Abstract
The evolutionarily conserved Wnt/β-catenin signaling pathway plays a fundamental role during metazoan development, regulating numerous processes including cell fate specification, cell migration, and stem cell renewal. Wnt ligand binding leads to stabilization of the transcriptional effector β-catenin and upregulation of target gene expression to mediate a cellular response. During larval development of the nematode Caenorhabditis elegans, Wnt/β-catenin pathways act in fate specification of two hypodermal cell types, the ventral vulval precursor cells (VPCs) and the lateral seam cells. Because little is known about targets of the Wnt signaling pathways acting during larval VPC and seam cell differentiation, we sought to identify genes regulated by Wnt signaling in these two hypodermal cell types. We conditionally activated Wnt signaling in larval animals and performed cell type–specific "mRNA tagging" to enrich for VPC and seam cell–specific mRNAs, and then used microarray analysis to examine gene expression compared to control animals. Two hundred thirty-nine genes activated in response to Wnt signaling were identified, and we characterized 50 genes further. The majority of these genes are expressed in seam and/or vulval lineages during normal development, and reduction of function for nine genes caused defects in the proper division, fate specification, fate execution, or differentiation of seam cells and vulval cells. Therefore, the combination of these techniques was successful at identifying potential cell type–specific Wnt pathway target genes from a small number of cells and at increasing our knowledge of the specification and behavior of these C. elegans larval hypodermal cells.
Keywords: Wnt signaling, mRNA tagging, seam cells, vulval precursor cells, C. elegans, differentiation
The proper development of metazoans is governed by changes in the transcriptional profile of various tissues and cell types over time, and several evolutionarily conserved signaling pathways function to stimulate the transcriptional changes necessary for changes in cell fate and behavior during development. The Wnt/β-catenin signaling pathway is one highly conserved extracellular signaling pathway widely used during metazoan development; however, Wnt signaling also acts in the maintenance of tissue homeostasis via regulation of stem cell renewal (Cadigan and Peifer 2009; Schuijers and Clevers 2012). Due to its widespread use, it is not surprising that the Wnt pathway is aberrantly activated in many diseases including cancer (Polakis 2012; Clevers and Nusse 2012). The Wnt/β-catenin pathway switches between active and inactive states based on the stability of a key transcription factor β-catenin (Cadigan 2012; Clevers and Nusse 2012) . In the absence of the Wnt ligand, cytoplasmic β-catenin is bound by the components of a “destruction complex” comprising Axin, kinases CK1α and GSK3, and the adenomatous polyposis coli (APC) protein, which phosphorylates β-catenin and marks it for proteosomal degradation. The binding of the Wnt ligand to its receptor results in the disruption of the destruction complex, thus stabilizing β-catenin and allowing its entry into the nucleus where it binds to a member of the TCF/LEF transcription factor family to activate expression of Wnt pathway target genes. Microarray and ChIP-based techniques have been used in different vertebrate model systems and tissue culture to identify a spectrum of target genes activated and inhibited by Wnt signaling, providing an understanding of cellular responses as they occur in vivo during normal development and in the disease state (see Vlad et al. 2008; Nusse 2013).
As in other organisms, Wnt signaling is also utilized many times during development of the nematode C. elegans, where it regulates cell fate specification, polarity, and migration in the embryo and larva. Unlike other organisms, there are two β-catenin–dependent Wnt pathways in C. elegans, the Wnt/BAR-1 canonical (WBC) pathway and the Wnt/β-catenin asymmetry pathway (for review, see Jackson and Eisenmann 2012; Sawa and Korswagen 2013). The mechanism of the WBC pathway, which functions in several larval developmental decisions, is similar to the well-described vertebrate Wnt/β-catenin signaling pathway. However, the WBA pathway, which functions in both the embryo and larva in asymmetric divisions requiring the adoption of distinct daughter fates, has a mechanism that may be unique to C. elegans. In this pathway, the asymmetric distribution of key pathway components in the mother and daughter cells, along with the simultaneous activation of a pathway involving TAK-1 and NLK homologs, leads to Wnt pathway activation in one daughter of an asymmetric division but not the other, resulting in adoption of distinct daughter cell fates. Our laboratory and others have shown that Wnt signaling regulates the development of two cell types that are part of the nematode hypodermis, the single-cell-layer outer covering of the animal (Chisholm and Hsiao 2012). The WBC pathway plays a role in the specification of the larval vulval precursor cells (VPCs), which divide to generate the C. elegans adult vulva, whereas the WBA pathway functions in the division and specification of specialized epithelial cells called seam cells.
The VPCs are six equipotent cells that are specified during the L2/L3 molt to adopt the 1°, 2°, or 3° cell fate based on integration of information from Ras, Notch, and Wnt signaling pathways (Sternberg 2005). Three VPCs adopt 1° and 2° fates and divide to generate cells that form the vulval opening. Reduction of WBC pathway activity causes fewer than three VPCs to adopt vulval fates, resulting in vulvaless (Vul), protruding vulva (Pvl), or egg laying defective (Egl) phenotypes (Eisenmann et al. 1998; Gleason et al. 2002, 2006). Conversely, overactivation of the WBC pathway causes more than three VPCs to adopt vulval fates, resulting in a multivulva (Muv) phenotype (Gleason et al. 2002; Gleason et al. 2006). Except for the Hox gene lin-39, no targets of the WBC pathway in VPC fate specification have been identified (Eisenmann et al. 1998)
The seam cells are lateral epithelial cells arranged in a single row running the length of the animal on both sides (Joshi et al. 2010; Chisholm and Hsiao 2012). Ten seam cells per side (H0–H2, V1–V6, and T) are born during embryogenesis but do not divide. Later, during each of the four larval stages (L1–L4), most seam cells divide asymmetrically in a stem cell–like manner to give rise to an anterior hypodermal daughter that differentiates and fuses with the surrounding skin and a posterior daughter that retains the seam cell fate and the ability to divide further (Joshi et al. 2010). During the second larval stage, six seam cells also divide symmetrically to generate two seam cell daughters, raising the number to 16 seam cells per side. After their final division in the fourth larval stage, the seam cells exit the cell cycle, differentiate, and fuse to form a long, single-cell syncytium that secretes a cuticular structure called the alae (Joshi et al. 2010). The asymmetric division of larval seam cells is regulated by the WBA pathway (Herman and Horvitz 1994; Whangbo et al. 2000; Herman 2001; Takeshita and Sawa 2005; Goldstein et al. 2006; Mizumoto and Sawa 2007; Huang et al. 2009c; Gleason and Eisenmann 2010; Ren and Zhang 2010; Banerjee et al. 2010). Reduction of the WBA pathway activity during larval life causes seam-fated daughters of a seam cell division to adopt hypodermal fates, resulting in fewer adult seam cells, whereas an increase in the WBA pathway activity causes hypodermal-fated daughters to adopt the seam cell fate, resulting in too many seam cells (Huang et al. 2009c; Gleason and Eisenmann 2010; Ren and Zhang 2010; Banerjee et al. 2010). We have recently shown that GATA factor–encoding genes egl-18 and elt-6 function as downstream targets of the WBA pathway required to retain the posterior seam cell daughter fate during these larval asymmetric seam cell divisions (Gorrepati et al. 2013).
Although the mechanisms of the two C. elegans β-catenin-dependent Wnt pathways differ, the common outcome is the regulation of target gene expression by a nuclear complex between a β-catenin (BAR-1 or SYS-1) and the sole C. elegans TCF homolog, POP-1. A key question is how formation of a β-catenin/POP-1 complex leads to distinct cellular responses in different tissues and cell types, such as the VPCs and seam cells. As a first step in addressing this question, the target genes activated by Wnt signaling in distinct processes must be identified. Previously, molecular genetic analyses have identified a few transcription factors functioning downstream of Wnt signaling in different C. elegans Wnt-mediated processes (Eisenmann et al. 1998; Jiang and Sternberg 1998; Maloof et al. 1999; Streit et al. 2002; Maduro et al. 2005; Shetty et al. 2005; Arata et al. 2006; Lam et al. 2006; Bertrand and Hobert 2009; Gorrepati et al. 2013). However, attempts to identify the broad range of Wnt signaling targets governing a particular process at a specific time in development using techniques such as microarray analysis or RNAseq have only been recently undertaken in C. elegans. Recently, our laboratory utilized global activation of Wnt signaling by a dominant activating BAR-1/β-catenin and microarray analysis to identify more than 100 potential Wnt pathway targets functioning in larval life, and revealed a previously unknown role for Wnt signaling in adult cuticle development (Jackson et al. 2014). Taking the opposite approach, van der Bent et al. (2014) used microarray analysis to identify more than 1000 genes differentially regulated between wild-type and bar-1 null mutant strains. In addition to these genomic methods, a bioinformatic search utilizing an extended POP-1/TCF binding site also was successful at identifying novel Wnt responsive genes (Bhambhani et al. 2014). However, all of these methods analyzed target genes at the level of the whole animal and may have missed cell type–specific targets of Wnt signaling functioning in only a few cells, such as the VPCs (six cells) and seam cells (32 cells).
The control of cell fate specification in the VPCs and the asymmetric division of the seam cells by Wnt signaling made these cell types attractive candidates for attempting to identify cell type–specific Wnt targets in the worm. To achieve this, we used heat shock inducible dominant variants of BAR-1/β-catenin and POP-1/TCF to activate or inhibit Wnt signaling, respectively, at a specific time point in larval development (Gleason et al. 2002; Korswagen et al. 2002; Gleason and Eisenmann 2010; Jackson et al. 2014). To selectively enrich for mRNAs expressed in seam cells and VPCs, we utilized the "mRNA tagging method," which has been successfully used in combination with microarray analysis to catalog gene expression patterns from small numbers of cells (Roy et al. 2002; Kunitomo et al. 2005; Pauli et al. 2006; Von Stetina et al. 2007a,b; Takayama et al. 2010). This novel combination of differential activation of Wnt signaling with the "mRNA tagging method" and microarray analysis allowed us to identify 239 Wnt responsive genes differentially regulated by Wnt signaling in the seam cells or VPCs. Preliminary characterization of 50 known and novel genes was pursued to validate their expression in these cell types and to determine any possible role in their development or behavior. We found seven Wnt target genes that affect seam syncytium and alae formation both as a consequence of an effect on total seam cell number (cki-1, cdk-4, kin-10, mlt-11, nhr-23) and also independent of it (bus-8, K10D6.2). Two other Wnt responsive genes (pak-1 and lin-1) were shown to function in VPC fate specification. This work validates the use of these different techniques in combination for the identification of cell type–specific signaling pathway targets in vivo in C. elegans and extends our previous work on the role of Wnt signaling in the regulation of seam cell and VPC fate specification by identifying new Wnt targets that control various aspects of the behavior of these cells during larval development.
Materials and Methods
Strains and alleles
Bristol N2 variety of C. elegans was used as the wild-type strain. All experiments were performed at 20° unless otherwise noted. The genes and alleles used in this work are described in Wormbase (Harris et al. 2010; Yook et al. 2012).
LGI: kin-10(ok1751); LGII: rrf-3(pk1426); LGIII: unc-119(ed3); LGIV: dpy-20(e1282), unc-30(e191); LGX: pak-1(ok448), osm-11(rt142)
Strains used:
deIs10: dpy-20(e1282); unc-30(e191); Is[bar-1e::flag3x::pab-1; unc-30(+); ajm-1::gfp]
huIs1: dpy-20(e1282); unc-30(e191); Is[hsp-16.2::delNTbar-1; dpy-20(+)] (Gleason et al. 2002)
deIs26: dpy-20(e1282); unc-30(e191); Is[hsp-16.2::delNTpop-1; dpy-20(+)]
sIs12963: [F09D12.1p::gfp]; dpy-5(e907) (McKay et al. 2003). This strain is referred to as grd-10::gfp in this work.
scm::gfp: unc-119(e2498) III; wIs51[scmp::gfp; unc-119(+)] (Gleason and Eisenmann 2010)
Generation of strains for mRNA tagging
The bar-1e enhancer element fused to the minimal pes-10 promoter was PCR amplified from the VPCe::pes-10::GFP vector (Natarajan et al. 2004) with primers containing KpnI restriction sites. This 1131-bp PCR product was cloned upstream of Flag3X-PAB-1 in the pSV15 vector (Von Stetina et al. 2007a). The resulting cloned vector pBJ1000 (50 ng/µl) was coinjected with transformation markers unc-30(+) (100 ng/µl of pSC11) and ajm-1::gfp (50 ng/µl of pJS191) into dpy-20(e1282) unc-30(e191); huIs1 worms using standard injection protocols (Mello and Fire 1995), and one line was selected for integration using gamma irradiation (3850 RADs). A transgenic line showing more than 95% transmission of the integrated array was backcrossed six times to generate dpy-20(e1282) unc-30(e191); deIs10 [VPCe::flag3x::pab-1;unc-30(+); ajm-1::gfp]. Transgenic worms bearing hs::ΔNTpop-1 were generated by injecting pHCK28 (50 ng/µl) (Korswagen et al. 2000) and the coinjection marker dpy-20(+) (100 ng/µl of pMH86) into dpy-20(e1362) unc-30(e191) worms as above. Array integration into the genome was performed in the same manner as described above to generate dpy-20(e1282) unc-30(e191); deIs26 [hs::ΔNTpop-1; dpy-20(+)]. The deIs10 worms were crossed with huIs1 and deIs26 animals to generate the experimental strains dpy-20(e1282) unc-30(e191); deIs10; huIs1 and dpy-20(e1282) unc-30(e191); deIs10; deIs26, respectively, which were used for mRNA tagging and microarray analysis.
Heat shock and mRNA tagging protocol
Mixed-stage embryos from gravid adults of experimental strains (deIs10; huIs1 and deIs10; deIs26) and control strains (N2 and deIs10; scm::gfp) were hatched overnight in 50 ml of M9. The synchronized L1s obtained were grown for 26 hr (L2/L3 molt) at 20°, heat shocked at 37° for 30 min, and then recovered for 1 hr at 20°. mRNA tagging was performed as previously described with a few modifications (Von Stetina et al. 2007b). Briefly, heat shocked and recovered animals were treated with 20% paraformaldehyde for 1 hr at 4°, sonicated utilizing a Branson 450 Sonifier (2 W output, 20% duty cycle, 6 pulses for 6 rounds), homogenized for 5 min using a 7-ml Dounce homogenizer, and centrifuged at 11,750 rpm for 20 min at 4° to pellet worm debris; 1 ml of worm lysate supplemented with 10 μl rRNAsin (Promega #N-2515) and 8 μl 200 mM ribonucleoside vanadyl complex (Sigma #R-3380) was added to 100 μl prewashed anti-FLAG beads (Sigma #F-2426) and rocked at 4° for 12–16 hr. Beads were washed six times in low-salt homogenization buffer (25 mM NaCl, 20 mM HEPES, 1 mM EGTA, 1 mM EDTA, 0.6 mg/ml Heparin, and 10% glycerol) at 4° with rocking in between. Reversal of crosslinked FLAG::PAB-1/mRNA complexes was accomplished by adding 125 μl of elution buffer (50 mM Tris, 10 mM EDTA and 1.3% SDS) and 10 μl rRNAsin and rolling at 65° for 30 min; beads were spun down to obtain the eluted mRNA. After a second elution, the eluates were pooled and mRNA was extracted using the Trizol method.
mRNA amplification and microarray analysis
Biological triplicates of mRNA samples obtained by mRNA tagging and coimmunoprecipitation from deIs10; huIs1, deIs10; deIs26, deIs10;scm::gfp, and N2 were subjected to DNase treatment. A total of 50 ng of mRNA was used as the starting sample for the RNA amplification process using the WT-Ovation Pico System (NuGEN #3300) to generate 3–6 μg of amplified cDNA. Fragmentation and biotin labeling of the amplified cDNA samples were performed out using the FLOvation cDNA Biotin Module V2 kit (NuGEN #4200). The labeled cDNA was hybridized to Affymetrix C. elegans GeneChip array (Affymetrix #900384) containing 22,500 probe sets targeting 21,150 unique C. elegans transcripts (Affymetrix 2004). The hybridization intensities obtained from each experiment were normalized using the Robust Multichip Average (RMA) analysis, and the detection calls and directional change calls were given based on Affymetrix’s MicroArray Suite 5 (MAS5) software analysis.
Putative Wnt pathway targets were selected based on the following criteria: (1) an average fold change of 1.5-fold or more in deIs10; huIs1 (Wnt pathway overactivation) compared to control (N2); (2) present and increased calls in at least two of three biological replicates; (3) ANOVA P ≤0.05; and (4) ratio of average fold change ≥1.5-fold between Wnt pathway overactivated (deIs10; huIs1) and underactivated (deIs10; deIs26) conditions. These criteria identified 238 Wnt targets to which kin-10 was added, although its P was more than 0.05 (P = 0.12), increasing the total list of identified targets to 239. The statistical significance of the overlap between our gene lists and those identified by others (Table 1) was calculated using a web-based hypergeometric distribution calculator designed by Jim Lund (University of Kentucky, Lexington, KY, USA) (http://nemates.org/MA/progs/overlap_stats.html).
Table 1. Conditional Wnt activation coupled with mRNA tagging enriches for cell type–specific transcripts and Wnt-regulated genes.
| Cell/Tissue Type | Reference | No. of Genes | No. of Common Genes | R-factor | Probability |
|---|---|---|---|---|---|
| Larval seam cells | Wormbase release 220 | 392 | 24 | 5.7 | 8.8×10−12 |
| Larval and adult vulval cells | Wormbase release 220 | 305 | 8 | 2.4 | 0.02 |
| Embryonic muscle | Fox et al. 2007 | 389 | 4 | 0.9 | 0.41 |
| Embryonic/larval body wall muscle | Spencer et al. 2011 | 123 | 0 | 0 | 0.26 |
| Embryonic/larval A class neurons | Spencer et al. 2011 | 108 | 0 | 0 | 0.31 |
| Embryonic/larval pan-neural | Spencer et al. 2011 | 104 | 0 | 0.9 | 0.31 |
| Embryonic/larval intestine | Spencer et al. 2011 | 298 | 0 | 0 | 0.04 |
| Global Wnt targets | Jackson et al. 2014 | 110 | 24 | 20.1 | 9.6×10−25 |
| bar-1(ga80) downregulated targets | van der Bent et al. 2014 | 710 | 25 | 3.3 | 2.3×10−7 |
| bar-1(ga80) upregulated targets | van der Bent et al. 2014 | 425 | 10 | 2.2 | 0.01 |
Shown is a comparison of our list of 239 seam/VPC Wnt-regulated genes identified by conditional Wnt pathway activation and mRNA tagging to sets of genes known to be expressed in the seam cells, vulval cells, and other embryonic and larval tissues, and to the list of Wnt-regulated genes identified previously using these reagents without tissue-specific transcript enrichment (global Wnt targets). The statistical significance of the overlap between two lists was calculated based on a hypergeometric distribution. A representation (R) factor >1 indicates more overlap than expected between two independent groups of genes, whereas an R factor <1 indicates less overlap than expected (Lund et al. 2002). Genes identified in this work that are known to be expressed in the seam cells or vulval cells are indicated in Table S1. Genes in common between this work and that of Jackson et al. (2014) are indicated in Table S2. Genes in common between this work and that of van der Bent et al. (2014) are indicated in Table S3.
qRT-PCR analysis
For qRT-PCR analysis, 1–1.5 μl cDNA (50 ng/μl) was mixed with 200 nM gene-specific forward and reverse primers and 12.5 μl of SYBR Green supermix in a final volume of 25 μl. PCR was performed on a BioRad iCycleriQ multicolor real-time PCR detection system (technical triplicates of each reaction were performed). The fold change of target genes was determined from the observed Ct (cycle threshold) values and calculated using the Pfaffl equation, (E target)ΔCt target (control-treated)/(E reference)ΔCt ref (control-treated) (Pfaffl 2001), with a reference gene of gpd-2. A minimum of three biological replicates of control and experimental samples were analyzed.
Transcriptional reporter construct
To create promoter::yfp transcriptional reporter constructs, the intergenic region between the ATG of the gene of interest and the next upstream gene was PCR amplified with gene-specific primers containing Gateway attachment sites and cloned into pBJ101, which contains a 2XNLS::YFP coding sequence and the unc-119(+) minigene (Jackson et al. 2014). Oligonucleotide sequences are provided in Supporting Information, Table S7. Each promoter::yfp construct (150 ng/µl) was coinjected with ajm-1::gfp (pJS191; 50 ng/µl) into unc-119(ed3) worms. Transgenic worms were detected based on the ability to move and expression of ajm-1::gfp. At least two independent lines were analyzed for each gene.
RNA interference
Sequence-verified RNAi clones used in this work were obtained from the Ahringer library (Kamath and Ahringer 2003) and the Vidal library (Rual et al. 2004a) (gifts from Dr. Iqbal Hamza of UMCP and Dr. Mark Wilson of JHU). RNAi on L1 and P0 animals was performed as described (Kamat et al. 2001; Liu et al. 2014). Worms of the appropriate age were analyzed on a Zeiss Axioplan 2 equipped with Nikon DXM1200 digital camera for imaging.
Immunostaining
Immunostaining of worms was performed as described (Duerr 2006). Anti-FLAG mouse monoclonal antibody M2 (Sigma #F3165) was used at 1/1000 for PAB-1 detection and MH27 mouse monoclonal antibody (Developmental Studies Hybridoma Bank) at 1/100 for detection of the junctional molecule AJM-1. Stained worms were mounted and were analyzed on a Zeiss Axioplan 2 equipped with a Nikon DXM1200 digital camera for imaging.
Results and Discussion
Conditional Wnt pathway activation, mRNA tagging, and microarray analysis identify 239 putative Wnt pathway target genes in the seam cells and VPCs
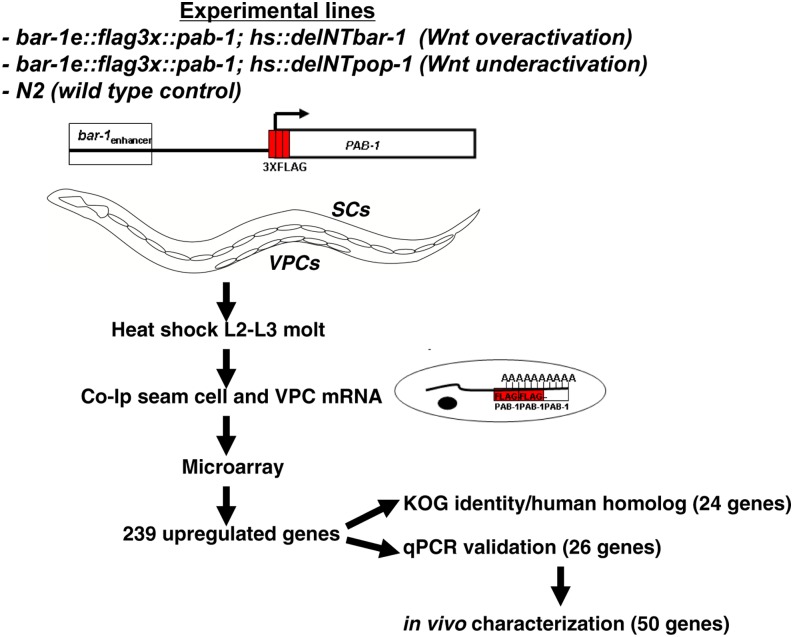
To identify genes regulated by Wnt signaling in the larval seam and vulval precursor cells, three methods were used: (1) conditional Wnt pathway activation or inhibition at a defined time during larval development; (2) mRNA tagging to enrich for seam and VPC transcripts; and (3) microarray analysis to identify differentially regulated genes (Figure 1). We conditionally activated Wnt signaling at the time of the L2/L3 molt using hs::delNTbar-1, which encodes a heat shock inducible variant of BAR-1/β-catenin lacking the amino terminal region required for degradation in the absence of signaling (Gleason and Eisenmann 2010; Jackson et al. 2014). Expression of this truncated variant at the L2/L3 molt causes an activated Wnt pathway phenotype in both the VPCs (Gleason et al. 2002) and seam cells (Gleason and Eisenmann 2010), and we previously used this reagent to identify Wnt signaling target genes by microarray analysis (Jackson et al. 2014). We conditionally inhibited Wnt signaling via expression of a heat shock inducible variant of POP-1/TCF that lacks the N-terminal β-catenin binding domain required for activation of target gene expression (hs::delNTpop-1). Expression of this dominant negative ΔNTPOP-1 has been shown to cause phenotypes consistent with reduced Wnt signaling in both neurons and seam cells during larval development (Korswagen et al. 2000; Gleason and Eisenmann 2010), and to reduce expression of Wnt target genes (Jackson et al. 2014). We activated or inhibited Wnt signaling by growing experimental (hs::delNTbar-1, hs::delNTpop-1) and control strains in triplicate to the L2/L3 molt, subjecting them to a single heat shock for 30 min at 37°, then collecting mRNA for analysis 1 hr after heat shock (Jackson et al. 2014).
Figure 1.
Conditional Wnt pathway activation, mRNA tagging, and microarray analysis to identify seam and VPC-specific Wnt targets. Outlined here is the procedure used to conditionally activate the Wnt pathway in L2/L3 worms by heat shock and enrich for transcripts expressed in the seam cells and VPCs using the mRNA tagging method. An SC+VPC-specific enhancer element from the bar-1 gene (bar-1e) was used to drive FLAG-tagged poly-A tail binding protein (PAB-1) in the seam cells and VPCs. Crosslinked FLAG-PAB-1-mRNA complexes were immunoprecipitated, enriching for transcripts from those cell types. Transcript profiles after heat shock from the three indicated strains were determined using a C. elegans genomic DNA microarray, and transcripts upregulated in response to Wnt pathway activation were identified. A subset of genes was subject to further in vivo characterization based on criteria of their identity or independent qPCR validation.
In our previous analysis of the transcriptional response to activated Wnt signaling in larvae, we characterized gene expression from the entire animal (Jackson et al. 2014). To enrich for transcripts from the small number of larval cells of interest to us, the seam cells (32 cells) and VPCs (6 cells), we used the "mRNA tagging" method (Roy et al. 2002). In this technique, a FLAG tagged version of the polyA binding protein PAB-1 is expressed using a cell type–specific enhancer and mRNAs are crosslinked to FLAG-PAB-1 in vivo and coimmunoprecipitated in vitro (Figure 1). The mRNA tagging method has been successfully used to generate inventories of genes expressed in various C. elegans tissues (Roy et al. 2002; Kunitomo et al. 2005; Pauli et al. 2006; Von Stetina et al. 2007a; Von Stetina et al. 2007b; Takayama et al. 2010). FLAG-PAB-1 expression in our experiments was under the control of an enhancer from the bar-1 gene that is active in larval seam cells and VPCs (bar-1e) (Natarajan et al. 2004) (Figure S1). mRNA bound to FLAG-PAB-1 was coimmunoprecipitated from Wnt activated and Wnt inhibited strains, and these seam and VPC enriched transcript pools were used for microarray analysis on Affymetrix C. elegans genome arrays as previously described (Jackson et al. 2014). Gene expression from the experimental strains was compared to that in control strains. After the application of several selection criteria (see Materials and Methods) we identified 239 genes upregulated in response to overexpression of the dominant BAR-1 protein, corresponding to ∼1% of the unique C. elegans transcripts represented on the genome array (Table S1). We refer to these genes as larval "Wnt pathway-regulated" or "Wnt target" genes.
If these genes are potential Wnt targets from the seam and vulval cells, we would predict overlap with genes known to be expressed or function in the larval seam cells or VPCs, and known or putative Wnt pathway target genes. As shown in Table 1, genes expressed in the larval seam cells were over-represented in our list of cell type–specific Wnt pathway–regulated genes (R-factor = 5.7; P = 8.8×10−12). Genes expressed in the vulval cells were also over-represented (P = 0.02), whereas genes expressed in embryonic and larval body wall muscle cells, neurons, and intestinal cells (Spencer et al. 2011) were not (R factor <1 in each case; Table 1). We also compared our list of genes with those Wnt-regulated genes identified previously using these reagents (Jackson et al. 2014). Twenty-four of the 110 genes activated by Wnt signaling in that study were present among the Wnt pathway–regulated genes identified here (R factor = 20.1; P = 9.6×10−25) (Table 1, Table S2). Recently, microarray analysis was also used to identify more than 1100 genes showing altered expression at the L4 stage in a strain carrying a loss-of-function mutation in the gene encoding the beta-catenin BAR-1 (van der Bent et al. 2014). We found 35 genes in common between these bar-1-regulated genes and our 239 genes; 25 of the genes were downregulated upon loss of BAR-1 (van der Bent et al. 2014) and upregulated upon expression of activated BAR-1 (this work), a significant overlap (Table 1, Table S3). Thirteen of these genes (col-138, col-38, col-49, col-71, C04F1.1, C18A11.4, dao-4, dpy-11, F25E5.2, grd-2, hog-1, R07B1.5, and T07G12.3) were downregulated at the L4 stage upon loss of bar-1 (van der Bent et al. 2014) and upregulated in both our previous work using overexpression of BAR-1 at the L2/L3 molt (Jackson et al. 2014) and in this work (Table S3); therefore, they are strong candidates for larval targets of the WBC pathway. Finally, of the small number of previously known direct Wnt pathway targets, our mRNA tagging approach identified the Hox gene mab-5, which is a predicted target of the WBC pathway in the seam cell V5.p (Hunter et al. 1999; Maloof et al. 1999) . Hox gene lin-39, a predicted target of the WBC pathway in the VPCs (Eisenmann et al. 1998; Hoier et al. 2000; Wagmaister et al. 2006), was upregulated 1.5-fold upon Wnt pathway overactivation, but failed to meet the target selection criterion of P ≤ 0.05. The other known Wnt signaling targets are expressed in other cell types or developmental stages, and are not predicted to be identified by our method. Taken together, these results suggest that combining conditional activation of the Wnt signaling pathway with mRNA tagging and microarray analysis was successful in identifying putative Wnt pathway target genes with a high representation of genes expressed in the seam cells and vulval cells.
Characterization of Wnt pathway-regulated genes
Analysis of the 239 Wnt pathway-regulated genes by DAVID (Huang et al. 2009a,b) identified 57 genes in four significantly enriched, functionally related groups: structural constituents of cuticle; transcription factors; signaling proteins; and protein kinases (Table S4). Thirteen of the 57 genes (∼24%) are expressed in the seam cells and/or the developing vulva (Yook et al. 2012) (Table S4). The largest class of enriched genes includes those encoding components of the worm’s outer surface, the cuticle, which is synthesized by hypodermal cells (Page and Johnstone 2007). In our previous analysis of Wnt-activated genes in the larva, we identified a subset of cuticle collagen genes that are normally expressed in the mid L4 stage, but that can be expressed earlier in response to ectopic Wnt signaling (Jackson et al. 2014). Reduction of function for these genes caused a defect in adult cuticle integrity (Jackson et al. 2014). The identification of a large number of col genes in the current assay confirms the result from the previous analysis and supports the hypothesis of a previously unknown role for Wnt signaling in adult cuticle synthesis (Jackson et al. 2014; van der Bent et al. 2014). We did not pursue further characterization of the cuticle component genes identified in this list of seam and VPC Wnt target genes.
To identify new Wnt pathway–regulated genes for further analysis, we took two approaches. First, we categorized the putative Wnt pathway target genes based on their KOG identity (EuKaryotic Orthologous Groups) (Yook et al. 2012) and occurrence of a known human ortholog (Shaye and Greenwald 2011). This method identified 124 genes that encode transcription factors, signaling molecules, kinases, enzymes, collagens, or other proteins. From these broad categories we selected 24 genes to examine further; nine were chosen because they had human homologs, and the others were selected in such a way to span a range of Wnt activation levels (they ranged from 1.5× to 23.5× increased expression). Second, we repeated the mRNA tagging experiment and performed qPCR to analyze gene expression levels for more than 100 of the putative target genes identified by microarray analysis (data not shown). Twenty-seven of the genes were upregulated in response to Wnt pathway activation when assayed by this method, and 12 of these were also downregulated in response to Wnt pathway inhibition (Table S5). We do not have an explanation for the low validation rate by qPCR; however, a similarly low validation rate was seen in our previous microarray analysis using these conditional activation/inhibition reagents (Jackson et al. 2014). Both of these studies used a low cutoff for inclusion in the data set, and we noted that the qPCR validation rate declines as one moves down the genes in rank order (L. Gorrepati, B. Jackson and D. Eisenmann, unpublished observations). Among the genes regulated by both activation and inhibition of Wnt signaling was the Wnt pathway negative regulatory component pry-1/Axin, which we previously showed is a Wnt-responsive target gene in C. elegans as in other species (Jackson et al. 2014). We chose 26 qPCR validated genes, including 17 genes that had no previous characterization, for further study. In this way we selected 50 seam cell/VPC Wnt pathway–regulated genes encoding previously known and novel gene products for further characterization (Table 2). In addition to these genes, we also identified the adjacent genes egl-18 and elt-6, which encode GATA transcription factors previously shown to be required for seam cell fate specification in the embryo (Koh and Rothman 2001). In a separate analysis (Gorrepati et al. 2013), we showed that egl-18 and elt-6 function downstream of Wnt signaling in the larva seam cells to maintain the progenitor cell fate during larval seam cell divisions (these genes are not described further here).
Table 2. Wnt-regulated genes from the seam cells and VPCs selected for further characterization.
| Gene Function | Gene | Encodes | Fold Change Wnt Overactivation | Expression | Reference |
|---|---|---|---|---|---|
| Signaling | che-14 | Dispatched homolog | 4.1 | Seam cells, vulva | Michaux et al. 2000 |
| Jarriault et al. 2008 | |||||
| grd-3 | Hedgehog-like protein | 2.5 | Seam cells | Aspock et al. 1999 | |
| Mallo et al. 2002 | |||||
| grd-14 | Hedgehog-like protein | 2.4 | Seam cells | Hao et al. 2006 | |
| McKay et al. 2003 | |||||
| hog-1 | Hedgehog-like protein | 3.1 | Other | Hao et al. 2006 | |
| old-2 | Protein tyrosine kinase | 6.7 | N.D. | ||
| osm-11 | DOS domain-containing ligand | 1.6 | Seam cells, VPCs | Komatsu et al. 2008 | |
| Singh et al. 2011 | |||||
| rab-8 | Ras GTPAse | 2.1 | Other | Meissner et al. 2011 | |
| rhgf-2 | Guanine nucleotide exchange factor | 2.1 | Other | Ruvinsky et al. 2007 | |
| McKay et al. 2003 | |||||
| Lin et al. 2012 | |||||
| Transcription regulation | ldb-1 | LIM domain-binding factor | 1.9 | Vulva, other | Dupuy et al. 2007 |
| Cassata et al. 2000 | |||||
| Wenick and Hobert 2004 | |||||
| Reece-Hoyes et al. 2007 | |||||
| Levin et al. 2012 | |||||
| lin-1 | Ets-domain-containing transcription factor | 1.9 | Other | Dupuy et al. 2007 | |
| McKay et al. 2003 | |||||
| Murray et al. 2012 | |||||
| mab-5 | Homeodomain transcription factor | 3.1 | P cells, other | Salser and Kenyon 1992 | |
| nhr-23 | Nuclear hormone receptor | 2.7 | Seam cells, hyp | Kostrouchova et al. 1998 | |
| Feng et al. 2012 | |||||
| nhr-74 | Nuclear hormone receptor | 4.8 | Seam cells | Miyabayashi et al. 1999 | |
| nhr-113 | Nuclear hormone receptor | 4.3 | Seam cells | Reece-Hoyes et al. 2007 | |
| tbx-2 | T-box transcription factor | 1.9 | Other | Miyahara et al. 2004 | |
| Roy Chowdhuri et al. 2006 | |||||
| Smith and Mango 2007 | |||||
| Enzymes | cdk-4 | Cyclin-dependent kinase | 3.9 | Seam cells, VPCs, other | Park and Krause 1999 |
| dhs-5 | Dehydrogenase | 2.8 | Hyp, other | Wang et al. 2006 | |
| pak-1 | p21-activated kinase | 1.6 | Seam cell T, VPCs, other | Chen et al. 1996 | |
| Lino and Yamamoto 1998 | |||||
| Goh et al. 2012 | |||||
| glna-2 | Glutaminase | 2.6 | Hyp, other | McKay et al. 2003 | |
| F26E4.5 | Tyrosine kinase | 3.5 | N.D. | ||
| bus-8 | Glycosyltransferase | 2.9 | Seam cells | Partridge et al. 2008 | |
| kin-10 | Casein kinase beta regulatory subunit | 1.5 | Other | Hu et al. 2006 | |
| Meissner et al. 2011 | |||||
| Unknown | nlp-25 | neuropeptide-like protein | 10.0 | Other | This work |
| T26E4.4 | Human FUT2 homolog | 13.3 | P cells, hyp | Jackson et al. 2014 | |
| C54C8.2 | Uncharacterized protein | 3.1 | Seam cells | Reece-Hoyes et al. 2007 | |
| Other | cki-1 | Cell cycle inhibitor | 3.8 | Seam cells, VPCs, other | Hong et al.1998 |
| Feng et al. 1999 | |||||
| Saito et al. 2004 | |||||
| Fukuyama et al. 2003 | |||||
| Fujita et al. 2007 | |||||
| col-49 | Collagen | 23.5 | Seam cells, P cells, hyp | Jackson et al. 2014 | |
| pry-1 | Scaffolding protein | 4.2 | Seam cells, VPCs, other | Korswagen et al. 2002 | |
| Mizumoto and Sawa 2007 | |||||
| rsp-4 | RNA processing protein | 1.7 | Other | Kawano et al. 2000 | |
| sqv-7 | Nucleotide sugar transporter | 2.2 | Seam cells, VPCs | Hwang and Horvitz 2002 | |
| McKay et al. 2003 | |||||
| Dupuy et al. 2007 | |||||
| twk-5 | K channel | 2.8 | No expression | This work | |
| add-1 | Cytoskeletal protein | 3.7 | Seam cells, hyp, other | McKay et al. 2003 | |
| Dupuy et al. 2007 | |||||
| Vukojevic et al. 2012 | |||||
| K10D6.2 | Membrane protein | 2.7 | Seam cells, other | Kunitomo et al. 2005 | |
| rhy-1 | Regulator of hypoxia-inducible factor | 3.6 | Vulva, hyp | Shen et al. 2006 | |
| Y71H2AL.1 | Calcium-binding protein | 1.7 | Other | Wagner et al. 2011 | |
| mlt-11 | Serine protease inhibitor | 1.8 | Seam cells, hyp | Frand et al. 2005 | |
| Unknown | C28H8.1 | Human gene BCL7A ortholog | 3 | N.D. | |
| nspe-1 | Nematode-specific peptide | 7.5 | Other | This work | |
| pepm-1 | PEPtidase M1 domain-containing protein | 2.3 | Vulva | Inoue et al. 2002 | |
| ttr-15 | TransThyretin-related protein | 2 | No expression | This work | |
| ttr-3 | TransThyretin-related protein | 3.6 | P cells, hyp | This work | |
| ttr-4 | TransThyretin-related protein | 3.6 | No expression | This work | |
| B0454.5 | Uncharacterized protein | 3.8 | P cells, hyp | This work | |
| C08E3.13 | Uncharacterized protein | 2.2 | vulva, other | Mochii et al. 1999 | |
| C14C11.7 | Uncharacterized protein | 4.5 | No expression | This work | |
| K10D6.3 | Uncharacterized protein | 5.2 | No expression | This work | |
| M04C9.1 | Uncharacterized protein | 5.4 | No expression | This work | |
| Y37E11B.7 | Uncharacterized protein | 7.7 | No expression | This work | |
| Y43C5A.3 | Uncharacterized protein | 10.0 | P cells, hyp | This work | |
| Y55F3AM.10 | Uncharacterized protein | 3.4 | No expression | This work |
Shown here are 50 seam/VPC Wnt pathway-regulated genes selected for further characterization grouped into broad classes based on the identity of the protein encoded by each gene. The fold change in expression in Wnt pathway overactivation condition observed by microarray analysis is shown. Also shown is each gene’s expression pattern (previously known or reported here). hyp indicates hypodermal expression not in the seam or vulva lineages (P cells, VPCs, vulva); other indicates known expression outside of hypodermal lineages
Expression analysis of putative seam and VPC Wnt target genes
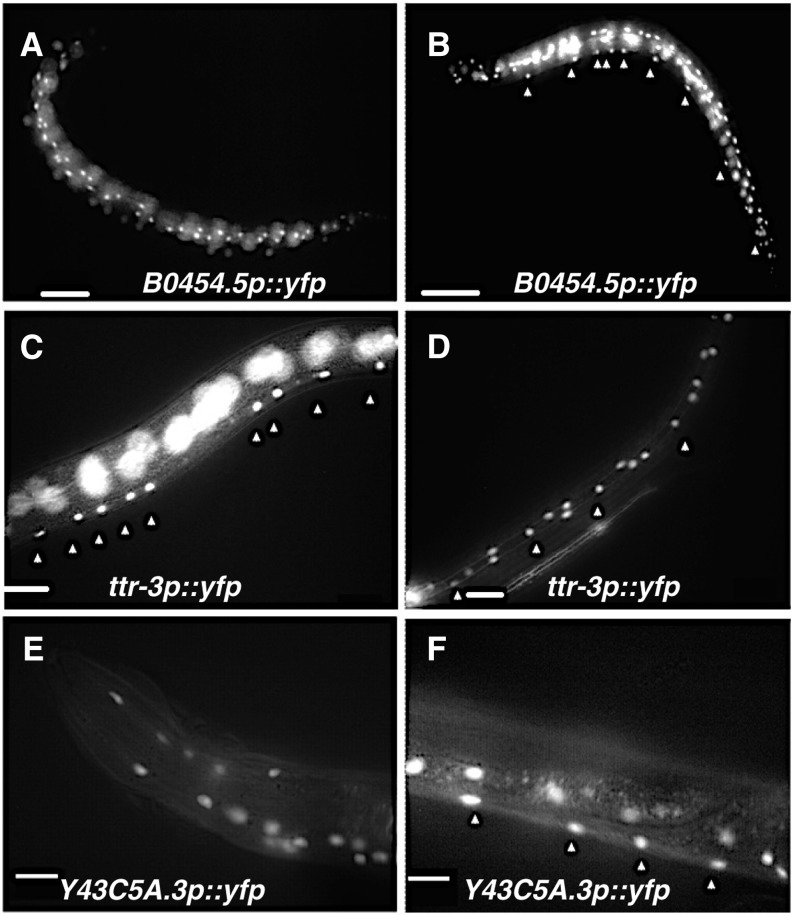
Spatial expression data is known for 32 of the 50 putative Wnt target genes: 10 are expressed in seam cells, 3 are expressed in P cells (precursors to VPCs) and/or hypodermis, seven are expressed in seam and vulva, three are expressed in the vulva, and one is expressed in vulva and hypodermis; (Yook et al. 2012) (Table 2). Eight genes have known expression in other tissues but not in the seam and vulva (Yook et al. 2012) (Table 2); however, four of them have vulval or seam phenotypes when their function is compromised (lin-1, kin-10, rsp-4, and hog-1) (Ferguson and Horvitz 1985; Longman et al. 2001; Kamath and Ahringer 2003; Rual et al. 2004; Zugasti et al. 2005), suggesting they may be expressed in those cell types. Of the 18 genes with no known expression data, we generated 14 promoter::yfp constructs in which the upstream region from the start codon of the gene of interest to the next upstream gene was fused to 2X::NLS::YFP coding sequences (this work; Jackson et al. 2014). Five of 14 genes (B0454.5, ttr-3, Y43C5A.3, col-49, T26E4.5) were expressed in the hypodermis, seam cells, or P cell descendents, whereas two genes, nlp-25 and nspe-1, showed expression in other tissues (Figure 2, Table 2) (Jackson et al. 2014). We failed to detect YFP expression for eight genes, suggesting the promoter::yfp constructs may not have included all possible regulatory elements (six of the eight genes were previously uncharacterized genes encoding novel proteins). In summary, of the 46 candidate genes examined, 63% showed expression in the seam or vulval lineages. Therefore, given their expression in the cell types of interest, and their upregulation in response to Wnt signaling, we consider these genes likely to be bona fide Wnt target genes in the seam cells and the vulval cells.
Figure 2.
Transcriptional reporter analysis reveals putative Wnt targets are expressed in seam cells, hypodermis, and P cell descendents. Shown here are promoter::YFP expressing worms. (A and B) The target gene B0454.5 is expressed in hypodermal cells (unlabeled) and P cell descendents (arrowheads in B) throughout larval life and in adults. (C and D) ttr-3::YFP is also expressed in the P cell descendents (arrowheads in C), seam cells (arrowheads in D), and hypodermal cells (unlabeled). (E and F) Expression of Y43C5A.3::YFP is shown in the hypodermal cells (E) and P cells (arrowheads in F). In (A) through (C), the out-of-focus fluorescence is from the seam and hypodermal cells, present in a different focal plane. Scale bars represent 50 μm in all figures.
Wnt targets genes function in seam cell and vulval development
The seam cells are lateral hypodermal cells extending in a single row from nose to tail that divide in a stem cell–like manner during each of the four larval stages to generate one daughter that retains the seam cell fate and the ability to divide further, and another daughter that differentiates and joins the syncytial skin of the growing worm (Joshi et al. 2010). The asymmetric division of the seam cells is known to be regulated by Wnt signaling (Gleason and Eisenmann 2010). After their final division in the L4 stage, the 16 seam cells on each side of the animal exit the cell cycle, terminally differentiate, and fuse with each other to form a single syncytial cell (Joshi et al. 2010). The differentiated seam cells secrete the specialized cuticular structure called alae (Joshi et al. 2010), which are visible as lateral parallel ridges running the length of the body in adult worms.
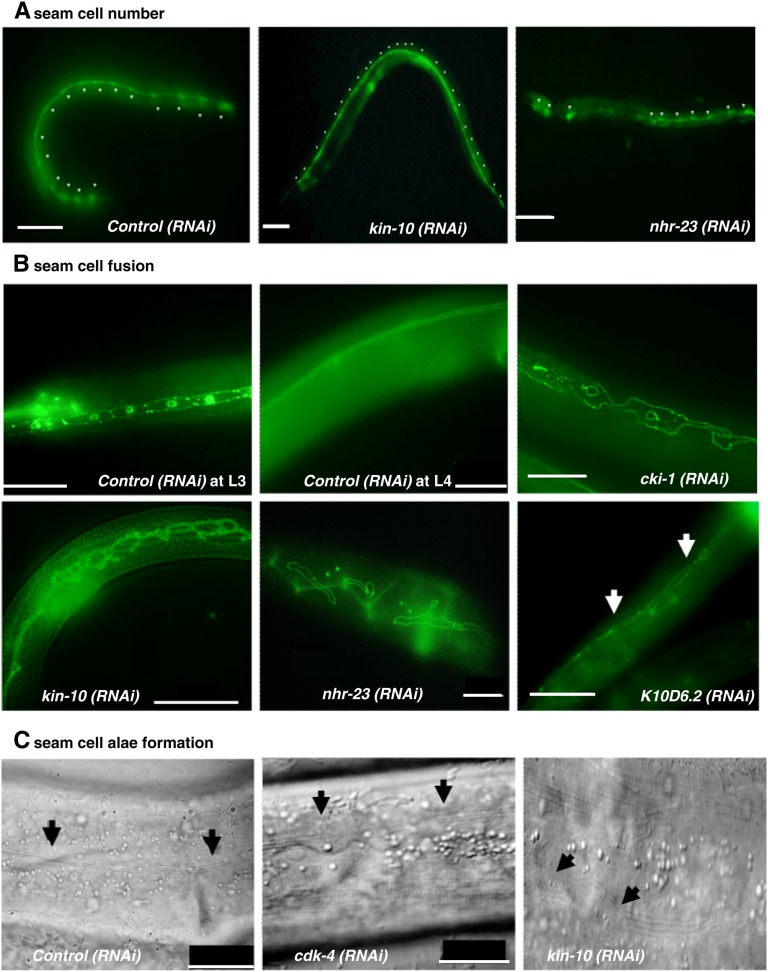
We examined the role of 50 putative Wnt target genes in larval seam cell development using RNAi interference (RNAi). To increase the likelihood of observing novel phenotypes, we used an rrf-3(pk1426) background, which is hypersensitive to RNA interference (Simmer et al. 2002). We monitored seam cell numbers in RNAi-treated animals using the transcriptional reporter grd-10p::gfp, which expresses GFP in the nucleus and cytoplasm of all seam cells from early larval stages to adult (Figure 3, Table 3) (McKay et al. 2003). Alterations in seam cell number could indicate a defect in adoption or maintenance of the seam cell fate, execution of the asymmetric seam cell divisions, or seam cell survival (Gleason and Eisenmann 2010; Gorrepati et al. 2013). We also examined RNAi-treated animals for defects in seam cell fusion by antibody staining for the adherens junction protein AJM-1, and for defects in adult alae formation (Figure 3, Table 4, Table 5) (Page and Johnstone 2007). Defects in these properties could indicate a defect in the specification, survival, or terminal differentiation of seam cells.
Figure 3.
Putative Wnt targets influence seam cell number, formation of seam syncytium, and cuticular alae. (A) Seam cells (asterisks) in RNAi control, kin-19(RNAi), and nhr-23(RNAi) young adult animals. All animals contain grd-10::gfp, which expresses GFP in the nucleus and cytoplasm of seam cells at all stages. RNAi control animals have 16 seam cells per side, whereas kin-10(RNAi) animals have additional seam cells and nhr-23(RNAi) animals have fewer seam cells. (B) After their last division in the L4 stage, seam cells fuse to form a single-cell syncytium that runs the length of the animal, which can be visualized by antibody staining for the junctional protein AJM-1. The first two panels show seam cells in RNAi control animals before (L3) and after homotypic fusion (late L4). At this later time, cki-1(RNAi) and kin-10 (RNAi) animals show ring-like malformations in the syncytium, nhr-23(RNAi) worms have gaps in the syncytium, and K10D6.2(RNAi) worms have gaps as well as unfused seam cells (arrow heads). (C) After their last division in the L4 stage, seam cells secrete a specialized cuticular structure called alae that is visualized as three to four parallel ridges running the length of the animal. The first panel shows a portion of the midbody cuticle of an RNAi control animal with intact alae. cdk-4(RNAi) animals show partial or incomplete alae (alae end at arrowheads with gap between), whereas kin-10(RNAi) worms exhibited misformed alae (arrow heads). Scale bars represent 50 μm in all figures.
Table 3. Reduction of function of five Wnt-regulated genes alters terminal seam cell number.
| Strain | n | Average Adult Seam Cell No. | Range |
|---|---|---|---|
| control(RNAi) | 246 | 15.6 | 13–16 |
| cdk-4(RNAi) | 35 | 11.9a | 7–16 |
| cki-1(RNAi) | 38 | 20.6a | 17–26 |
| kin-10(RNAi) | 42 | 23.9a | 16–36 |
| mlt-11(RNAi) | 43 | 10.5a | 7–13 |
| nhr-23(RNAi) | 53 | 13.3a | 9–16 |
All animals contain rrf-3(pk1426) and seam cell–specific marker grd-10::gfp; GFP+ cells were counted in young adult worms. The number of adult seam cells per side in control RNAi (feeding vector alone) and animals treated with RNAi against the indicated Wnt regulated genes is shown.
P ≤ 0.001 (unpaired t-test) compared to control (RNAi).
Table 4. Wnt pathway components and Wnt-regulated genes influence seam syncytium formation.
| Strain | n | Normal (%) | Gaps (%) | Unfused (%) | Rings (%) |
|---|---|---|---|---|---|
| control(RNAi) | 74 | 96 | 4 | 0 | 0 |
| bus-8(RNAi) | 72 | 64 | 26b | 8b | 1 |
| cdk-4(RNAi) | 36 | 53 | 47b | 0 | 0 |
| cki-1(RNAi) | 33 | 12 | 85b | 0 | 3 |
| K10D6.2(RNAi) | 56 | 0 | 70b | 27b | 3b |
| kin-10(ok1751) | 51 | 39 | 2 | 0 | 59b |
| kin-10(RNAi) | 51 | 16 | 0 | 0 | 84b |
| mlt-11(RNAi) | 41 | 10 | 76b | 2 | 12b |
| nhr-23(RNAi) | 30 | 3 | 77b | 17b | 3a |
Antibody staining against AJM-1 protein was used to visualize the seam syncytium in rrf-3(pk1426) animals treated with RNAi against the indicated Wnt-regulated genes. Normal indicates fused seam cells appearing as parallel tracks running the entire length of the animal; partial indicates gaps; unfused indicates presence of unfused seam cells; and rings indicates ring-shaped misformations along the seam syncytium. The percentage of animals with the indicated seam syncytium defects is shown.
P ≤ 0.05.
P ≤ 0.001 (Fisher’s exact test) compared to control (feeding vector alone).
Table 5. Wnt pathway components and Wnt-regulated genes influence adult alae formation.
| Strain | n | Normal (%) | Partial (%) | Absent (%) | Rings (%) |
|---|---|---|---|---|---|
| control(RNAi) | 40 | 100 | 0 | 0 | 0 |
| bus-8(RNAi) | 37 | 8 | 30b | 62b | 0 |
| cdk-4(RNAi) | 31 | 36 | 45b | 19b | 0 |
| cki-1(RNAi) | 26 | 26 | 70b | 4 | 0 |
| K10D6.2(RNAi) | 58 | 38 | 58b | 7a | 0 |
| kin-10(ok1751) | 52 | 0 | 10b | 6b | 84b |
| kin-10(RNAi) | 31 | 6 | 7b | 0 | 87b |
| mlt-11(RNAi) | 41 | 2 | 20b | 78b | 0 |
| nhr-23(RNAi) | 34 | 0 | 24b | 76b | 0 |
rrf-3(pk1426) animals were treated with RNAi against the indicated Wnt-regulated genes and examined for defective adult alae formation. The percentage of worms with the indicated types of alae defects is shown. Normal indicates three parallel ridges running along the length of the animal; partial indicates gaps; absent indicates lack of alae; and rings indicates ring-like misformations along the length of the alae.
P ≤ 0.05.
P ≤ 0.001 (Fisher’s exact test) compared to control (feeding vector alone).
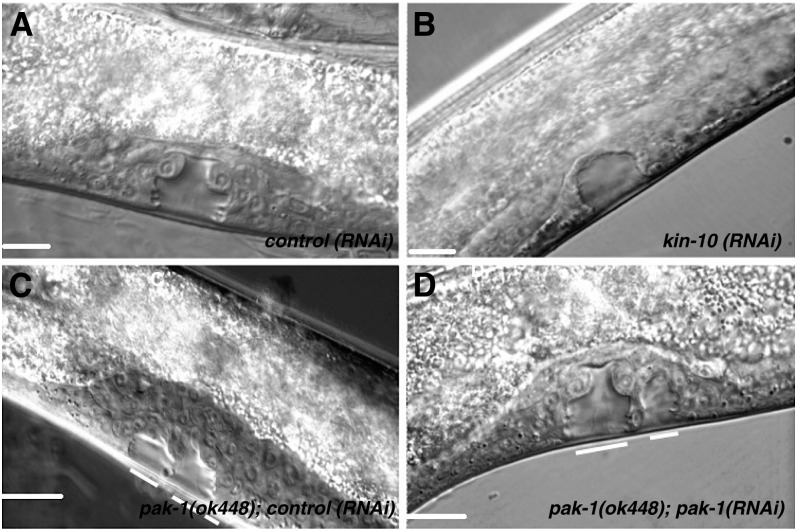
We also tested the involvement of the putative Wnt target genes in vulval precursor cell (VPC) fate specification by observing L4 vulval formation in RNAi-treated animals (Figure 4, Table 6). Wnt signaling is known to function in the specification or maintenance of the VPC fate, as well as in the asymmetric division of the VPCs P5.p and P7.p (Eisenmann et al. 1998; Eisenmann and Kim 2000; Gleason et al. 2002, 2006; Green et al. 2008). Abnormalities in vulval morphology observed at the L4 stage could indicate a defect in VPC fate specification or fate execution, vulval cell formation, or vulval morphogenesis (Eisenmann and Kim 2000).
Figure 4.
Reduction-of-function of putative Wnt targets affects L4 vulva formation. (A) In the control, three VPCs adopting primary and secondary fates in the L3 stage divide further and undergo morphogenesis, resulting in a Christmas tree–shaped L4 vulva. (B) When fewer than three VPCs adopt vulval fates, an underinduced phenotype can be seen as observed in kin-10(RNAi) worms. (C and D) The polarity of P7.p is reversed in Wnt signaling mutants resulting in a Bivulva phenotype. A similar phenotype was observed in pak-1(RNAi) worms and in pak-1(ok448) mutants. Scale bars represent 50 μm.
Table 6. Wnt-regulated genes required for proper L4 vulva formation.
| Strain | n | Normal (%) | Underinduced (%) | Overinduced (%) | Bivulva (%) | Other (%) |
|---|---|---|---|---|---|---|
| control (RNAi) | 81 | 98 | 0 | 0 | 0 | 2 |
| lin-1(RNAi) | 35 | 6 | 0 | 91b | 0 | 3 |
| kin-10(RNAi) | 45 | 4 | 85b | 4b | 0 | 7b |
| pak-1(ok448) | 61 | 80 | 5 | 0 | 13b | 2 |
| pak-1(RNAi) | 63 | 90 | 2 | 0 | 8a | 0 |
The top shows the percentage of rrf-3(pk1426) animals showing L4 vulval defects upon RNAi treatment for lin-1 and kin-10. Normal indicates the characteristic Christmas tree–shaped L4 vulva formed when three VPCs adopt primary and secondary fates and divide further. Underinduced indicates improper L4 vulva formed when fewer than three VPCs adopt vulval fates. Overinduced indicates multiple vulval invaginations that form when more than three VPCs adopt vulval fates. Bivulva indicates a reversal in the polarity of the VPC P7.p resulting in an extra invagination.
P ≤ 0.05.
P ≤ 0.001 (Fisher’s exact test) compared to control (RNAi).
Of the 50 genes examined, five genes (cdk-4, cki-1, kin-10, mlt-11, and nhr-23) showed alterations in adult seam cell number (Table 3), seven genes (bus-8, cdk-4, cki-1, kin-10, mlt-11, nhr-23, and K10D6.2) showed defects in seam syncytium and alae formation (Table 4, Table 5), and three genes (lin-1, kin-10, and pak-1) had defects in L4 vulva formation (Table 6). We briefly describe these nine genes below.
Wnt target genes that affect seam cell fate specification and terminal differentiation
cdk-4 (cyclin-dependent kinase 4):
CDK-4 forms a complex with cyclin D1 (CYD-1 in C. elegans) that regulates progress from the G1 to S phase of the cell cycle (van den Heuvel 2005). In C. elegans, CDK-4 is expressed during larval life in many cells, including the Pn.p cells, VPCs, and seam cells, and cdk-4 RNAi-treated animals have defects in cell cycle progression for many postembryonic lineages (Park and Krause 1999). As expected for a positive cell cycle regulator, we found that cdk-4(RNAi) animals had fewer seam cells as adults (11.9 SCs/side, compared to an average of 15.6 in control RNAi animals) (Table 3); 47% of cdk-4(RNAi) worms displayed gaps in the seam syncytium (Table 4), whereas 64% of treated animals had missing or abnormal cuticular alae (Table 5, Figure 3K). These latter phenotypes are most likely due to the decrease in seam cell number observed in these animals, leading to gaps in the linear array of seam cells along the body axis. In humans, the cyclin D1 gene is a transcriptional target of Wnt signaling (Shtutman et al. 1999; Tetsu and McCormick 1999); however, the worm cyd-1 gene did not pass our criteria for selection of Wnt pathway targets. These results suggest that C. elegans may use a different mechanism to activate cell proliferation in response to Wnt signaling, perhaps by upregulation of expression of cdk-4 rather than its cyclin partner.
cki-1 (cyclin-dependent kinase inhibitor 1):
Curiously, the cki-1 gene was also found as a potential Wnt target gene in our analysis. The cyclin-dependent kinase inhibitor CKI-1 is homologous to the mammalian Cip/Kip family of CKI proteins, which inhibit CDKs functioning in the G1 to S phase transition (Clayton et al. 2008). Loss of function of cki-1 results in extra cell divisions in the embryo as well as in many postembryonic cell lineages, including the VPCs and seam cells (Hong et al. 1998; Fukuyama et al. 2003; Nimmo et al. 2005; Kagoshima et al. 2007; Clayton et al. 2008; Buck et al. 2009). As expected, we observed an increased seam cell number in cki-1(RNAi)-treated adults (20.6 SCs/side) (Table 3). Curiously, although cki-1(RNAi) worms had more seam cells, more than 85% of animals had gaps in the seam syncytium or showed misformed syncytium (Table 4, Figure 3F). In addition, the adult alae were missing or only partially formed in 74% of adult worms (Table 5). These phenotypes suggest a defect in terminal differentiation of the seam cells. Although cell differentiation in general was not found to be affected by loss of CKI-1 (van den Heuvel 2005), our results indicate that reduction of function for cki-1 may interfere with the ability of the seam cells to terminally differentiate properly, perhaps due to failure to exit the cell cycle at the proper time of development. It is currently unclear whether cdk-4 and cki-1 are both targets of the Wnt pathway in the same cells at the same time in development; further analysis will be needed to resolve this apparent incongruous result.
kin-10 (casein kinase II beta regulatory subunit):
kin-10 encodes an ortholog of the human casein kinase II beta regulatory subunit encoded by CSNK2B. In C. elegans, kin-10 is widely expressed throughout development, although specific expression in seam cells and VPCs was not reported (Hu et al. 2006). KIN-10 is required for male mating, general viability, and function of the RNA interference pathway (Kim et al. 2005; Hu et al. 2006). Interestingly, we observed an increase in terminal seam cell number in kin-10(RNAi) animals (Table 3, Figure 3B), and >80% of these animals had misformed seam syncytium and alae (Table 4, Table 5, Figure 3, G and L). These defects were also seen in animals containing the (ok1751) mutation, which is a large internal deletion in the kin-10 gene (Yook et al. 2012) (Table 4, Table 5). Finally, as observed previously in a genome-wide RNAi screen (Simmer et al. 2003), kin-10(RNAi) worms had vulval defects, with 85% of animals having an underinduced phenotype, indicating a possible previously unidentified role for casein kinase II in vulval development (Table 6, Figure 4B).
In Xenopus and mice, casein kinase 2 has been shown to be a positive modulator of Wnt/β-catenin signaling and was activated upon Wnt stimulation in murine cell culture (Song et al. 2003; Dominguez et al. 2004). Some components of the Wnt pathway such as Dvl, APC, TCF, and β-catenin are in vitro substrates of CK2 (Dominguez et al. 2009). Curiously, our results show that the kin-10(RNAi) phenotype in the seam cells is the same as that caused by overactivation of the WBA pathway (an increase in seam cell number) (Gleason and Eisenmann 2010), whereas the kin-10(RNAi) phenotype in the vulval cells is the same as that caused by reduction of function of the WBC pathway (underinduction) (Eisenmann et al. 1998; Gleason et al. 2006). It is possible that kin-10 may be a target of the WBA pathway in the seam cells and a target of the WBC pathway in the VPCs, and may modulate the function of these pathways in different ways in these two cell types. Finally, the defects in seam cell terminal differentiation seen in kin-10(RNAi) animals could be a consequence of the increase in seam cell number or KIN-10 could have separate functions in seam cell division/specification and differentiation. Additional experiments will be needed to characterize the role of KIN-10 in these Wnt-mediated processes.
nhr-23 (nuclear hormone receptor 23):
nhr-23 encodes an orphan nuclear hormone receptor (NHR) that is expressed in the seam cells and the major hypodermal syncytium hyp7 throughout larval development, with expression peaking in the mid larval stage (Kostrouchova et al. 1998). Like its fly counterpart DHR3, NHR-23 is involved in molting and may be a regulator of other genes acting in the process (Kostrouchova et al. 1998, 2001). We found that RNAi treatment against nhr-23 starting from the L1 stage resulted in animals with severe molting defects, as reported previously (Kostrouchova et al. 1998, 2001). However, we also observed that the seam cell number in nhr-23(RNAi) animals was reduced (13.3 SCs/side) (Table 3, Figure 3C), and almost 100% of nhr-23(RNAi) animals had defects in seam cell syncytium formation and alae formation (Table 4, Table 5, Figure 3H), indicating a previously unexplored role for NHR-23 in these processes. A "branched" alae defect in nhr-23(RNAi) animals was previously noted, although a defect in seam cell number was not seen (Kostrouchova et al. 1998, 2001). Since Wnt pathway mutants do not show molting defects (J. Gleason, L. Gorrepati and D. Eisenmann, unpublished observations), it is possible that Wnt signaling may be only one component of the regulation of nhr-23 expression in the larva, such that reduction of Wnt signaling alone may not be sufficient to cause the molting phenotypes observed in nhr-23(RNAi) worms.
mlt-11 (molting defective 11):
The process of molting requires spatially coordinated and sequential functioning of transcription factors, signaling molecules, proteases, protease inhibitors, and other factors (Frand et al. 2005; Monsalve and Frand 2012). The mlt-11 gene, which encodes a serine protease inhibitor, was identified in an RNAi screen for molting defects (Frand et al. 2005). mlt-11 is expressed in the seam cells and hypodermis throughout larval development, with a peak of expression in the seam cells right before each larval molt, coinciding with its predicted role in protecting the new cuticle from proteases that are required to degrade the old cuticle (Frand et al. 2005). Interestingly, in addition to molting defects, we found that mlt-11(RNAi) animals had a decreased number of seam cells as adults (10.5 SCs/side) (Table 3), and a large percentage of mlt-11(RNAi) animals showed defects in seam syncytium and alae formation (Table 4, Table 5). This effect on seam cell number was surprising given the proposed extracellular function of MLT-11 in the molting process. We examined 12 other genes identified in molting screens for defects in seam cell number and found that reduction of function for mlt-7 also caused a decrease in adult cells expressing three different markers of seam cell fate (Table S6). mlt-7 encodes a heme peroxidase expressed in the hypodermis (including lateral and ventral cells) required for cross-linking of cuticle collagens (Thein et al. 2009). Intriguingly, the effect of mlt-7 and mlt-11 reduction of function on seam cell number suggests that there may be feedback regulation from the extracellular molting process to the division behavior of the seam cells, as was suggested earlier (Frand et al. 2005). Finally, it should be noted that mlt-11 is predicted to function downstream of NHR-23, as its expression was affected in nhr-23(RNAi) animals (Frand et al. 2005). Therefore, it is possible that mlt-11 was identified in this analysis because it is a downstream target of nhr-23, which is upregulated upon Wnt pathway overactivation, and that the effect of nhr-23(RNAi) on seam cell number is due to the regulation of mlt-11 by NHR-23.
Wnt target genes that affect seam cell fate terminal differentiation only
bus-8 (bacterially unswollen 8):
BUS-8 is a glycosyl transferase that functions in N-linked glycosylation of proteins (Partridge et al. 2008). BUS-8 acts in the embryo during ventral closure when epithelial cells fuse and migrate to enclose the body (Partridge et al. 2008). Postembryonically, BUS-8 is expressed exclusively in the seam cells, where it functions in molting and in maintaining cuticle integrity (Gravato-Nobre et al. 2005; Partridge et al. 2008). Consistent with previous results, we observed that treatment of rrf-3 worms with bus-8 RNAi from the L1 stage resulted in animals with molting defects, some of which developed normally until adulthood and laid eggs that did not hatch. We found that terminal seam cell number in bus-8(RNAi) animals was unaffected (data not shown); however, 34% of bus-8(RNAi) animals showed gaps in the seam syncytium or had unfused seam cells (Table 4), and 92% of bus-8(RNAi) of animals had either partial or no alae (Table 5). These results indicate a previously unidentified role for this protein glycosylation factor in seam cell terminal differentiation. Because bar-1 loss-of-function mutants do not have seam cell terminal differentiation defects of this type (J. Gleason, L. Gorrepati, and D. Eisenmann, unpublished results), this again suggests that regulation by Wnt signaling may represent only one component of bus-8 expression, such that loss of Wnt signaling does not phenocopy the bus-8 mutant seam terminal differentiation phenotype.
K10D6.2:
K10D6.2, which encodes a claudin-like transmembrane protein, was identified as a gene expressed in C. elegans sensory neurons by the mRNA tagging method, and it was also found to be strongly expressed in the seam cells during larval life (Kunitomo et al. 2005; Simske and Hardin 2011). In two genome-wide screens, RNAi for K10D6.2 resulted in embryonic lethality (Maeda et al. 2001; Rual et al. 2004). As with bus-8, K10D6.2(RNAi) animals showed no significant change in adult seam cell number (data not shown), but 100% of adults had defects in formation of the adult seam syncytium, with unfused seam cells visible (Table 4, Figure 3I), and 65% of K10D6.2(RNAi) adult animals had partial or no alae (Table 5). Even in younger K10D6.2(RNAi) larvae, seam cells failed to contact each other to form a linear array, exhibiting gaps along the length of the animal (data not shown). These RNAi results indicate the K10D6.2 gene product likely functions in seam cell terminal differentiation, perhaps in the homotypic fusion of the seam cells.
Wnt target genes that act in vulval development
pak-1 (p21-activated kinase 1):
pak-1 encodes a p21 activated kinase homolog required for proper axonal guidance (Lucanic et al. 2006), which is expressed in several tissues, including the seam cell T and its descendants, and the VPCs (Iino and Yamamoto 1998; Goh et al. 2012). In addition to its role in the specification of VPC fates, Wnt signaling also regulates the polarity of the descendants of secondary-fated VPC, P7.p (Inoue et al. 2004; Deshpande et al. 2005; Green et al. 2007, 2008). Consistent with a role in that process, we found that 8% of pak-1(RNAi) animals had a Bivulva phenotype indicative of P7.p polarity defects (Table 6). This phenotype was slightly stronger in pak-1(ok448) animals, which carry a large deletion in the pak-1 gene (Harris et al. 2010; Yook et al. 2012) (Table 6). Curiously, others have reported that only ∼1% of pak-1(ok448) mutant worms exhibited defects in P7.p polarity, but that pak-1(ok448) could suppress the defects in P7.p polarity and reporter gene expression exhibited by a mutation in lin-17, which encodes a member of the Frizzled family of Wnt receptors (Goh et al. 2012). Therefore, it was proposed that PAK-1 negatively regulates the Wnt signaling pathway acting in P7.p polarity during vulval development (Goh et al. 2012). Our results show that loss of function of pak-1 by RNAi or mutation results in a weak Bivulva phenotype similar to that caused by reduction of Wnt signaling, suggesting that PAK-1 plays a positive role downstream of Wnt signaling in P7.p. It is not clear why there is a difference in results, although differences in strain background could be possible. PAK-1 was recently shown to regulate the phosphorylation of β-catenin in colon cancer cells (Zhu et al. 2012). It is possible that pak-1 may be a downstream target of Wnt signaling in vulval development that is required for phosphorylation of Wnt signaling components or other proteins necessary for successful orientation of P7.p polarity.
lin-1 (Lineage abnormal 1):
lin-1 encodes an Ets-domain transcription factor that acts downstream of RTK/Ras signaling in the VPC P6.p during vulval development (Beitel et al. 1995). Loss of function of lin-1 causes induction of VPCs, resulting in a multivulva phenotype, suggesting LIN-1 prevents these VPCs from adopting induced vulval cell fates in the absence of RTK/Ras signaling (Beitel et al. 1995; Sternberg 2005). Consistent with this, we found that 91% of lin-1(RNAi) animals had ectopic vulval invaginations characteristic of a multivulva phenotype (Table 6). Because the overinduced vulval phenotype caused by loss of lin-1 activity is the opposite of the underinduced phenotype resulting from reduction of Wnt pathway activity (Eisenmann et al. 1998; Eisenmann and Kim 2000; Gleason et al. 2006), it was intriguing to identify lin-1 as a possible Wnt pathway target. qPCR showed that endogenous lin-1 transcript levels increased slightly upon Wnt pathway activation and decreased slightly upon Wnt pathway inhibition (Table S5). Therefore, we hypothesize that Wnt signaling may be one of several components that maintain lin-1 expression in the VPCs before induction, such that while overexpression of ΔNTBAR-1 upregulates lin-1 expression, reduction of Wnt signaling, such as in a bar-1(ga80) mutant, does not reduce lin-1 expression enough to cause an overinduction phenotype.
Conclusion
Forward genetic analysis in C. elegans has proven very useful in identifying signaling pathways regulating many developmental processes (Greenwald 2005). However, the same forward genetic techniques have often proven much less effective at identifying the targets of these pathways that actually effect the transcription changes underlying the biological responses elicited by pathway activation. A possible explanation for this result is that inactivation of a single target gene may be unlikely to cause a strong phenotype like that caused by loss of core signaling pathway components. Therefore, genomic approaches may be useful for identifying the downstream targets of signaling pathways acting during development.
We previously showed that Wnt signaling plays a role in cell fate specification of hypodermal seam and vulval precursor cells during C. elegans larval development (Eisenmann et al. 1998; Gleason et al. 2002, 2006; Gleason and Eisenmann 2010). However, few direct targets of the Wnt pathway in these cells have been identified (Jackson and Eisenmann 2012). To attempt to identify Wnt pathway target genes in C. elegans, we used a gain-of-function approach, in which we conditionally expressed a gain-of-function BAR-1 beta catenin protein at the time of the L2/L3 molt, followed by microarray analysis to identify differentially regulated genes. In our initial approach, we analyzed changes in gene expression from the whole animal and identified more than 100 genes that showed increased expression upon delNTBAR-1 expression (Jackson et al. 2014).
Here, we combined this previous approach with the "mRNA tagging" method (Roy et al. 2002) to enrich for transcripts from the seam and vulval precursor cells. We believe this combination of approaches has been successful at finding cell type–specific targets of the Wnt signaling pathway because we identified many genes known to be expressed in or function in the seam or vulval precursor cells, and we showed that nine of the genes identified by this method have loss-of-function phenotypes consistent with a function in these cell types. Several of these genes were known to function in hypodermal cell division or to have phenotypes affecting the seam or vulval precursor cells; however, a number of genes (bus-8, cki-1, K10D6.2, kin-10, mlt-11, nhr-23) were shown to have novel phenotypes in one or both of these cell types upon reduction of gene function. Further, in work reported elsewhere, we showed that the GATA factor encoding genes egl-18 and elt-6 identified by this method are targets of the Wnt pathway in the seam cells and are required for the maintenance of the seam cell "progenitor" fate in one daughter of the larval seam cell asymmetric divisions (Gorrepati et al. 2013). Given the success of this method in identifying Wnt target genes acting in the larval hypodermal cells, an approach like this could be useful for identifying target genes regulated by other signaling pathways that function in C. elegans development, if conditional pathway activation using suitable reagents is possible.
We chose the approach of conditionally expressing a gain-of-function beta catenin variant over the more simple approach of analyzing gene expression in a Wnt pathway loss-of-function strain as a way to bypass any indirect effects on gene expression that might result from cell fate transformations due to loss of Wnt pathway activity (Jackson et al. 2014). Intriguingly, a recent report showed that a large number of genes show alterations in expression at the L4 stage in a strain containing a bar-1 loss-of-function mutation (van der Bent et al. 2014). The fact that a small set of genes showed reduced expression upon loss of bar-1 (van der Bent et al. 2014) and increased expression upon expression of activated BAR-1 (Jackson et al. 2014; this work) identifies these gene as strong candidates for bona fide BAR-1-regulated or Wnt-regulated target genes. Therefore, the use of complementary loss-of-function and gain-of-function approaches, when possible, may strongly highlight a subset of genes as targets of a signaling pathway for further in-depth biological investigation.
Supplementary Material
Acknowledgments
We thank S. Von Stetina, J. D. Watson, D. M. Miller, B. M. Jackson, J. E. Gleason, I. Hamza, A. Golden, and C. Wolkow for helpful advice and sharing of reagents, strains, and unpublished information. We thank members of the Eisenmann lab for advice and support. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440), and by the National BioResource Project of Japan. This work was partially supported by NSF grant IBN-0131485 and NIH grant GM65424 (to D.M.E.). This work was supported, in part, by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.017715/-/DC1
Microarray data accession number: GSE52747.
Communicating editor: M. Walhout
Literature Cited
- Arata Y., Kouike H., Zhang Y., Herman M. A., Okano H., et al. , 2006. Wnt signaling and a Hox protein cooperatively regulate psa-3/Meis to determine daughter cell fate after asymmetric cell division in C. elegans. Dev. Cell 11: 105–115. [DOI] [PubMed] [Google Scholar]
- Aspöck G., Kagoshima H., Niklaus G., Bürglin T. R., 1999. Caenorhabditis elegans has scores of hedgehog-related genes: sequence and expression analysis. Genome Res. 9: 909–923. [DOI] [PubMed] [Google Scholar]
- Banerjee D., Chen X., Lin S. Y., Slack F. J., 2010. kin-19/casein kinase Ialpha has dual functions in regulating asymmetric division and terminal differentiation in C. elegans epidermal stem cells. Cell Cycle 9: 4748–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitel G. J., Tuck S., Greenwald I., Horvitz H. R., 1995. The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes Dev. 9: 3149–3162. [DOI] [PubMed] [Google Scholar]
- Bertrand V., Hobert O., 2009. Wnt asymmetry and the terminal division of neuronal progenitors. Cell Cycle 8: 1973–1974. [DOI] [PubMed] [Google Scholar]
- Bhambhani C., Ravindranath A. J., Mentink R. A., Chang M. V., Betist M. C., et al. , 2014. Distinct DNA binding sites contribute to the TCF transcriptional switch in C. elegans and Drosophila. PLoS Genet. 10: e1004133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck S. H., Chiu D., Saito R. M., 2009. The cyclin-dependent kinase inhibitors, cki-1 and cki-2, act in overlapping but distinct pathways to control cell cycle quiescence during C. elegans development. Cell Cycle 8: 2613–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan K. M., Peifer M., 2009. Wnt signaling from development to disease: insights from model systems. Cold Spring Harb. Perspect. Biol. 1: a002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan K. M., 2012. TCFs and Wnt/beta-catenin signaling: more than one way to throw the switch. Curr. Top. Dev. Biol. 98: 1–34. [DOI] [PubMed] [Google Scholar]
- Cassata G., Röhrig S., Kuhn F., Hauri H. P., Baumeister R., et al. , 2000. The Caenorhabditis elegans Ldb/NLI/Clim orthologue ldb-1 is required for neuronal function. Dev. Biol. 226: 45–56. [DOI] [PubMed] [Google Scholar]
- Chen W., Chen S., Yap S. F., Lim L., 1996. The Caenorhabditis elegans p21-activated kinase (CePAK) colocalizes with CeRac1 and CDC42Ce at hypodermal cell boundaries during embryo elongation. J. Biol. Chem. 271: 26362–26368. [DOI] [PubMed] [Google Scholar]
- Chisholm A. D., Hsiao T. I., 2012. The Caenorhabditis elegans epidermis as a model skin. I: development, patterning, and growth. Wiley Interdiscip. Rev. Dev. Biol. 1: 861–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton J. E., van den Heuvel S. J., Saito R. M., 2008. Transcriptional control of cell-cycle quiescence during C. elegans development. Dev. Biol. 313: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H., Nusse R., 2012. Wnt/beta-catenin signaling and disease. Cell 149: 1192–1205. [DOI] [PubMed] [Google Scholar]
- Deshpande R., Inoue T., Priess J. R., Hill R. J., 2005. lin-17/Frizzled and lin-18 regulate POP-1/TCF-1 localization and cell type specification during C. elegans vulval development. Dev. Biol. 278: 118–129. [DOI] [PubMed] [Google Scholar]
- Dominguez I., Mizuno J., Wu H., Song D. H., Symes K., et al. , 2004. Protein kinase CK2 is required for dorsal axis formation in Xenopus embryos. Dev. Biol. 274: 110–124. [DOI] [PubMed] [Google Scholar]
- Dominguez I., Sonenshein G. E., Seldin D. C., 2009. Protein kinase CK2 in health and disease: CK2 and its role in Wnt and NF-kappaB signaling: linking development and cancer. Cell. Mol. Life Sci. 66: 1850–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr, J. S., 2006 Immunohistochemistry (June 19, 2006), in WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.105.1, http://www.wormbook.org. 10.1895/wormbook.1.105.1 [DOI]
- Dupuy D., Bertin N., Hidalgo C. A., Venkatesan K., Tu D., et al. , 2007. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat. Biotechnol. 25: 663–668. [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Kim S. K., 2000. Protruding vulva mutants identify novel loci and Wnt signaling factors that function during Caenorhabditis elegans vulva development. Genetics 156: 1097–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann D. M., Maloof J. N., Simske J. S., Kenyon C., Kim S. K., 1998. The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development 125: 3667–3680. [DOI] [PubMed] [Google Scholar]
- Feng H., Zhong W., Punkosdy G., Gu S., Zhou L., et al. , 1999. CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat. Cell Biol. 1: 486–492. [DOI] [PubMed] [Google Scholar]
- Feng H., Craig H. L., Hope I. A., 2012. Expression pattern analysis of regulatory transcription factors in Caenorhabditis elegans. Methods Mol. Biol. 786: 21–50. [DOI] [PubMed] [Google Scholar]
- Ferguson E. L., Horvitz H. R., 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110: 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox R. M., Watson J. D., Von Stetina S. E., McDermott J., Brodigan T. M., et al. , 2007. The embryonic muscle transcriptome of C. elegans. Genome Biol. 8: R188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frand A. R., Russel S., Ruvkun G., 2005. Functional genomic analysis of C. elegans molting. PLoS Biol. 3: e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Takeshita H., Sawa H., 2007. Cyclin E and CDK2 repress the terminal differentiation of quiescent cells after asymmetric division in C. elegans. PLoS ONE 2: e407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama M., Gendreau S. B., Derry W. B., Rothman J. H., 2003. Essential embryonic roles of the CKI-1 cyclin-dependent kinase inhibitor in cell-cycle exit and morphogenesis in C elegans. Dev. Biol. 260: 273–286. [DOI] [PubMed] [Google Scholar]
- Gleason J. E., Eisenmann D. M., 2010. Wnt signaling controls the stem cell-like asymmetric division of the epithelial seam cells during C. elegans larval development. Dev. Biol. 348: 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason J. E., Korswagen H. C., Eisenmann D. M., 2002. Activation of Wnt signaling bypasses the requirement for RTK/Ras signaling during C. elegans vulval induction. Genes Dev. 16: 1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason J. E., Szyleyko E. A., Eisenmann D. M., 2006. Multiple redundant Wnt signaling components function in two processes during C. elegans vulval development. Dev. Biol. 298: 442–457. [DOI] [PubMed] [Google Scholar]
- Goh K. Y., Ng N. W., Hagen T., Inoue T., 2012. p21-Activated kinase interacts with Wnt signaling to regulate tissue polarity and gene expression. Proc. Natl. Acad. Sci. USA 109: 15853–15858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B., Takeshita H., Mizumoto K., Sawa H., 2006. Wnt signals can function as positional cues in establishing cell polarity. Dev. Cell 10: 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrepati L., Thompson K. W., Eisenmann D. M., 2013. C. elegans GATA factors EGL-18 and ELT-6 function downstream of Wnt signaling to maintain the progenitor fate during larval asymmetric divisions of the seam cells. Development 140: 2093–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravato-Nobre M. J., Nicholas H. R., Nijland R., O’Rourke D., Whittington D. E., et al. , 2005. Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics 171: 1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. L., Inoue T., Sternberg P. W., 2007. The C. elegans ROR receptor tyrosine kinase, CAM-1, non-autonomously inhibits the Wnt pathway. Development 134: 4053–4062. [DOI] [PubMed] [Google Scholar]
- Green J. L., Inoue T., Sternberg P. W., 2008. Opposing Wnt pathways orient cell polarity during organogenesis. Cell 134: 646–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald, I., 2005 Introduction to signal transduction (September 9, 2005), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.20.1, http://www.wormbook.org. 10.1895/wormbook.1.20.1 [DOI]
- Hao L., Johnsen R., Lauter G., Baillie D., Bürglin T. R., 2006. Comprehensive analysis of gene expression patterns of hedgehog-related genes. BMC Genomics 7: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T. W., Antoshechkin I., Bieri T., Blasiar D., Chan J., et al. , 2010. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 38: D463–D467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman M. A., 2001. C. elegans POP-1/TCF functions in a canonical Wnt pathway that controls cell migration and in a noncanonical Wnt pathway that controls cell polarity. Development 128: 581–590. [DOI] [PubMed] [Google Scholar]
- Herman M. A., Horvitz H. R., 1994. The Caenorhabditis elegans gene lin-44 controls the polarity of asymmetric cell divisions. Development 120: 1035–1047. [DOI] [PubMed] [Google Scholar]
- Hoier E. F., Mohler W. A., Kim S. K., Hajnal A. F., 2000. The Caenorhabditis elegans APC-related gene apr-1 is required for epithelial cell migration and Hox gene expression. Genes Dev. 14: 874–886. [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Roy R., Ambros V., 1998. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development 125: 3585–3597. [DOI] [PubMed] [Google Scholar]
- Hu J., Bae Y. K., Knobel K. M., Barr M. M., 2006. Casein kinase II and calcineurin modulate TRPP function and ciliary localization. Mol. Biol. Cell 17: 2200–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B. T., Lempicki R. A., 2009a Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B. T., Lempicki R. A., 2009b Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huang X., Tian E., Xu Y., Zhang H., 2009c The C. elegans engrailed homolog ceh-16 regulates the self-renewal expansion division of stem cell-like seam cells. Dev. Biol. 333: 337–347. [DOI] [PubMed] [Google Scholar]
- Hunter C. P., Harris J. M., Maloof J. N., Kenyon C., 1999. Hox gene expression in a single Caenorhabditis elegans cell is regulated by a caudal homolog and intercellular signals that inhibit Wnt signaling. Development 126: 805–814. [DOI] [PubMed] [Google Scholar]
- Hwang H.-Y., Horvitz H. R., 2002. The SQV-1 UDP-glucuronic acid decarboxylase and the SQV-7 nucleotide-sugar transporter may act in the Golgi apparatus to affect Caenorhabditis elegans vulval morphogenesis and embryonic development. Proc. Natl. Acad. Sci. USA 99: 14218–14223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino Y., Yamamoto M., 1998. Expression pattern of the C. elegans P21-activated protein kinase, CePAK. Biochem. Biophys. Res. Commun. 245: 177–184. [DOI] [PubMed] [Google Scholar]
- Inoue T., Oz H. S., Wiland D., Gharib S., Deshpande R., et al. , 2004. C. elegans LIN-18 is a Ryk ortholog and functions in parallel to LIN-17/Frizzled in Wnt signaling. Cell 118: 795–806. [DOI] [PubMed] [Google Scholar]
- Inoue T. D. R. Sherwood, G. Aspock, J.A. Butler, B. P. Gupta et al, 2002. Gene expression markers for Caenorhabditis elegans vulval cells. Mech. Dev. 119: S203–S209. [DOI] [PubMed] [Google Scholar]
- Jackson, B. M., P. Abete-Luzi, M. W. Krause, and D. M. Eisenmann, 2014 Use of an activated beta-catenin to identify Wnt pathway target genes in Caenorhabditis elegans, including a subset of collagen genes expressed in late larval development. G3 (Bethesda) 4: 733–747. [DOI] [PMC free article] [PubMed]
- Jackson B. M., Eisenmann D. M., 2012. Beta-catenin-dependent Wnt signaling in C. elegans: Teaching an old dog a new trick. Cold Spring Harb. Perspect. Biol. 4: a007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S., Schwab Y., Greenwald I., 2008. A Caenorhabditis elegans model for epithelial-neuronal transdifferentiation. Proc. Natl. Acad. Sci. USA 105: 3790–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L. I., Sternberg P. W., 1998. Interactions of EGF, Wnt and HOM-C genes specify the P12 neuroectoblast fate in C. elegans. Development 125: 2337–2347. [DOI] [PubMed] [Google Scholar]
- Joshi P. M., Riddle M. R., Djabrayan N. J., Rothman J. H., 2010. Caenorhabditis elegans as a model for stem cell biology. Dev. Dyn. 239: 1539–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T., Fujita M., Sakamoto H., 2000. Unique and redundant functions of SR proteins, a conserved family of splicing factors, in Caenorhabditis elegans development. Mech. Dev. 95: 67–76. [DOI] [PubMed] [Google Scholar]
- Kagoshima H., Shigesada K., Kohara Y., 2007. RUNX regulates stem cell proliferation and differentiation: insights from studies of C. elegans. J. Cell. Biochem. 100: 1119–1130. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J., 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. K., Gabel H. W., Kamath R. S., Tewari M., Pasquinelli A., et al. , 2005. Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167. [DOI] [PubMed] [Google Scholar]
- Koh K., Rothman J. H., 2001. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development 128: 2867–2880. [DOI] [PubMed] [Google Scholar]
- Komatsu H., Chao M. Y., Larkins-Ford J., Corkins M. E., Somers G. A., et al. , 2008. OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. PLoS Biol. 6: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korswagen H. C., Herman M. A., Clevers H. C., 2000. Distinct beta-catenins mediate adhesion and signalling functions in C. elegans. Nature 406: 527–532. [DOI] [PubMed] [Google Scholar]
- Korswagen H. C., Coudreuse D. Y., Betist M. C., van de Water S., Zivkovic D., et al. , 2002. The Axin-like protein PRY-1 is a negative regulator of a canonical Wnt pathway in C. elegans. Genes Dev. 16: 1291–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrouchova M., Krause M., Kostrouch Z., Rall J. E., 1998. CHR3: a Caenorhabditis elegans orphan nuclear hormone receptor required for proper epidermal development and molting. Development 125: 1617–1626. [DOI] [PubMed] [Google Scholar]
- Kostrouchova M., Krause M., Kostrouch Z., Rall J. E., 2001. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 98: 7360–7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitomo H., Uesugi H., Kohara Y., Iino Y., 2005. Identification of ciliated sensory neuron-expressed genes in Caenorhabditis elegans using targeted pull-down of poly(A) tails. Genome Biol. 6: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam N., Chesney M. A., Kimble J., 2006. Wnt signaling and CEH-22/tinman/Nkx2.5 specify a stem cell niche in C. elegans. Curr. Biol. 16: 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M., Hashimshony T., Wagner F., Yanai I., 2012. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev. Cell 22: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Lin L., Tran T., Hu S., Cramer T., Komuniecki R., et al. , 2012. RHGF-2 is an essential Rho-1 specific RhoGEF that binds to the multi-PDZ domain scaffold protein MPZ-1 in Caenorhabditis elegans. PLoS ONE 7: e31499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. J., Reece-Hoyes J. S., Walhout A. J., Eisenmann D. M., 2014. Multiple transcription factors directly regulate Hox gene lin-39 expression in ventral hypodermal cells of the C. elegans embryo and larva, including the hypodermal fate regulators LIN-26 and ELT-6. BMC Dev. Biol. 14: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman D., McGarvey T., McCracken S., Johnstone I. L., Blencowe B. J., et al. , 2001. Multiple interactions between SRm160 and SR family proteins in enhancer-dependent splicing and development of C. elegans. Curr. Biol. 11: 1923–1933. [DOI] [PubMed] [Google Scholar]
- Lucanic M., Kiley M., Ashcroft N., L’Etoile N., Cheng H. J., 2006. The Caenorhabditis elegans P21-activated kinases are differentially required for UNC-6/netrin-mediated commissural motor axon guidance. Development 133: 4549–4559. [DOI] [PubMed] [Google Scholar]
- Lund J., Tedesco P., Duke K., Wang J., Kim S. K., et al. , 2002. Transcriptional profiling of aging in C. elegans. Curr. Biol. 12: 1566–1573. [DOI] [PubMed] [Google Scholar]
- Maeda I., Kohara Y., Yamamoto M., Sugimoto A., 2001. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11: 171–176. [DOI] [PubMed] [Google Scholar]
- Maduro M. F., Kasmir J. J., Zhu J., Rothman J. H., 2005. The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development. Dev. Biol. 285: 510–523. [DOI] [PubMed] [Google Scholar]
- Mallo G. V., Kurz C. L., Couillault C., Pujol N., Granjeaud S., et al. , 2002. Inducible antibacterial defense system in C. elegans. Curr. Biol. 12: 1209–1214. [DOI] [PubMed] [Google Scholar]
- Maloof J. N., Whangbo J., Harris J. M., Jongeward G. D., Kenyon C., 1999. A Wnt signaling pathway controls hox gene expression and neuroblast migration in C. elegans. Development 126: 37–49. [DOI] [PubMed] [Google Scholar]
- McKay S. J., Johnsen R., Khattra J., Asano J., Baillie D. L., et al. , 2003. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 68: 159–169. [DOI] [PubMed] [Google Scholar]
- Mello C., Fire A., 1995. DNA transformation. Methods Cell Biol. 48: 451–482. [PubMed] [Google Scholar]
- Meissner B., Rogalski T., Viveiros R., Warner A., Plastino L., et al. , 2011. Determining the sub-cellular localization of proteins within Caenorhabditis elegans body wall muscle. PLoS ONE 6: e19937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux G., Gansmuller A., Hindelang C., Labouesse M., 2000. CHE-14, a protein with a sterol-sensing domain, is required for apical sorting in C. elegans ectodermal epithelial cells. Curr. Biol. 10: 1098–1107. [DOI] [PubMed] [Google Scholar]
- Miyabayashi T., Palfreyman M. T., Sluder A. E., Slack F., Sengupta P., 1999. Expression and function of members of a divergent nuclear receptor family in Caenorhabditis elegans. Dev. Biol. 215: 314–331. [DOI] [PubMed] [Google Scholar]
- Miyahara K., Suzuki N., Ishihara T., Tsuchiya E., Katsura I., 2004. TBX2/TBX3 transcriptional factor homologue controls olfactory adaptation in Caenorhabditis elegans. J. Neurobiol. 58: 392–402. [DOI] [PubMed] [Google Scholar]
- Mizumoto K., Sawa H., 2007. Two betas or not two betas: regulation of asymmetric division by beta-catenin. Trends Cell Biol. 17: 465–473. [DOI] [PubMed] [Google Scholar]
- Mochii M., Yoshida S., Morita K., Kohara Y., Ueno N., 1999. Identification of transforming growth factor-beta- regulated genes in Caenorhabditis elegans by differential hybridization of arrayed cDNAs. Proc. Natl. Acad. Sci. USA 96: 15020–15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalve G. C., Frand A. R., 2012. Toward a unified model of developmental timing: A “molting” approach. Worm 1: 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. I., Boyle T. J., Preston E., Vafeados D., Mericle B., et al. , 2012. Multidimensional regulation of gene expression in the C. elegans embryo. Genome Res. 22: 1282–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan L., Jackson B. M., Szyleyko E., Eisenmann D. M., 2004. Identification of evolutionarily conserved promoter elements and amino acids required for function of the C. elegans beta-catenin homolog BAR-1. Dev. Biol. 272: 536–557. [DOI] [PubMed] [Google Scholar]
- Nimmo R., Antebi A., Woollard A., 2005. mab-2 encodes RNT-1, a C. elegans Runx homologue essential for controlling cell proliferation in a stem cell-like developmental lineage. Development 132: 5043–5054. [DOI] [PubMed] [Google Scholar]
- Nusse, R., 2013 The Wnt Homepage, http://www.stanford.edu/group/nusselab/cgi-bin/wnt/.
- Page, A. P., and I. L. Johnstone, 2007 The cuticle (March 19, 2007), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.138.1, http://www.wormbook.org. 10.1895/wormbook.1.138.1 [DOI]
- Park M., Krause M. W., 1999. Regulation of postembryonic G(1) cell cycle progression in Caenorhabditis elegans by a cyclin D/CDK-like complex. Development 126: 4849–4860. [DOI] [PubMed] [Google Scholar]
- Partridge F. A., Tearle A. W., Gravato-Nobre M. J., Schafer W. R., Hodgkin J., 2008. The C. elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev. Biol. 317: 549–559. [DOI] [PubMed] [Google Scholar]
- Pauli F., Liu Y., Kim Y. A., Chen P. J., Kim S. K., 2006. Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development 133: 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P., 2012. Drugging Wnt signalling in cancer. EMBO J. 31: 2737–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Zhang H., 2010. Wnt signaling controls temporal identities of seam cells in Caenorhabditis elegans. Dev. Biol. 345: 144–155. [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes J. S., Shingles J., Dupuy D., Grove C. A., Walhout A. J., et al. , 2007. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics 8: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P., Stuart J., Lund J., Kim S., 2002. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature 418: 975–979. [DOI] [PubMed] [Google Scholar]
- Roy Chowdhuri S., Crum T., Woollard A., Aslam S., Okkema P. G., 2006. The T-box factor TBX-2 and the SUMO conjugating enzyme UBC-9 are required for ABa-derived pharyngeal muscle in C. elegans. Dev. Biol. 295: 664–677. [DOI] [PubMed] [Google Scholar]
- Rual J. F., Ceron J., Koreth J., Hao T., Nicot A. S., et al. , 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14: 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I., Ohler U., Burge C. B., Ruvkun G., 2007. Detection of broadly expressed neuronal genes in C. elegans. Dev. Biol. 302: 617–626. [DOI] [PubMed] [Google Scholar]
- Saito R. M., Perreault A., Peach B., Satterlee J. S., van den Heuvel S., 2004. The CDC-14 phosphatase controls developmental cell-cycle arrest in C. elegans. Nat. Cell Biol. 6: 777–783. [DOI] [PubMed] [Google Scholar]
- Salser S. J., Kenyon C., 1992. Activation of a C. elegans Antennapedia homologue in migrating cells controls their direction of migration. Nature 355: 255–258. [DOI] [PubMed] [Google Scholar]
- Sawa, H., and H. C. Korswagen, 2013 Wnt signaling in C. elegans (December 9, 2013), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.7.2, http://www.wormbook.org. 10.1895/wormbook.1.7.2 [DOI] [PMC free article] [PubMed]
- Schuijers J., Clevers H., 2012. Adult mammalian stem cells: the role of Wnt, Lgr5 and R-spondins. EMBO J. 31: 2685–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye D. D., Greenwald I., 2011. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS ONE 6: e20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Shao Z., Powell-Coffman J. A., 2006. The Caenorhabditis elegans rhy-1 gene inhibits HIF-1 hypoxia-inducible factor activity in a negative feedback loop that does not include vhl-1. Genetics 174: 1205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty P., Lo M. C., Robertson S. M., Lin R., 2005. C. elegans TCF protein, POP-1, converts from repressor to activator as a result of Wnt-induced lowering of nuclear levels. Dev. Biol. 285: 584–592. [DOI] [PubMed] [Google Scholar]
- Shtutman M., Zhurinsky J., Simcha I., Albanese C., D’Amico M., et al. , 1999. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 96: 5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Moorman C., van der Linden A. M., Kuijk E., van den Berghe P. V., et al. , 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S. P., Nonet M. L., et al. , 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12: 1317–1319. [DOI] [PubMed] [Google Scholar]
- Simske J. S., Hardin J., 2011. Claudin family proteins in Caenorhabditis elegans. Methods Mol. Biol. 762: 147–169. [DOI] [PubMed] [Google Scholar]
- Singh K., Chao M. Y., Somers G. A., Komatsu H., Corkins M. E., et al. , 2011. C. elegans Notch signaling regulates adult chemosensory response and larval molting quiescence. Curr. Biol. 21: 825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. A., Mango S. E., 2007. Role of T-box gene tbx-2 for anterior foregut muscle development in C. elegans. Dev. Biol. 302: 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D. H., Dominguez I., Mizuno J., Kaut M., Mohr S. C., et al. , 2003. CK2 phosphorylation of the armadillo repeat region of beta-catenin potentiates Wnt signaling. J. Biol. Chem. 278: 24018–24025. [DOI] [PubMed] [Google Scholar]
- Spencer W. C., Zeller G., Watson J. D., Henz S. R., Watkins K. L., et al. , 2011. A spatial and temporal map of C. elegans gene expression. Genome Res. 21: 325–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg, P. W., 2005 Vulval development (June, 25 2005), WormBook, ed. The C. elegans Research Community Wormbook, /10.1895/wormbook.1.6.1, http://www.wormbook.org. 10.1895/wormbook.1.6.1 [DOI] [PMC free article] [PubMed]
- Streit A., Kohler R., Marty T., Belfiore M., Takacs-Vellai K., et al. , 2002. Conserved regulation of the Caenorhabditis elegans labial/Hox1 gene ceh-13. Dev. Biol. 242: 96–108. [DOI] [PubMed] [Google Scholar]
- Takayama J., Faumont S., Kunitomo H., Lockery S. R., Iino Y., 2010. Single-cell transcriptional analysis of taste sensory neuron pair in Caenorhabditis elegans. Nucleic Acids Res. 38: 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita H., Sawa H., 2005. Asymmetric cortical and nuclear localizations of WRM-1/beta-catenin during asymmetric cell division in C. elegans. Genes Dev. 19: 1743–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsu O., McCormick F., 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398: 422–426. [DOI] [PubMed] [Google Scholar]
- Thein M. C., Winter A. D., Stepek G., McCormack G., Stapleton G., et al. , 2009. Combined extracellular matrix cross-linking activity of the peroxidase MLT-7 and the dual oxidase BLI-3 is critical for post-embryonic viability in Caenorhabditis elegans. J. Biol. Chem. 284: 17549–17563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel, S., 2005 Cell-cycle regulation (September 21, 2005), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.28.1, http://www.wormbook.org. 10.1895/wormbook.1.28.1 [DOI] [PMC free article] [PubMed]
- van der Bent M. L., Sterken M. G., Volkers R. J., Riksen J. A., Schmid T., et al. , 2014. Loss-of-function of beta-catenin bar-1 slows development and activates the Wnt pathway in Caenorhabditis elegans. Sci. Rep. 4: 4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad A., Rohrs S., Klein-Hitpass L., Muller O., 2008. The first five years of the Wnt targetome. Cell. Signal. 20: 795–802. [DOI] [PubMed] [Google Scholar]
- Von Stetina S. E., Fox R. M., Watkins K. L., Starich T. A., Shaw J. E., et al. , 2007a UNC-4 represses CEH-12/HB9 to specify synaptic inputs to VA motor neurons in C. elegans. Genes Dev. 21: 332–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina S. E., Watson J. D., Fox R. M., Olszewski K. L., Spencer W. C., et al. , 2007b Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol. 8: R135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukojevic V., Gschwind L., Vogler C., Demougin P., de Quervain D. J., et al. , 2012. A role for α-adducin (ADD-1) in nematode and human memory. EMBO J. 31: 1453–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagmaister J. A., Gleason J. E., Eisenmann D. M., 2006. Transcriptional upregulation of the C. elegans Hox gene lin-39 during vulval cell fate specification. Mech. Dev. 123: 135–150. [DOI] [PubMed] [Google Scholar]
- Wagner J., Allman E., Taylor A., Ulmschneider K., Kovanda T., et al. , 2011. A calcineurin homologous protein is required for sodium-proton exchange events in the C. elegans intestine. Am. J. Physiol. Cell Physiol. 301: C1389–C1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhao J., Corsi A. K., 2006. Identification of novel target genes of CeTwist and CeE/DA. Dev. Biol. 293: 486–498. [DOI] [PubMed] [Google Scholar]
- Wenick A. S., Hobert O., 2004. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev. Cell 6: 757–770. [DOI] [PubMed] [Google Scholar]
- Whangbo J., Harris J., Kenyon C., 2000. Multiple levels of regulation specify the polarity of an asymmetric cell division in C. elegans. Development 127: 4587–4598. [DOI] [PubMed] [Google Scholar]
- Yook K., Harris T. W., Bieri T., Cabunoc A., Chan J., et al. , 2012. WormBase 2012: more genomes, more data, new website. Nucleic Acids Res. 40: D735–D741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Wang Y., Huang B., Liang J., Ding Y., et al. , 2012. A Rac1/PAK1 cascade controls beta-catenin activation in colon cancer cells. Oncogene 31: 1001–1012. [DOI] [PubMed] [Google Scholar]
- Zugasti O., Rajan J., Kuwabara P. E., 2005. The function and expansion of the Patched- and Hedgehog-related homologs in C. elegans. Genome Res. 15: 1402–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.