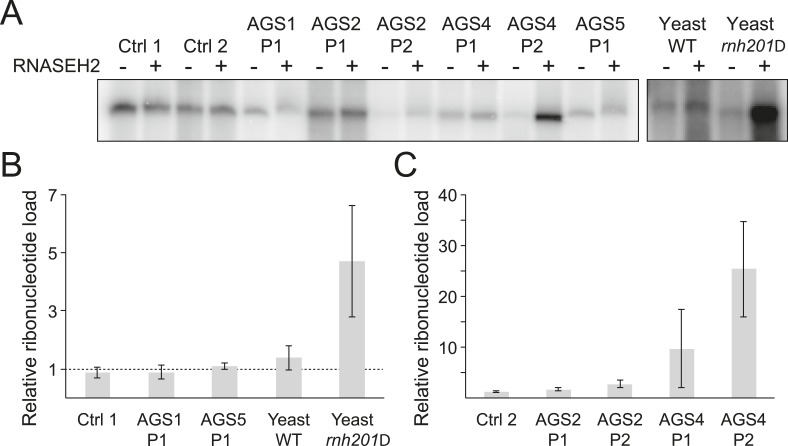
While it is likely that dysfunction in AGS enzymes results in an accumulation of incompletely metabolized immunogenic nucleic acid species (Crow and Rehwinkel, 2009), the identity and source of these molecules remain unclear. RNase H2 generally degrades RNA:DNA hybrids (Cerritelli and Crouch, 2009) and is specifically responsible for removing single ribonucleotides that are misincorporated during DNA replication (Reijns et al., 2012). We therefore determined whether ribonucleotide accumulation is a common feature of AGS. For this, we treated genomic DNA from AGS and control primary fibroblasts with purified human RNase H2 and measured the presence of resulting nicks by DNA polymerase I-dependent nick translation in the presence of [α-32P] dCTP. Genomic DNA from wild-type and RNase H2A-deficient (rnh201Δ) yeast cells that readily incorporate ribonucleotides (Nick McElhinny et al., 2010) was used as a control. Relative to wild-type, rnh201Δ yeast cells showed a fourfold to fivefold increase in ribonucleotide accumulation (Figure 2A,B), consistent with prior results (Nick McElhinny et al., 2010). Cells from two patients mutated in RNASEH2B (AGS2) showed a twofold to threefold increase in labeling (Figure 2A,C). Cells from two patients mutated in the catalytic RNase H2A subunit (AGS4) showed a pronounced 10–25-fold increase in ribonucleotides (Figure 2A,C). Thus, as observed in yeast (Nick McElhinny et al., 2010) and mouse (Reijns et al., 2012) models, a reduction of human RNase H2 activity leads to elevated levels of ribonucleotides in genomic DNA. By contrast, no significant increase in ribonucleotide loads could be detected in patients mutated in TREX1 (AGS1) or SAMHD1 (AGS5) (Figure 2A,B). Thus, while aberrant accumulation of ribonucleotides may contribute to the severity of the disease in RNase H2-defective AGS patients, it is unlikely to represent a common form of disease-causing nucleic acids in AGS.
Figure 2. Incorporation of ribonucleotides in genomic DNA is only observed in RNase H2-deficient AGS fibroblasts.
(A) Genomic DNA was treated with RNase H2 to reveal the presence of ribonucleotides as single-strand nicks. An untreated control was used to reveal background single-stranded breaks in genomic DNA preparations. The intensity of labeling after gel electrophoresis was measured. (B, C) Relative ribonucleotide loads are reported as fold-increase of radiolabel incorporation of RNase H2-treated over untreated sample. At least three independent replicates were performed and the error bars indicate SEM.
RNA:DNA hybrids accumulate in AGS patients
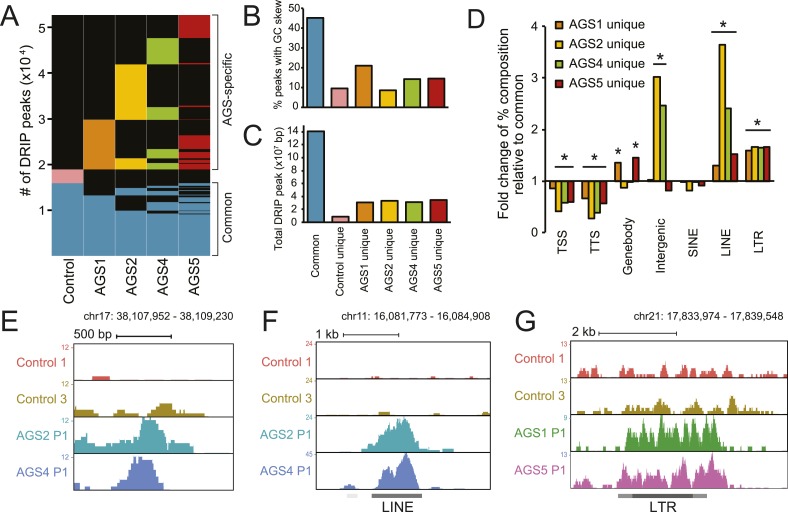
To identify other species of RNA:DNA hybrids that may be accumulating in AGS, we performed DRIP-seq, a technique originally developed to profile R-loop formation genome-wide (Ginno et al., 2012). R-loops are long RNA:DNA hybrid structures that form co-transcriptionally upon re-annealing of the RNA transcript to the template DNA strand, forcing the non-template strand into a single-stranded state. R-loops are enriched at the 5′- and 3′-ends of human genes where they play roles in epigenetic control and transcription regulation (Ginno et al., 2012, 2013). DRIP-seq revealed that AGS and control fibroblasts share a total of 15,897 DRIP peaks representing approximately 141 megabases of genomic space (Figure 3A,C, Figure 3—figure supplement 1A,B). Consistent with prior studies, 32% of these peaks mapped to promoters or gene ends, which represents a strong enrichment over expected genomic distribution and further indicates that gene ends correspond to R-loop formation hotspots (Ginno et al., 2013). Our high-coverage DRIP-seq data also revealed that 56% of the R-loop signal mapped onto gene bodies, suggesting that RNA–DNA entanglements during transcription are prevalent (Figure 3—figure supplement 1C). By contrast, only 15.6% of common R-loop peaks mapped to intergenic regions, consistent with a predominant genic origin for R-loop formation (Figure 3—figure supplement 1C). Overall, common DRIP peaks showed strong overlap with GC skew (Figure 3B), a key sequence determinant that favors co-transcriptional R-loop formation (Ginno et al., 2013). The observation that AGS samples display R-loop formation over expected R-loop forming, GC-skewed, regions indicates that canonical R-loop formation is not significantly altered in AGS patients (Figure 3—figure supplement 1A,B).
Figure 3. AGS fibroblasts accumulate RNA:DNA hybrids.
(A) All genomic loci overlapping with a DRIP peak in at least one sample are stacked vertically; the position of each peak in a stack is constant horizontally across samples. Each patient subtype or control occupies a vertical bar, as labeled. Each bar corresponds to merged data sets from two independent samples. Common peaks (i.e., form in control and at least one AGS sample) are represented in blue. Control-unique DRIP peaks are shown in pink; lack of DRIP signal over a given peak in any sample is shown as black. AGS-unique peaks are colored orange, yellow, green, and red in AGS1, 2, 4, and 5, respectively. Brackets on the right side demarcate common and AGS-specific peaks, respectively. (B, C) Graphs showing the % overlap between DRIP peaks and blocks of GC skew (B); and the total size of DRIP peaks in each category (C). Color codes are as described for (A). (D) Enrichment or depletion of AGS-unique DRIP peaks over different genomic features is shown relative to common DRIP peaks. * indicates p < 0.002 and fold change >20% relative to common peaks. (E–G) Representative examples of AGS-specific DRIP peaks over an intergenic region (E), a truncated long interspersed nuclear elements (LINE) element (F) and a truncated long terminal repeats (LTR) element (G).
Figure 3—figure supplement 1. Canonical R-loop genomic patterns are not affected in AGS fibroblasts.
Despite the relative agreement between AGS and control R-loop profiles over a broad set of common peaks, a large excess of DRIP peaks (33,781 in total) was observed specifically in AGS cells (Figure 3A). These novel peaks of RNA:DNA hybrids were in many cases unique to each patient sub-type. While a small number of DRIP peaks were also unique to the control sample, AGS-specific DRIP peaks in each AGS subtype were both threefold to fourfold greater in numbers, and when combined, occupied a three to four times larger genomic space than control-specific peaks (Figure 3C). On average, AGS-specific DRIP peaks occupied an additional 23.8 megabases of DNA sequence. This indicates that all AGS subtypes are burdened by higher loads of RNA:DNA hybrids in their genomes.
Unlike common DRIP peaks, AGS-specific DRIP peaks were threefold to sevenfold less likely to overlap with GC skew (Figure 3B), suggesting that they may originate according to a non-canonical mechanism. Additionally, AGS-specific DRIP peaks were depleted at transcription start sites (TSSs) and transcription termination sites (TTSs) (Figure 3D), where R-loops are typically observed (Ginno et al., 2013). In contrast, AGS2- and AGS4-specific peaks were significantly enriched over intergenic portions of the human genome (Figure 3D,E). AGS1- and AGS5-specific DRIP peaks were instead enriched over gene body regions (Figure 3D, Figure 3—figure supplement 1D). Interestingly, all AGS-specific DRIP peaks were significantly enriched in repeat classes corresponding to long interspersed nuclear elements (LINE) and long terminal repeats (LTR) retrotransposons (Figure 3D,F,G, Figure 3—figure supplement 1D). Analysis of the overlap of AGS-specific DRIP peaks with human-specific, retrotransposition-competent, LINE-1 elements failed to reveal any significant trend (data not shown). Therefore, whether the reported enrichment of AGS-specific DRIP peaks over LINE and LTR repeats carries biological significance or is simply a reflection of the increased repeat content of intergenic and intronic space remains to be determined. Altogether, our results indicate that AGS mutations in TREX1, RNASEH2A, RNASEH2B, and SAMHD1 are associated with the accumulation of RNA:DNA hybrids over repeat-rich intergenic and gene body regions.
AGS patients show pronounced genome-wide DNA hypomethylation
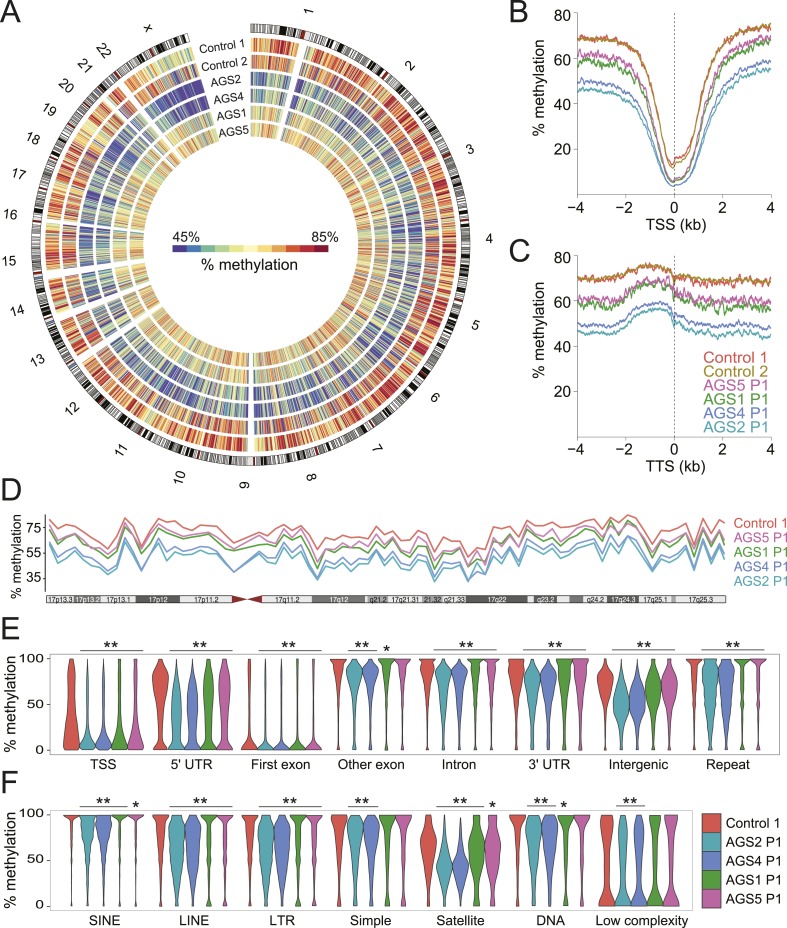
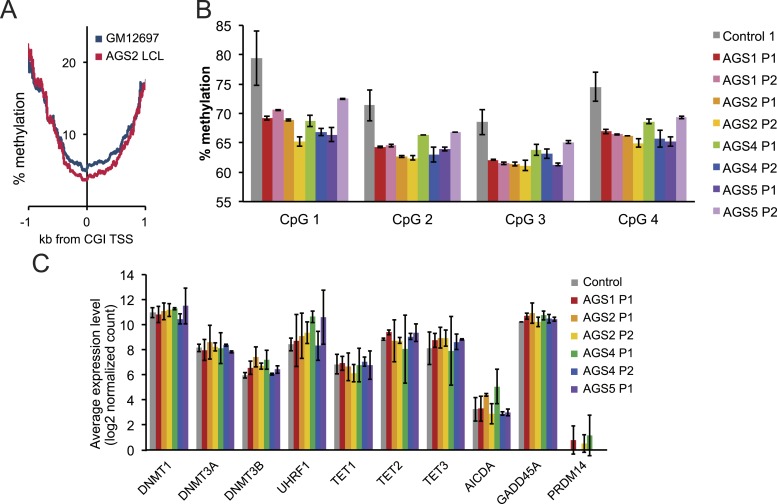
The observation that RNA:DNA hybrids accumulate over intergenic regions is surprising given that they are normally maintained in a transcriptionally quiescent state owing to the deposition of silencing epigenetic marks such as DNA methylation (Yoder et al., 1997). To determine if DNA methylation patterns were altered in AGS, we performed MethylC-seq (Lister et al., 2009) to profile DNA methylation at low coverage (3.21–7.82 X coverage) across the same set of primary fibroblasts studied above (Supplementary file 1). All AGS cells displayed significant, global, DNA hypomethylation (Figure 4A). AGS2 and 4 cells showed profound DNA hypomethylation, with a ∼20% reduction in genomic methylation levels overall. AGS1 and 5 showed a more moderate, but still highly significant, 5–10% reduction in DNA methylation. This reduction affected TSSs and TTSs (Figure 4B,C; Figure 4—figure supplement 1A) and spread along the lengths of entire chromosomes (Figure 4D), significantly impacting nearly all genomic compartments, including genic, intergenic, and repeat regions (Figure 4E). Analysis of repeat classes further revealed that LINE and LTR elements were significantly hypomethylated in all AGS subtypes (Figure 4F). Pyrosequencing assays targeting human-specific LINE-1 elements also revealed a small but significant decrease in DNA methylation over 4 CpG sites carried at the LINE-1 5′-UTR promoter (Figure 4—figure supplement 1B). SINE elements, satellite repeats, and other repeat types were significantly hypomethylated in AGS2 and 4 but not always in AGS1 and 5 (Figure 4F). This profound reduction in DNA methylation was not caused by any detectable deregulation in the expression of genes encoding for the DNA methylation machinery or of components of a putative DNA de-methylation system (Figure 4—figure supplement 1C).
Figure 4. Genome-wide DNA hypomethylation in AGS fibroblasts.
(A) Circos plot depicting DNA methylation along human chromosomes. Each tick mark is colored according to the average percent methylation across a 2 Mb genomic region (see color legend). (B, C) Metaplots of DNA methylation around transcription start site (TSS) (B) and transcription termination site (TTS) (C). (D) Percent methylation along chromosome 17. (E, F) Violin plots depicting methylation levels at different genomic (E) and repeat (F) regions. p-values are relative to control. **p < 1e-9, *p < 0.01.
Figure 4—figure supplement 1. Methylation profiles of AGS and control cells.
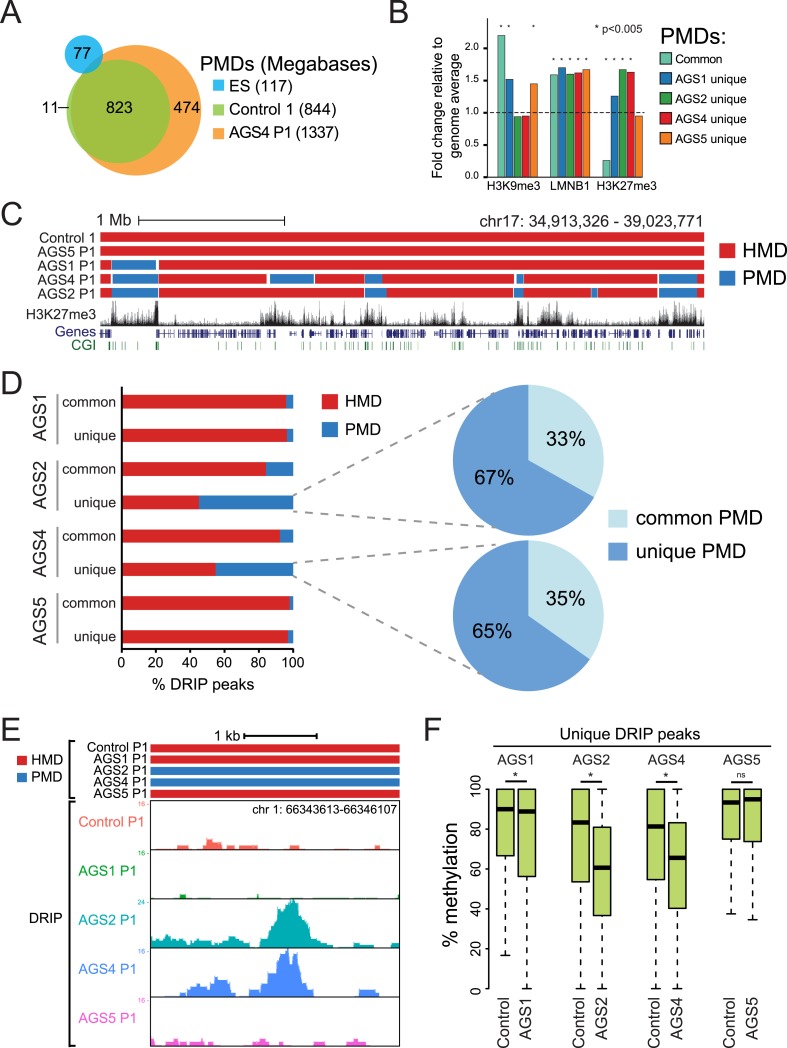
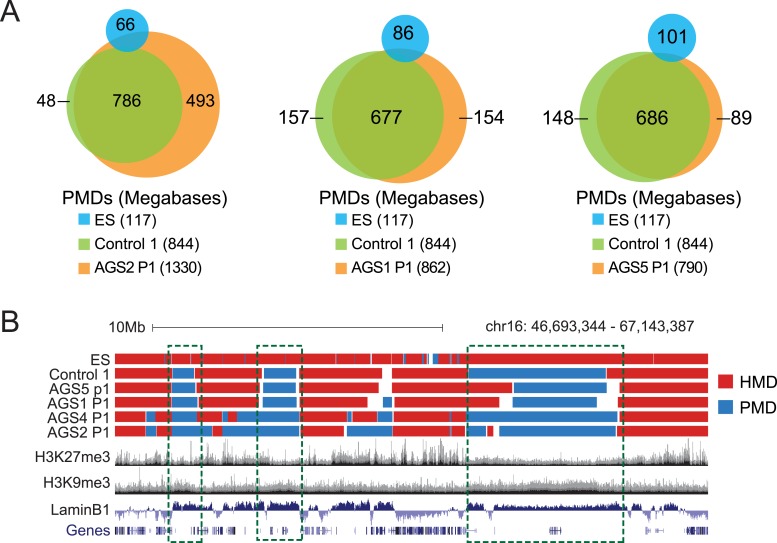
To further characterize the DNA hypomethylation observed in AGS cells, we employed a Hidden Markov Model (HMM) to annotate highly and partially methylated domains (HMDs and PMDs, respectively) (Schroeder et al., 2011). PMDs in control fibroblasts were generally shared by AGS fibroblasts but not by embryonic stem (ES) cells (Figure 5A; Figure 5—figure supplement 1A), consistent with the notion that they represent fibroblast-specific PMDs. AGS2 and AGS4 fibroblasts displayed broader PMDs than the control cells, while the size of PMD regions in AGS1 and AGS5 cells was relatively unchanged (Figure 5A; Figure 5—figure supplement 1A). Analysis of the chromatin features associated with PMDs showed that common PMDs resided within silenced genomic regions enriched for Lamin-B1, a component of the nuclear lamina, and the silencing mark histone H3 Lysine 9 trimethylation (H3K9me3), as described previously (Berman et al., 2012). Common PMDs were also strongly depleted for the H3K27me3 mark, a modification associated with facultative heterochromatin and silencing over developmental genes (Figure 5B, Figure 5—figure supplement 1B). Interestingly, AGS-specific PMDs, while still associated with Lamin-B1 and to some extent with H3K9me3, were enriched for H3K27me3, particularly over regions corresponding normally to large H3K27me3 blocks (Figure 5B,C). This suggests that the DNA hypomethylation observed in AGS is particularly associated with regions of silenced, condensed chromatin.
Figure 5. AGS-specific PMDs are enriched over H3K27me3-marked genomic regions.
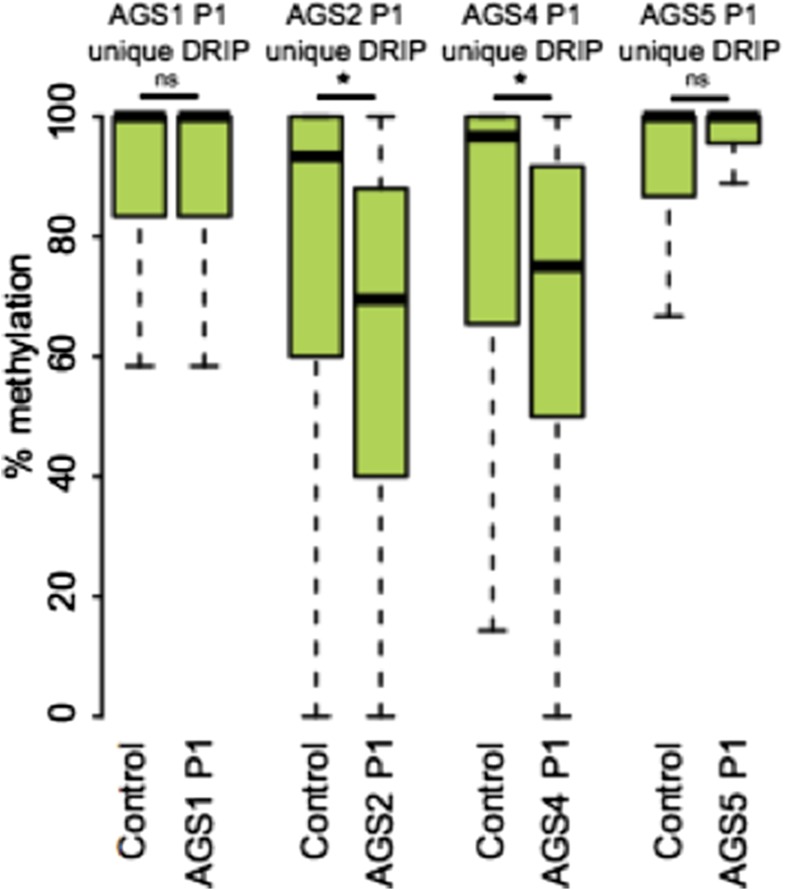
(A) Venn diagram displaying partially methylated domains (PMDs) in AGS4 and control fibroblasts compared to embryonic stem (ES) cells. (B) Location analysis of PMDs over regions marked by H3K9me3, Lamin-B1, and H3K27me3, shown as fold change relative to genome average (represented by the dotted horizontal line). Stars indicate the p-value of the deviation from the genome average. (C) Genome browser screenshot showing overlap of AGS-specific PMDs with H3K27me3. (D) Percent DRIP peaks (common or AGS-unique) in highly methylated domains (HMDs) and PMDs. The pie charts break down AGS2/4 PMDs to common and unique PMDs. (E) Genome browser screenshot showing overlap between AGS2/4 PMDs with AGS2/4 DRIP-seq peaks. (F) Percent DNA methylation of AGS-unique DRIP peaks measured in control and AGS fibroblasts (as indicated at the bottom of the graph). *p < 7.18e-11, ns = not significant.
Figure 5—figure supplement 1. HMDs and PMDs in AGS fibroblasts.
We next determined if the regions that became hypomethylated also corresponded to regions that accumulated RNA:DNA hybrids in DRIP-seq analysis. In AGS1 and AGS5 fibroblasts, which showed only moderate DNA hypomethylation and limited additional PMDs, DRIP peaks, whether common or AGS-unique, mostly landed in HMD regions (Figure 5D). This is consistent with AGS1- and AGS5-specific DRIP peaks being enriched over gene body regions (Figure 3D) since gene body regions are typically highly methylated (Lister et al., 2009). In sharp contrast, nearly half of the AGS-unique DRIP peaks in AGS2 and AGS4 fibroblasts landed on PMD regions while only 10–15% of common DRIP peaks matched to PMDs in these two samples. Furthermore, two-thirds of these PMD-contained AGS-unique DRIP peaks matched to AGS-specific PMDs (Figure 5D,E). Consistent with this, the DNA methylation levels measured over AGS1-, AGS2-, and AGS4- (but not AGS5) unique DRIP peaks were significantly lower in their own respective samples compared to the DNA methylation levels of that same regions measured in control (Figure 5F). Thus, our data show a remarkable agreement between regions that accumulate RNA:DNA hybrids in the AGS2, AGS4, and to a lesser extent AGS1 subtypes and regions that undergo DNA hypomethylation.
RNASEH2 depletion triggers DNA hypomethylation
To determine if the loss of DNA methylation observed in AGS patient cells could be directly due to AGS mutations, we focused on RNASEH2A since AGS4 fibroblasts displayed profound DNA hypomethylation (Figure 4). We employed a lentivirus-mediated CRISPR/Cas method to knockout exon 6 in RNASEH2A (RNASEH2A KO) in a HeLa cell line harboring a silent green fluorescent protein (GFP) reporter gene (HeLa-GFP) (Poleshko et al., 2010). As a control, we treated the same cell line with a CRISPR vector containing a scramble guide sequence (scramble). Effective gene knockout was validated by PCR genotyping (not shown) and by Western blots (Figure 6A). To further validate the knockout, we measured the DNA damage response in RNASEH2A KO and scramble cells using an antibody against gamma H2AX. As expected from previous reports (Reijns et al., 2012; Gunther et al., 2015), we observed higher levels of gamma H2AX in RNASEH2A KO cells (Figure 6B), indicative of an activated DNA damage signaling.
Figure 6. CRISPR/Cas-mediated RNASEH2A knockout induces DNA hypomethylation.
(A) Western blot using an anti-RNase H2A antibody shows successful gene knockout in HeLa-GFP cells. Scramble cells were generated with a CRISPR vector carrying a scrambled guide RNA sequence. Tubulin was used as a loading control. (B) Immunocytochemistry confirms elevated DNA damage response in RNASEH2A KO cells; DAPI (blue), gamma H2AX antibody (green). (C) Bright field and green fluorescent protein (GFP) microscopy images of RNASEH2A KO and scramble control cells. Knockout of the RNASEH2A gene triggers the reactivation of the silent GFP reporter in HeLa-GFP cells. (D) (Top) Western blot showing GFP expression in HeLa-GFP cells (RNASEH2A KO and scramble); (bottom) bar graph quantification of Western blot. (E) Bisulfite methylation sequencing for two different LINE-1 loci in RNASEH2A KO and scramble control cells. Black and white circles represent methylated and unmethylated CpG sites, respectively. Missing bubbles indicate that a CpG site is absent from the sequence of that particular molecule. The coordinates for each fragment analyzed are indicated at top. (F) Quantification of percent methylation at each CpG sites surveyed in panel E. p-value was calculated using a paired Wilcoxon test with the alternative hypothesis that RNASEH2A KO is less methylated than scramble.
Compared to scramble, RNASEH2A KO cells showed a clear increase in the number and brightness of GFP-positive cells (Figure 6C). GFP expression in RNASEH2A KO cells was 14-fold higher than in scramble cells, as measured by Western blot (Figure 6D), demonstrating reactivation of the GFP reporter gene. To survey DNA methylation levels in RNASEH2A KO and scramble cells, we focused on two 5′-UTR portions common to LINE-1 elements and performed DNA methylation sequencing after sodium bisulfite treatment. RNASEH2A KO cells showed significant DNA hypomethylation relative to scramble (Figure 6E,F). This suggests a direct effect of RNASEH2A deficiency in triggering DNA hypomethylation and gene reactivation at least for the loci studied here.
Discussion
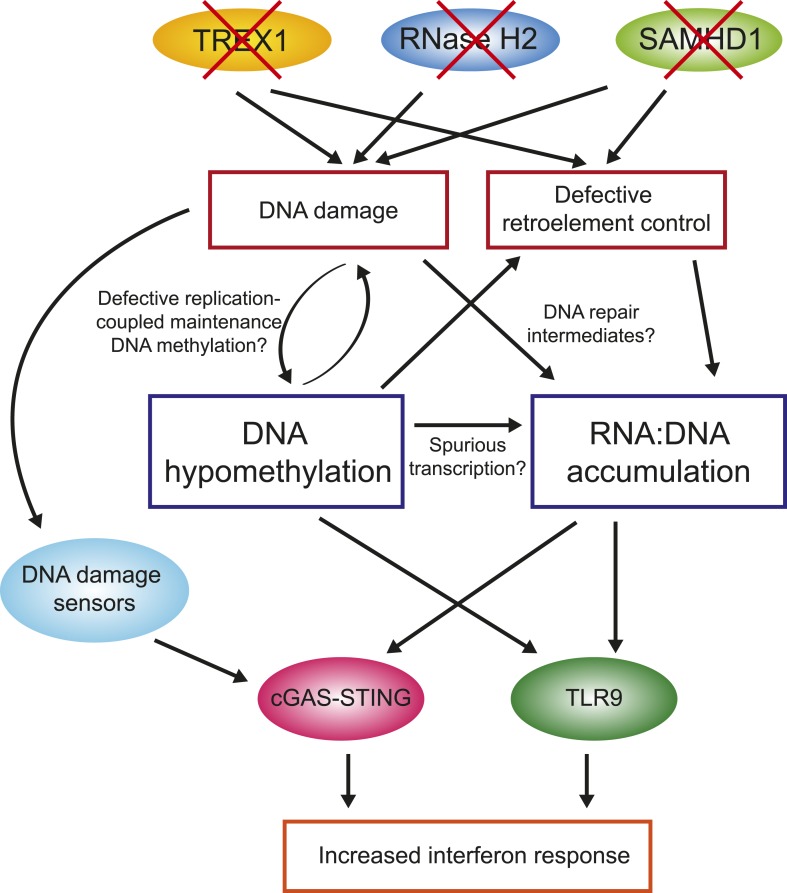
One of the key observations from this study is that primary fibroblasts from all four AGS subtypes studied here experienced global loss of DNA methylation. While additional samples encompassing other AGS mutations subtypes will need to be studied to confirm this trend, this observation nonetheless reinforces the similarities between AGS and SLE, although the DNA hypomethylation reported here is stronger than that previously reported in SLE. In AGS, DNA hypomethylation was found to affect every genomic compartment, including genic and intergenic regions, unique and repeated sequences (Figure 4). In AGS2 and AGS4, where the effect was most marked, the DNA hypomethylation led to the formation of AGS-specific PMDs in addition to those normally observed in fibroblasts (Lister et al., 2009). Similar to previously characterized PMDs in somatic tissues and cancer samples, common fibroblasts PMDs described here overlapped with inactive, late-replicating, heterochromatic regions associated with H3K9me3 and Lamin-B1 (Hawkins et al., 2010; Berman et al., 2012). Interestingly, AGS-specific PMDs, but not common PMDs, were also enriched over regions marked by H3K27me3, a mark of facultative heterochromatin associated with developmental silencing (Figure 5). This suggests that silent, condensed genomic regions, may in fact be intrinsically prone to becoming PMDs, and that this tendency is made worse by AGS2 and AGS4 mutations. Our data further indicate that RNase H2 defects directly drive these epigenetic perturbations given that we could recapitulate DNA hypomethylation at a subset of LINE-1 sequences and trigger the reactivation of a silent GFP reporter by knocking out RNASEH2A (Figure 6). While the precise mechanism responsible for DNA hypomethylation in AGS remains to be elucidated, it is worth noting that defects in TREX1, RNase H2, and SAMHD1 all trigger the DNA damage response and are associated with increased genomic instability (Yang et al., 2007; Reijns et al., 2012; Clifford et al., 2014; Kretschmer et al., 2015). RNase H2 in fact associates with the DNA replication fork through the PCNA (proliferating cell nuclear antigen) clamp (Bubeck et al., 2011) and TREX1 was found to translocate to the nucleus upon replication stress where it might be involved in processing aberrant DNA replication intermediates (Yang et al., 2007). It is therefore possible that the replication stress caused by AGS mutations indirectly results in defects in replication-coupled, DNMT1-mediated DNA methylation maintenance (Figure 7) (Law and Jacobsen, 2010). In support of this notion, a recent report shows that the PCNA clamp unloads from the lagging strand of stalled DNA replication forks (Yu et al., 2014). Given that the DNMT1 maintenance DNA methyltransferase binds to PCNA (Leonhardt et al., 1992) and requires PCNA binding for efficient activity (Schermelleh et al., 2007), it is plausible that replication stress may lead to inefficient DNA methylation maintenance, particularly in silent, compact regions of the genome such as heterochromatic regions. An analogous mechanism was proposed to account for how defective DNA replication may trigger epigenetic instability by uncoupling DNA synthesis from histone recycling (Sarkies et al., 2010). Interestingly, 5-aza-2′-deoxycytidine, a drug best known as a DNA-demethylating agent, in fact causes genome-wide DNA damage (Karpf et al., 1999; Palii et al., 2008; Orta et al., 2013). Finally, pronounced DNA hypomethylation is also known to cause genomic instability (Jackson-Grusby et al., 2001; Chen et al., 2007; Liao et al., 2015) suggesting a possible positive feedback loop that may link and amplify defects in either pathways (Figure 7). It will be interesting to further investigate the connections between DNA replication stress, DNA hypomethylation, and epigenetic instability in the context of AGS or of other diseases associated with epigenetic aberrations such as cancers.
Figure 7. Overall model by which AGS1-5 mutations may lead to DNA hypomethylation, RNA:DNA hybrid accumulation, and IFN-stimulated immune response.
See ‘Discussion’ for details.
Understanding if and how DNA hypomethylation contributes to autoimmunity in AGS and SLE is more than ever a priority. One possibility is that DNA hypomethylation results in altered gene expression profiles. However, our RNA-seq characterization only identified a modest number of up- and down-regulated genes that were enriched for ontologies reflecting the biology of the disease itself, such as immunomodulatory genes (Figure 1). There is therefore no evidence for a global alteration of transcriptional profiles as a result of the DNA hypomethylation in AGS. This is consistent with the general absence of direct connections between DNA hypomethylation and gene reactivation (Bestor et al., 2015), including in extremely hypomethylated mouse ES cells (Li et al., 1992; Tsumura et al., 2006). It remains possible, however, that DNA hypomethylation may enable the transcription of normally silent and methylated retrotransposable elements. DNA methylation is indeed the primary silencing force ensuring retrotransposon control in somatic cells (Walsh et al., 1998; Yoder and Bestor, 1998). If correct, this could lead to the reactivation of endogenous retroviral-like particles, which if detected by innate immune sensors, could in turn induce the anti-viral IFNα cytokine, a hallmark of AGS and SLE (Volkman and Stetson, 2014). Here, we showed that the DNA hypomethylation observed in AGS samples affected LTR, LINE, and to a lower extent, SINE repeated DNA elements (Figure 4). However, methylation pyrosequencing focused on human-specific LINE-1s showed that the extent of DNA hypomethylation at these elements, while significant, was minimal and that they in fact remained mostly methylated (Figure 4—figure supplement 1B). Likewise mining of our RNA-seq data sets or targeted real-time reverse-transcription PCR (RT-qPCR) assays aimed at the LINE-1 mRNA transcript failed to provide convincing evidence for LINE-1 transcriptional reactivation (data not shown). Finally, Western blots aimed at detecting the LINE-1 Orf1p protein were negative for AGS fibroblasts even though the protein was easily detected in Ntera2 control cells (data not shown). Altogether, while we can't rule out that LINE-1, or other retroelements, undergo reactivation in AGS cells, particularly in other cell types or at earlier time points during development, the evidence suggests that the DNA hypomethylation observed in AGS primary fibroblasts is not accompanied by LINE-1 reactivation.
The second major observation from this study is that all AGS subtypes tested here exhibit large accumulation of RNA:DNA hybrid species (Figure 3). RNA:DNA hybrids therefore represent candidate immunogenic nucleic acids in AGS. In contrast, ribonucleotide incorporation in genomic DNA was only observed in RNase H2-mutated samples (Figure 2). Our DRIP-seq profiling reveals that AGS2- and AGS4-specific RNA:DNA hybrids map to the intergenic space, away from traditional R-loop hotspots including genes, promoters, and terminators. AGS1- and AGS5-specific RNA:DNA hybrids were by contrast enriched over gene body regions (Figure 3D). All AGS-unique RNA:DNA hybrid peaks showed a lower overlap with GC skew, an important sequence determinant of R-loop formation (Figure 3B), suggesting that these hybrids may occur by a mechanism distinct from the well-accepted ‘thread back’ R-loop mechanism (Aguilera and Garcia-Muse, 2012). AGS-specific DRIP peaks were enriched in LINE-1 and LTR-containing sequences. This enrichment is intriguing in light of the proposed roles of SAMHD1, RNase H2, and TREX1 in the control of retroelement transposition (Stetson et al., 2008; Zhao et al., 2013a; Volkman and Stetson, 2014), although, as discussed above, we did not find convincing evidence for transposon reactivation here. Alternatively, it is possible that enrichment of AGS-specific RNA:DNA hybrids over LINE-1-rich regions simply reflects the fact that they map preferentially to intergenic and intronic regions. One possible clue as to the origin of AGS-specific DRIP peaks stems from the fact that a large fraction of such peaks in AGS2 and AGS4 samples overlap with AGS-specific PMDs in these two subtypes (Figure 5). Direct measurements in fact show that AGS-specific DRIP peaks are significantly hypomethylated in AGS1, AGS2, and AGS4 (Figure 5). Thus, the two main features observed in AGS samples, excessive RNA:DNA hybrid formation and DNA hypomethylation, often overlap, strongly supporting a link between both changes in cis. One possible explanation for this overlap is that regions experiencing DNA hypomethylation as a result of AGS mutations may undergo spurious transcription that lead to co-transcriptional R-loop formation (Figure 7). Another non-exclusive possibility is based on the proposition that PMD regions may result from DNA damage and inefficient maintenance DNA methylation (see above). It was recently described that RNA transcripts may serve as a template for the repair of double-strand DNA breaks upon hybridization to DNA via Rad52, a key member of the homologous recombination pathway. Interestingly, RNase H2 activity was shown to block the formation of these RNA:DNA hybrids (Keskin et al., 2014). It is therefore possible that some of the DNA breaks caused by AGS mutations and replication stress are repaired according to a pathway prone to RNA:DNA hybrid formation (Figure 7). While speculative, this model may also account for the lack of overlap with GC skew since those hybrids are thought to be mediated by Rad52 and not to arise co-transcriptionally. Experiments aimed at testing this proposal will be important.
One key unanswered question concerns the manner in which RNA:DNA hybrids trigger an immune response. The cGAS-STING pathway activates the interferon response upon sensing of not only cytosolic DNA (Sun et al., 2013), but also RNA:DNA hybrids (Mankan et al., 2014). Interestingly, several studies reported that multiple dsDNA break repair proteins can sense damaged DNA and induce a STING-mediated type I IFN response (Zhang et al., 2011; Kondo et al., 2013; Hartlova et al., 2015), thereby highlighting previously underappreciated links between the DNA damage response and innate immune signaling. Besides STING, it is worth noting that the endosomal receptor TLR9 has also been identified as an RNA:DNA hybrid sensor (Rigby et al., 2014) in addition to its canonical role in sensing unmethylated CpG-rich DNA (Hemmi et al., 2000). It is therefore possible that AGS mutations-induced DNA hypomethylation also contributes to the increased interferon response in AGS (Figure 7). Interestingly, treatment of colon adenocarcinoma with 5-aza-2′-deoxycytidine, which causes DNA damage and DNA demethylation, triggers a prominent interferon response (Karpf et al., 1999). Future work will be necessary to dissect the pathways by which unmethylated DNA and RNA:DNA hybrids may be sensed to trigger the innate immune response characteristic of AGS, and by extension, SLE patients.
Materials and methods
Detection of incorporated ribonucleotides by nick translation assays
Detection of incorporated ribonucleotides was performed as previously described (Hiller et al., 2012). Briefly, 200 ng of human primary fibroblasts or yeast genomic DNA (wild-type and RNASEH2-deficient) was treated with 20 nM of purified recombinant human RNase H2 (Loomis et al., 2014) or water in RNase H2 reaction buffer (50 mM Tris-HCl pH 8, 60 mM KCl, 10 mM MgCl2, 0.01% BSA (Bovine Serum Albumin), 0.01% Triton) at 37°C for 1 hr. 20 μM of unlabeled dATP, dGTP and dTTP, plus 3.7 × 105 Bq [α-32P]-dCTP (PerkinElmer, Santa Clara, CA) and 5 U of Escherichia coli DNA polymerase I (New England Biolabs, Inc., Ipswich, MA) were added and the reaction was incubated at 16°C for 30 min and was run on a 1% TAE (Tris-acetate-EDTA) agarose gel. Visualization was performed using a Storm PhosphorImager, and bands were quantified using ImageQuant (GE Healthcare, United Kingdom). Relative ribonucleotide loads were calculated by dividing the radiolabel incorporation in the RNase H2-treated sample over the untreated sample. Experiments were performed at least in triplicate.
DRIP-seq
DRIP-seq was performed on primary fibroblasts as previously described (Ginno et al., 2012). Sequencing reads were trimmed with FastqMcf (Aronesty, 2011) before mapping to the hg19 reference genome using BWA 0.6.1 (Li and Durbin, 2009). Peak calling was first performed by MACS 1.4.2 (Zhang et al., 2008) using input library as control. DRIP peaks were further assigned onto restriction fragments using BEDtools (Quinlan and Hall, 2010). For control and each AGS subtype, DRIP peaks from two independent samples were merged into one sample for downstream analysis. All overlap analysis was performed using BEDTools (Quinlan and Hall, 2010). GC skew annotation was according to low stringency SkewR peaks as previously described (Ginno et al., 2013). The enrichment or depletion of AGS-unique DRIP peaks over different genomic regions was measured as fold-change of percent base-pair overlap of AGS-unique DRIP peaks relative to common DRIP peaks. The statistical significance of enrichment or depletion of AGS-unique DRIP peaks over a specific genomic feature was measured as follows: for each AGS-unique peak, 500 shuffled peaks of equal lengths were extracted from the common peak set and their overlap with various genomic features was recorded as % length overlap. The significance of the difference in overlap between observed (AGS unique) and expected (shuffled) was calculated using empirical p-values according to the Monte Carlo method.
Whole-genome bisulfite sequencing (MethylC-seq)
2 μg of genomic DNA was sheared by sonication down to 100–500 bp in size. Sequencing libraries were constructed as described before (Schroeder et al., 2011). We then performed bisulfite treatment using EZ DNA methylation-direct kit (Zymo Research, Irvine, CA) following manufacturer's instructions. 50 ng of bisulfite-treated DNA was amplified for 15 cycles using Pfu Cx Turbo Hotstart DNA polymerase (Agilent Technologies, Santa Clara, CA), and the quality of the library was checked on a 2100 Agilent Bioanalyzer prior to sequencing on an Illumina HiSeq 2000. Sequencing reads were trimmed as described in DRIP-seq. Mapping was performed using Bismark 0.7.7 (Krueger and Andrews, 2011), and percent methylation was called using custom Perl script, combind_strand_meth.pl. C to T conversion rate was determined as the ratio of converted C in CHG and CHH context relative to the total number of CHG and CHH. CpG coverage was calculated as the average number of reads over CpG sites in the genome. Both C to T conversion rate and CpG coverage are reported in Supplementary file 1. To ensure good coverage and eliminate PCR bias, CpG sites with coverage below 4× or more than 99.9th percentile were discarded. Circos plot was generated using Circos 0.62–1 (Krzywinski et al., 2009). TSS and TTS metaplots were generated using custom Perl script, wig_to_metaplot_lowmem.pl. Methylation levels at different genomic regions were calculated BEDtools (Quinlan and Hall, 2010). Wilcoxon paired test was performed using R with the alternative hypothesis that the AGS samples are less methylated than the control. PMDs and HMDs were called using StochHMM, a HMM-based software (Lott and Korf, 2014) as previously described (Schroeder et al., 2013) with modifications: PMDs were trained using random 25-kb regions with 25–55% methylation and HMDs were trained using random 25-kb regions with 60–100% methylation. H3K27me3 and H3K9me3 data sets were obtained from ENCODE normal human dermal fibroblasts (NHDF-Ad). The lamin B1 data set was obtained from Gene Expression Omnibus (GSE49341).
Reduced representation bisulfite sequencing
Reduced representation bisulfite sequencing (RRBS) library was prepared according to the Myers lab protocol (Varley et al., 2013) prior to sequencing on an Illumina HiSeq 2000. Sequencing reads were trimmed with Trim Galore 0.3.3 (Krueger, 2014) and mapped to hg19 genome using Bismark 0.7.7 (Krueger and Andrews, 2011). TSS metaplot was generated using wig_to_metaplot_lowmem.pl.
RNA-seq
RNA-seq libraries were constructed using the Illumina TruSeq kit prior to sequencing on an Illumina HiSeq 2000. Raw reads were trimmed as before and mapped to the hg19 genome using TopHat 2.0.5 (Trapnell et al., 2009). Read counts for each gene were calculated using HTSeq. Pearson's correlation between each pair of biological replicates was calculated by comparing every gene's log2 normalized read count (Supplementary file 4). Differential gene expression was identified using DESeq (Anders and Huber, 2010) using fold change > 2 and FDR (false discovery rate) < 0.1. Pathway analysis was performed using DAVID (Huang da et al., 2009).
Data access
DRIP-seq, MethylC-seq, RRBS, and RNA-seq data are available at the Gene Expression Omnibus (GEO) database, under the accession number GSE57353. Custom Perl scripts are available at https://github.com/ywlim/Perl.
RT-qPCR
Total RNA was extracted from 80% confluent primary fibroblasts using TRI reagent (Life Technologies, Grand Island, NY) and Direct-zol RNA miniprep kit (Zymo Research). RNA was reverse transcribed to first strand cDNA using iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad, Hercules, CA). cDNA was cleaned up using DNA clean and concentrator (Zymo Research) and resuspended in 10 μl water. For RT-qPCR, 5 μl of 1:50 dilution of the cDNA was used per well in a 20 μl reaction, along with 2 μl of 10 μM primer sets and 10 μl of SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). Reactions were run in duplicate at least on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) with the following protocol: 95°C (2 min), 40 cycles of 95°C (10 s) and 60°C (15 s). Quantification was calculated using the CFX Manager software (Bio-Rad). Primer sequences are listed on Supplementary file 3.
CRISPR/Cas knockout of RNASEH2A
RNASEH2A guide RNA sequence was designed using E-CRISP (http://www.e-crisp.org/) and the corresponding oligonucleotides (top: CACCGTTGGATACTGATTATGGCTC; bottom: AAACGAGCCATAATCAGTATCCAAC) and scramble oligonucleotides (top: CACCGGCACTACCAGAGCTAACTCA; bottom: AAACTGAGTTAGCTCTGGTAGTGCC) were purchased from Life Technologies. The oligonucleotides were annealed and ligated into lentiCRISPR v2 (Addgene plasmid 52961) at the BsmBI restriction site. The resulting lentiCRISPR-RNASEH2A and lentiCRISPR-scramble plasmids were transformed into DH10B cells and purified using Qiagen Plasmid Midi Kit. To produce lentivirus carrying the CRISPR plasmids, 5 μg lentiCRISPR-RNASEH2A or lentiCRISPR-scramble, 4 μg psPAX2, and 1.5 μg of pMD2.G were transfected into HEK293T cells on a 10-cm plate using Turbofect (Life Technologies) following manufacturer's protocol. Viral supernatant was collected 48 hr later, filter sterilized with a 0.45-μm filter (Millipore, Billerica MA), and 1 ml of it was used to infect HeLa-GFP cells on a 6-well plate. 1 μg/ml puromycin was added 24 hr later. 22 days later, HeLa-GFP-RNASEH2A KO and HeLa-GFP-scramble cells were imaged using GFP microscopy. Whole-cell protein was extracted and Western blot was performed using antibodies against RNase H2A (1:1000, ProSci) or GFP (1:500, UC Davis/NIH NeuroMab Facility, Davis, CA), and tubulin (1:1000, Sigma–Aldrich, St. Louis, MO). Immunocytochemistry was performed on the same cells using antibody against γH2AX (1:150, Abcam, United Kingdom).
LINE-1 bisulfite sequencing
600 ng of genomic DNA was bisulfite converted using EZ DNA methylation-direct kit (Zymo Research) and eluted in 30 μl elution buffer. 1 μl of it was then PCR amplified using two sets of LINE-1 primers (Supplementary file 3) (95°C for 4 min, 20 cycles of 95°C, 55°C and 72°C for 30 s each, 72°C for 4 min). PCR products were gel purified and cloned into pDRIVE cloning vector using Qiagen PCR cloning kit (Qiagen, Valencia, CA) and the resulting vectors were transformed into DH10B cells. Miniprep was performed using QIAprep Spin Miniprep Kit (Qiagen) and 16 clones were sequenced for each LINE-1 locus. Percent methylation at each CG site was the ratio of unconverted (CG) vs the sum of unconverted and converted (TG) molecules.
Acknowledgements
We thank Dr Yanick Crow and Dr Gillian Rice for providing AGS patient samples, Dr Jodi Nunnari for the RNASEH2-deficient yeast cells, Dr Giovanni Capranico for the psPAX2 and pMD2.G plasmids, Dr David Segal for the HeLa cell line harboring silent GFP reporter gene, Dr John Moran for helpful discussions, and members of the Chedin lab for useful comments. This work was funded by grants from the Hartwell Foundation and the National Institutes of Health (NIH R01 GM094299) to FC. YWL was supported in part by a pre-doctoral training grant (5T32GM007377) and a Dissertation Year fellowship from UC Davis. This work used the Vincent J Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grants:
Hartwell Foundation to Frédéric Chédin.
National Institute of General Medical Sciences (NIGMS) NIH R01 GM094299 to Frédéric Chédin.
National Institute of General Medical Sciences (NIGMS) 5T32GM007377 to Yoong Wearn Lim.
Additional information
Competing interests
The authors declare that no competing interests exist.
Author contributions
YWL, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
LAS, Acquisition of data, Drafting or revising the article.
XX, Acquisition of data, Drafting or revising the article.
SRH, Analysis and interpretation of data, Drafting or revising the article.
FC, Conception and design, Analysis and interpretation of data, Drafting or revising the article.
Additional files
Genotype and high-throughput sequencing information for DRIP-seq, MethylC-seq, and RNA-seq.
Significantly up- and down-regulated genes in Aicardi–Goutières syndrome fibroblasts and their gene ontology analysis.
Primer sequences for real-time reverse-transcription PCR and bisulfite sequencing.
Pearson's correlation between each pair of RNA-seq biological replicates.
Major dataset
The following dataset was generated:
Lim YW, 2015, DRIP-seq, RNA-seq and MethylC-seq datasets, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57353, Publicly available at the NCBI Gene Expression Omnibus (Accession no: GSE57353)
References
- Absher DM, Li X, Waite LL, Gibson A, Roberts K, Edberg J, Chatham WW, Kimberly RP. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLOS Genetics. 2013;9:e1003678. doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronesty E. Command-line tools for processing biological sequencing data. 2011. [Google Scholar]
- Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestar E, Esteller M, Richardson BC. The epigenetic face of systemic lupus erythematosus. The Journal of Immunology. 2006;176:7143–7147. doi: 10.4049/jimmunol.176.12.7143. [DOI] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, van den Berg D, Laird PW. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nature Genetics. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH, Edwards JR, Boulard M. Notes on the role of dynamic DNA methylation in mammalian development. Proceedings of the National Academy of Sciences of USA. 2015;112:6796–6799. doi: 10.1073/pnas.1415301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubeck D, Reijns MA, Graham SC, Astell KR, Jones EY, Jackson AP. PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Research. 2011;39:3652–3666. doi: 10.1093/nar/gkq980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. The FEBS Journal. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hevi S, Gay F, Tsujimoto N, He T, Zhang B, Ueda Y, Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nature Genetics. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- Clifford R, Louis T, Robbe P, Ackroyd S, Burns A, Timbs AT, Wright Colopy G, Dreau H, Sigaux F, Judde JG, Rotger M, Telenti A, Lin YL, Pasero P, Maelfait J, Titsias M, Cohen DR, Henderson SJ, Ross MT, Bentley D, Hillmen P, Pettitt A, Rehwinkel J, Knight SJ, Taylor JC, Crow YJ, Benkirane M, Schuh A. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood. 2014;123:1021–1031. doi: 10.1182/blood-2013-04-490847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ. Type I interferonopathies: a novel set of inborn errors of immunity. Annals of the New York Academy of Sciences. 2011;1238:91–98. doi: 10.1111/j.1749-6632.2011.06220.x. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, Corry PC, Cowan FM, Frints SG, Klepper J, Livingston JH, Lynch SA, Massey RF, Meritet JF, Michaud JL, Ponsot G, Voit T, Lebon P, Bonthron DT, Jackson AP, Barnes DE, Lindahl T. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nature Genetics. 2006a;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, Mckeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nature Genetics. 2006b;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Human Molecular Genetics. 2009;18:R130–R136. doi: 10.1093/hmg/ddp293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lim YW, Lott PL, Korf I, Chedin F. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Research. 2013;23:1590–1600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Molecular Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- Gunther C, Kind B, Reijns MA, Berndt N, Martinez-Bueno M, Wolf C, Tungler V, Chara O, Lee YA, Hubner N, Bicknell L, Blum S, Krug C, Schmidt F, Kretschmer S, Koss S, Astell KR, Ramantani G, Bauerfeind A, Morris DL, Cunninghame Graham DS, Bubeck D, Leitch A, Ralston SH, Blackburn EA, Gahr M, Witte T, Vyse TJ, Melchers I, Mangold E, Nothen MM, Aringer M, Kuhn A, Luthke K, Unger L, Bley A, Lorenzi A, Isaacs JD, Alexopoulou D, Conrad K, Dahl A, Roers A, Alarcon-Riquelme ME, Jackson AP, Lee-Kirsch MA. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. The Journal of Clinical Investigation. 2015;125:413–424. doi: 10.1172/JCI78001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlova A, Erttmann SF, Raffi FA, Schmalz AM, Resch U, Anugula S, Lienenklaus S, Nilsson LM, Kroger A, Nilsson JA, Ek T, Weiss S, Gekara NO. DNA damage Primes the type I interferon system via the cytosolic DNA sensor STING to Promote anti-Microbial innate immunity. Immunity. 2015;42:332–343. doi: 10.1016/j.immuni.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Hiller B, Achleitner M, Glage S, Naumann R, Behrendt R, Roers A. Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. The Journal of Experimental Medicine. 2012;209:1419–1426. doi: 10.1084/jem.20120876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nature Genetics. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O'Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreno L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Research. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries MA, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics. 2011;6:593–601. doi: 10.4161/epi.6.5.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpf AR, Peterson PW, Rawlins JT, Dalley BK, Yang Q, Albertsen H, Jones DA. Inhibition of DNA methyltransferase stimulates the expression of signal transducer and activator of transcription 1, 2, and 3 genes in colon tumor cells. Proceedings of the National Academy of Sciences of USA. 1999;96:14007–14012. doi: 10.1073/pnas.96.24.14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin H, Shen Y, Huang F, Patel M, Yang T, Ashley K, Mazin AV, Storici F. Transcript-RNA-templated DNA recombination and repair. Nature. 2014;515:436–439. doi: 10.1038/nature13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, Komatsu K, Akira S, Kawai T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proceedings of the National Academy of Sciences of USA. 2013;110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer S, Wolf C, Konig N, Staroske W, Guck J, Hausler M, Luksch H, Nguyen LA, Kim B, Alexopoulou D, Dahl A, Rapp A, Cardoso MC, Shevchenko A, Lee-Kirsch MA. SAMHD1 prevents autoimmunity by maintaining genome stability. Annals of the Rheumatic Diseases. 2015;74:e17. doi: 10.1136/annrheumdis-2013-204845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F. Trim Galore [Online] 2014. Available: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ [Accessed April 28, 2014] [Google Scholar]
- Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Research. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Reviews. Genetics. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, de Silva U, Bailey SL, Witte T, Vyse TJ, Kere J, Pfeiffer C, Harvey S, Wong A, Koskenmies S, Hummel O, Rohde K, Schmidt RE, Dominiczak AF, Gahr M, Hollis T, Perrino FW, Lieberman J, Hubner N. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nature Genetics. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier HU, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-P. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Karnik R, Gu H, Ziller MJ, Clement K, Tsankov AM, Akopian V, Gifford CA, Donaghey J, Galonska C, Pop R, Reyon D, Tsai SQ, Mallard W, Joung JK, Rinn JL, Gnirke A, Meissner A. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nature Genetics. 2015;47:469–478. doi: 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YW. 2015. DRIP-seq, RNA-seq and MethylC-seq datasets. NCBI Gene Expression Omnibus GSE57353. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE57353.
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis EW, Sanz LA, Chedin F, Hagerman PJ. Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLOS Genetics. 2014;10:e1004294. doi: 10.1371/journal.pgen.1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott PC, Korf I. StochHMM: a flexible hidden Markov model tool and C++ library. Bioinformatics. 2014;30:1625–1626. doi: 10.1093/bioinformatics/btu057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankan AK, Schmidt T, Chauhan D, Goldeck M, Honing K, Gaidt M, Kubarenko AV, Andreeva L, Hopfner KP, Hornung V. Cytosolic RNA: DNA hybrids activate the cGAS-STING axis. The EMBO Journal. 2014;33:2937–2946. doi: 10.15252/embj.201488726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick Mcelhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nature Chemical Biology. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orta ML, Calderon-Montano JM, Dominguez I, Pastor N, Burgos-Moron E, Lopez-Lazaro M, Cortes F, Mateos S, Helleday T. 5-Aza-2′-deoxycytidine causes replication lesions that require Fanconi anemia-dependent homologous recombination for repair. Nucleic Acids Research. 2013;41:5827–5836. doi: 10.1093/nar/gkt270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palii SS, van Emburgh BO, Sankpal UT, Brown KD, Robertson KD. DNA methylation inhibitor 5-Aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Molecular and Cellular Biology. 2008;28:752–771. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleshko A, Einarson MB, Shalginskikh N, Zhang R, Adams PD, Skalka AM, Katz RA. Identification of a functional network of human epigenetic silencing factors. The Journal of Biological Chemistry. 2010;285:422–433. doi: 10.1074/jbc.M109.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quddus J, Johnson KJ, Gavalchin J, Amento EP, Chrisp CE, Yung RL, Richardson BC. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. The Journal of Clinical Investigation. 1993;92:38–53. doi: 10.1172/JCI116576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenscroft JC, Suri M, Rice GI, Szynkiewicz M, Crow YJ. Autosomal dominant inheritance of a heterozygous mutation in SAMHD1 causing familial chilblain lupus. American Journal of Medical Genetics. Part A. 2011;155A:235–237. doi: 10.1002/ajmg.a.33778. [DOI] [PubMed] [Google Scholar]
- Reijns MAM, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, Devenney PS, Sexton D, Grimes G, Holt IJ, Hill RE, Taylor MS, Lawson KA, Dorin JR, Jackson AP. Enzymatic removal of ribonucleotides from DNA is Essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G, Newman WG, Dean J, Patrick T, Parmar R, Flintoff K, Robins P, Harvey S, Hollis T, O'Hara A, Herrick AL, Bowden AP, Perrino FW, Lindahl T, Barnes DE, Crow YJ. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. American Journal of Human Genetics. 2007a;80:811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B, Baxter P, Benko WS, Bergmann C, Bertini E, Biancheri R, Blair EM, Blau N, Bonthron DT, Briggs T, Brueton LA, Brunner HG, Burke CJ, Carr IM, Carvalho DR, Chandler KE, Christen HJ, Corry PC, Cowan FM, Cox H, D'Arrigo S, Dean J, de Laet C, de Praeter C, Dery C, Ferrie CD, Flintoff K, Frints SG, Garcia-Cazorla A, Gener B, Goizet C, Goutieres F, Green AJ, Guet A, Hamel BC, Hayward BE, Heiberg A, Hennekam RC, Husson M, Jackson AP, Jayatunga R, Jiang YH, Kant SG, Kao A, King MD, Kingston HM, Klepper J, van der Knaap MS, Kornberg AJ, Kotzot D, Kratzer W, Lacombe D, Lagae L, Landrieu PG, Lanzi G, Leitch A, Lim MJ, Livingston JH, Lourenco CM, Lyall EG, Lynch SA, Lyons MJ, Marom D, Mcclure JP, Mcwilliam R, Melancon SB, Mewasingh LD, Moutard ML, Nischal KK, Ostergaard JR, Prendiville J, Rasmussen M, Rogers RC, Roland D, Rosser EM, Rostasy K, Roubertie A, Sanchis A, Schiffmann R, Scholl-Burgi S, Seal S, Shalev SA, Corcoles CS, Sinha GP, Soler D, Spiegel R, Stephenson JB, Tacke U, Tan TY, Till M, Tolmie JL, Tomlin P, Vagnarelli F, Valente EM, Van Coster RN, Van der Aa N, Vanderver A, Vles JS, Voit T, Wassmer E, Weschke B, Whiteford ML, Willemsen MA, Zankl A, Zuberi SM, Orcesi S, Fazzi E, Lebon P, Crow YJ. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. American Journal of Human Genetics. 2007b;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BC, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, Mcdermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nature Genetics. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Del Toro Duany Y, Jenkinson EM, Forte GM, Anderson BH, Ariaudo G, Bader-Meunier B, Baildam EM, Battini R, Beresford MW, Casarano M, Chouchane M, Cimaz R, Collins AE, Cordeiro NJ, Dale RC, Davidson JE, de Waele L, Desguerre I, Faivre L, Fazzi E, Isidor B, Lagae L, Latchman AR, Lebon P, Li C, Livingston JH, Lourenco CM, Mancardi MM, Masurel-Paulet A, Mcinnes IB, Menezes MP, Mignot C, O'Sullivan J, Orcesi S, Picco PP, Riva E, Robinson RA, Rodriguez D, Salvatici E, Scott C, Szybowska M, Tolmie JL, Vanderver A, Vanhulle C, Vieira JP, Webb K, Whitney RN, Williams SG, Wolfe LA, Zuberi SM, Hur S, Crow YJ. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nature Genetics. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Forte GM, Szynkiewicz M, Chase DS, Aeby A, Abdel-Hamid MS, Ackroyd S, Allcock R, Bailey KM, Balottin U, Barnerias C, Bernard G, Bodemer C, Botella MP, Cereda C, Chandler KE, Dabydeen L, Dale RC, de Laet C, de Goede CG, Del Toro M, Effat L, Enamorado NN, Fazzi E, Gener B, Haldre M, Lin JP, Livingston JH, Lourenco CM, Marques W, jr, Oades P, Peterson P, Rasmussen M, Roubertie A, Schmidt JL, Shalev SA, Simon R, Spiegel R, Swoboda KJ, Temtamy SA, Vassallo G, Vilain CN, Vogt J, Wermenbol V, Whitehouse WP, Soler D, Olivieri I, Orcesi S, Aglan MS, Zaki MS, Abdel-Salam GM, Vanderver A, Kisand K, Rozenberg F, Lebon P, Crow YJ. Assessment of interferon-related biomarkers in Aicardi-Goutieres syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurology. 2013;12:1159–1169. doi: 10.1016/S1474-4422(13)70258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, Jenkinson EM, Bacino CA, Battini R, Bertini E, Brogan PA, Brueton LA, Carpanelli M, de Laet C, de Lonlay P, Del Toro M, Desguerre I, Fazzi E, Garcia-Cazorla A, Heiberg A, Kawaguchi M, Kumar R, Lin JP, Lourenco CM, Male AM, Marques W, jr, Mignot C, Olivieri I, Orcesi S, Prabhakar P, Rasmussen M, Robinson RA, Rozenberg F, Schmidt JL, Steindl K, Tan TY, van der Merwe WG, Vanderver A, Vassallo G, Wakeling EL, Wassmer E, Whittaker E, Livingston JH, Lebon P, Suzuki T, Mclaughlin PJ, Keegan LP, O'Connell MA, Lovell SC, Crow YJ. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nature Genetics. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B. Effect of an inhibitor of DNA methylation on T cells. II. 5-Azacytidine induces self-reactivity in antigen-specific T4+ cells. Human Immunology. 1986;17:456–470. doi: 10.1016/0198-8859(86)90304-6. [DOI] [PubMed] [Google Scholar]
- Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33:1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- Rigby RE, Webb LM, Mackenzie KJ, Li Y, Leitch A, Reijns MA, Lundie RJ, Revuelta A, Davidson DJ, Diebold S, Modis Y, Macdonald AS, Jackson AP. RNA: DNA hybrids are a novel molecular pattern sensed by TLR9. The EMBO Journal. 2014;33:542–558. doi: 10.1002/embj.201386117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkies P, Reams C, Simpson LJ, Sale JE. Epigenetic instability due to defective replication of structured DNA. Molecular Cell. 2010;40:703–713. doi: 10.1016/j.molcel.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Haemmer A, Spada F, Rosing N, Meilinger D, Rothbauer U, Cardoso MC, Leonhardt H. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Research. 2007;35:4301–4312. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DI, Blair JD, Lott P, Yu HO, Hong D, Crary F, Ashwood P, Walker C, Korf I, Robinson WP, Lasalle JM. The human placenta methylome. Proceedings of the National Academy of Sciences of USA. 2013;110:6037–6042. doi: 10.1073/pnas.1215145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DI, Lott P, Korf I, Lasalle JM. Large-scale methylation domains mark a functional subset of neuronally expressed genes. Genome Research. 2011;21:1583–1591. doi: 10.1101/gr.119131.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, Nakayama J, Okano M. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes To Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Tuller T, Atar S, Ruppin E, Gurevich M, Achiron A. Common and specific signatures of gene expression and protein-protein interactions in autoimmune diseases. Genes and Immunity. 2013;14:67–82. doi: 10.1038/gene.2012.55. [DOI] [PubMed] [Google Scholar]
- Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, Absher DM, Wold BJ, Myers RM. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Research. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman HE, Stetson DB. The enemy within: endogenous retroelements and autoimmune disease. Nature Immunology. 2014;15:415–422. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature Genetics. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Yang YG, Lindahl T, Barnes DE. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Bestor TH. A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Human Molecular Genetics. 1998;7:279–284. doi: 10.1093/hmg/7.2.279. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends in Genetics. 1997;13:335–340. doi: 10.1016/S0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Yu C, Gan H, Han J, Zhou ZX, Jia S, Chabes A, Farrugia G, Ordog T, Zhang Z. Strand-specific analysis shows protein binding at replication Forks and PCNA unloading from lagging strands when Forks Stall. Molecular Cell. 2014;56:551–563. doi: 10.1016/j.molcel.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung RL, Quddus J, Chrisp CE, Johnson KJ, Richardson BC. Mechanism of drug-induced lupus. I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. The Journal of Immunology. 1995;154:3025–3035. [PubMed] [Google Scholar]
- Zhang X, Brann TW, Zhou M, Yang J, Oguariri RM, Lidie KB, Imamichi H, Huang DW, Lempicki RA, Baseler MW, Veenstra TD, Young HA, Lane HC, Imamichi T. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. The Journal of Immunology. 2011;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao M, Sawalha AH, Richardson B, Lu Q. Impaired DNA methylation and its mechanisms in CD4(+)T cells of systemic lupus erythematosus. Journal of Autoimmunity. 2013;41:92–99. doi: 10.1016/j.jaut.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Zhao K, Du J, Han X, Goodier JL, Li P, Zhou X, Wei W, Evans SL, Li L, Zhang W, Cheung LE, Wang G, Kazazian HH, Jr, Yu XF. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell Reports. 2013a;4:1108–1115. doi: 10.1016/j.celrep.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Zhou H, Su SB. A critical role for interleukin-1beta in the progression of autoimmune diseases. International Immunopharmacology. 2013b;17:658–669. doi: 10.1016/j.intimp.2013.08.012. [DOI] [PubMed] [Google Scholar]