Abstract
Precise genome editing by the Cas9 nuclease depends on exogenously provided templates for homologous recombination. Here, we compare oligonucleotides with short homology and circular DNA molecules with extensive homology to genomic targets as templates for homology-based repair of CRISPR/Cas9 induced double-strand breaks. We find oligonucleotides to be templates of choice for introducing small sequence changes into the genome based on editing efficiency and ease of use. We show that polarity of oligonucleotide templates greatly affects repair efficiency: oligonucleotides in the sense orientation with respect to the target gene are better templates. In addition, combining a gene loss-of-function phenotype screen with detection of integrated fluorescent markers, we demonstrate that targeted knock-ins in Caenorhabditis elegans also can be achieved by homology-independent repair.
Keywords: genome editing, CRISPR, Cas9, NHEJ, C. elegans
The repurposing of the Streptococcus pyogenes Cas9 nuclease (Jinek et al. 2012) has transformed genome editing in many organisms, including C. elegans [reviewed by Waaijers and Boxem (2014)]. This system consists, at its simplest, of an invariant Cas9 nuclease and a small chimeric guide RNA (sgRNA) containing 20 nucleotides of complementarity to the target DNA sequence (Jinek et al. 2012; 2013; Cong et al. 2013). The only absolute sequence requirement is the presence of the so-called protospacer adjacent motif (5′ NGG 3′) immediately downstream of the region of sgRNA complementarity in the genomic target (Jinek et al. 2012). Cas9-induced, double-stranded DNA breaks can be repaired either by nonhomologous end-joining (NHEJ) to introduce mutations in the genomic locus, or the repair can be templated by exogenous DNA, resulting in precise, homologous recombination-based, changes (Jinek et al. 2012; 2013; Cong et al. 2013; Mali et al. 2013).
Multiple approaches to obtaining C. elegans mutants by CRISPR/Cas9 have been published, yet they have not been methodically compared to maximize numbers of mutants obtained and minimize effort. We set out to compare different homologous recombination templates and resulting apparent recombinants, with the goal of being able to recommend preferred strategies.
We explored efficiency of homologous repair using commonly used templates—oligonucleotides with small regions of homology and circular DNA with extensive homology to the target site—because such a comparison has not yet been reported. Here we show that oligonucleotides are comparable with double-stranded plasmids as templates for homologous repair, but that there exists a polarity preference that affects their efficiency. We also show that single-stranded circular DNA can serve as a template for homologous repair after CRISPR/Cas9-induced double-stranded breaks. Finally, we demonstrate insertions of large constructs into precise sites in the C. elegans genome based on NHEJ.
Materials and Methods
Plasmids
The PU6::unc-119 sgRNA plasmid (Friedland et al. 2013) was modified so that the NotI site was inserted between the K09B11.12 U6 promoter and the sgRNA backbone, deleting the unc-119-specific sgRNA sequences. This plasmid was termed pIK111. Gene-specific 20-nt sgRNA sequences were subsequently cloned in. First, an oligonucleotide of the form 5′ AATTGCAAATCTAAATGTTT(20 nt sgRNA-specific sequence) GTTTTAGAGCTAGAAATAGC 3′ was hybridized with its reverse and complementary oligonucleotide. The hybrid is then cloned into NotI-digested pIK111 by Gibson assembly (Gibson et al. 2009).
The Peft-3::Cas9::tbb-2 3′UTR, PU6::sgRNA plasmid pDD162 (Dickinson et al. 2013) was similarly modified to facilitate cloning of sgRNAs, if desired, resulting in plasmid pIK155.
The PU6::sgRNA (F+E) plasmid, pIK198 (#65629; Addgene), where the Cas9 binding region of the sgRNA was extended and a PolIII terminator removed (Chen et al. 2013a), was created by Gibson assembly of an IDT gBlock into a pUC57 plasmid digested with EcoRI. The same backbone was used by Ward (2015) in C. elegans; however, the U6 promoter used in that study (R07E5.16) differs from the one in PU6::unc-119, pIK111 and pIK198 (K09B11.12). Gene-specific, 20-nt sgRNA sequences subsequently were cloned in. An oligonucleotide of the form 5′ AATTGCAAATCTAAATGTTT (20-nt sgRNA-specific sequence) GTTTAAGAGCTATGCTGGAA 3′ was hybridized with its reverse and complementary oligonucleotide. The hybrid is cloned into NotI-digested pIK198 by Gibson assembly (Gibson et al. 2009).
The nonhomologous end joining templates for unc-22 and lin-41 were created by inserting hybridized oligonucleotides containing the gene-specific sgRNA, 4 bp upstream and 6 bp downstream from its endogenous genomic locus and 20 bp homology arms, into the EcoRI site of pIK127 (Peft-3::gfp::h2b::tbb-2 3′UTR) (#65631; Addgene) or BglII site of pIK137 (Peft-3::gfp::h2b::tbb-2 3′UTR, C. briggsae unc-119) (#65632; Addgene) by Gibson assembly.
Phagemid templates for sqt-1 and lin-12 experiments were created by Gibson assembly of polymerase chain reaction (PCR) products from recombinant animals from oligonucleotide-templated experiments into pBluescript SK+. The sqt-1 phagemid has homology arms of 1.3 and 1.7 kb, respectively. The lin-12 phagemid has homology arms of 1.5 kb each.
Oligonucleotides
All oligonucleotide homologous recombination templates were ordered from (Integrated DNA Technologies) IDT and purified by polyacrylamide gel electrophoresis. Sequences for homologous recombination template oligonucleotides for sqt-1 and lin-12 modification are presented in Figure 1A. The sense repair oligonucleotide for dpy-10(cn64) was AF-ZF-827 (Arribere et al. 2014); the antisense oligo was its reverse and complement, oIK770.
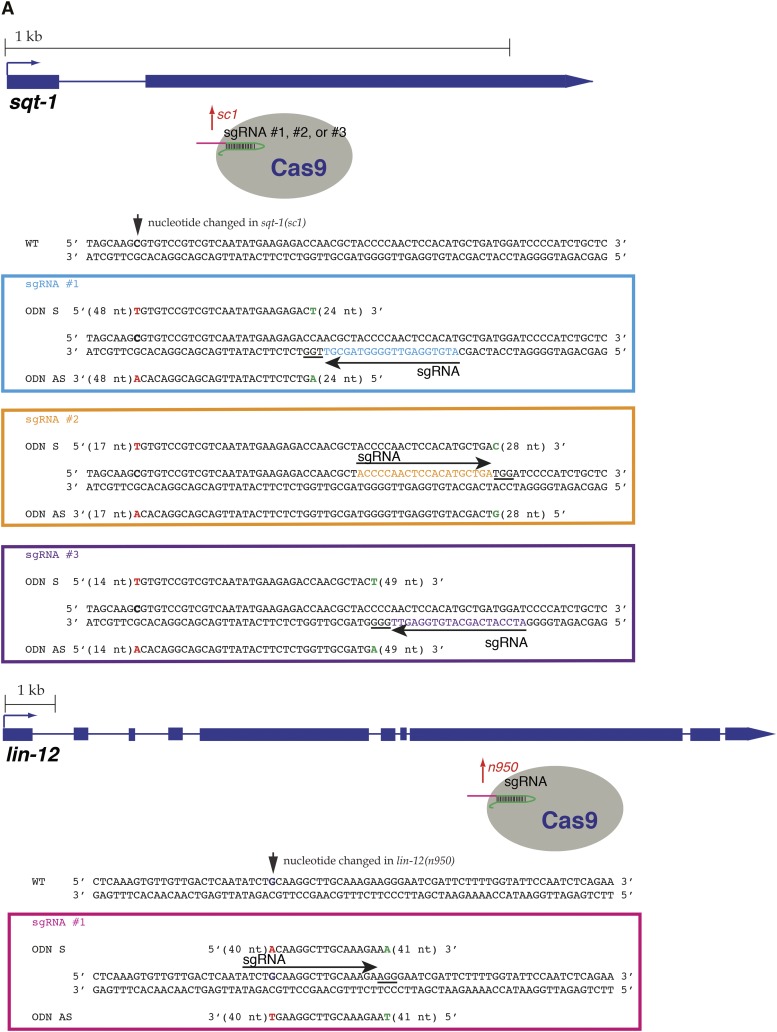
Figure 1.
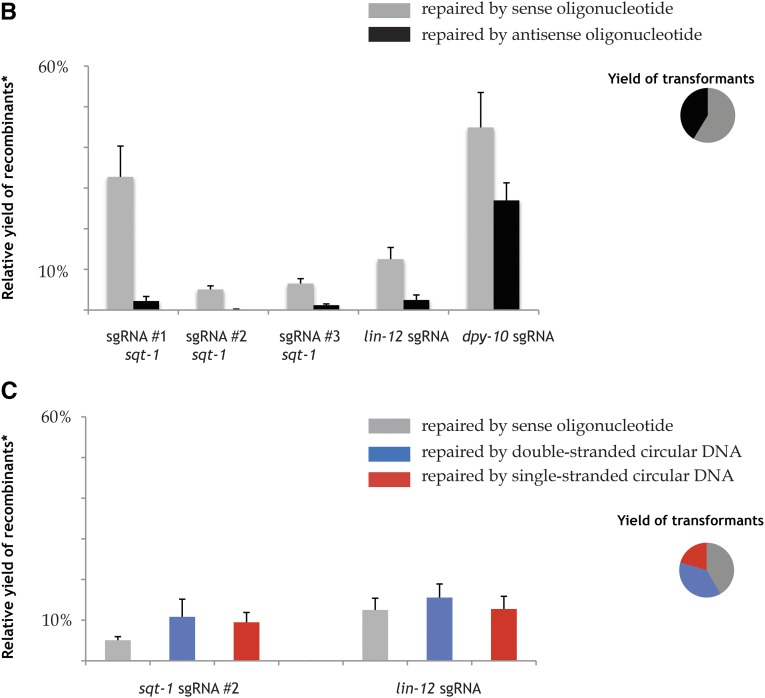
Homologous repair frequencies upon CRISPR/Cas9-induced double-strand breaks through oligonucleotides, double- and single-stranded circular DNA. (A) Sequences of three sgRNAs targeting sqt-1 and one sgRNA targeting lin-12, and DNA oligonucleotides (ODN) used as homologous recombination templates. 20-nt sgRNA target binding sites are shown as arrows next to the strand each is homologous to. Protospacer adjacent motif nucleotides are underlined. The nucleotide whose change results in a dominant mutation is shown in bold on the coding strand of the gene (C in sqt-1, G in lin-12). All oligonucleotides are 99-mers and contain the causative mutation, in red (T in sqt-1, A in lin-12) and a silent mutation, in green. For each oligonucleotide, the graphic shows the sequence of the two mutations and nucleotides between them, and gives the number of nucleotides in either recombination arm of the oligonucleotide. (B) Efficiency of repair by DNA oligonucleotides in the sense (gray) or antisense directions (black) in experiments using Cas9 and each sgRNA in turn. *Relative yield of recombinants in each experiment is calculated by dividing the number of mutant F1s with heritable mutations from each experiment by the number of green fluorescent protein (GFP)-positive animals resulting from the experiment, as a measure of microinjection efficiency. Error bars represent SEM (n = 3 separate experiments for each category). The pie chart represents the yield of GFP positive animals in 15 experiments using sense and 15 experiments using antisense oligonucleotides, respectively (1824 vs. 1283 animals). (C) Efficiency of repair by DNA oligonucleotides in the sense direction (gray), double-stranded (blue), and single-stranded circular DNA (red) in experiments using Cas9 and one sgRNA targeting sqt-1 and lin-12, respectively. *Relative yield of recombinants in each experiment is calculated by dividing the number of mutant F1s with heritable mutations from each experiment by the number of GFP-positive animals resulting from experiment, as a measure of micoinjection efficiency. Error bars represent SEM (n = 3 separate experiments for each category). The ± 95% CI intervals were 5.0 ± 3.4, 10.8 ± 17.0, and 9.5 ± 9.5% for sqt-1 sgRNA #2 oligonucleotides, double-stranded and single-stranded circular DNA templates, and 12.5 ± 11.3, 15.6 ± 13.1, and 12.7 ± 12.2% for lin-12 sgRNA oligonucleotides, double-stranded and single-stranded circular DNA templates, respectively. The pie chart represents the yield of GFP-positive animals in 6 experiments using sense oligonucleotides (784 animals), 6 experiments using double-stranded DNA (713 animals), and 6 experiments using single-stranded DNA (387 animals).
Injection mixes
All plasmids used for injection were purified by Nucleobond Xtra midi and Finalizer plus (740410.50 and 740520.20; MACHEREY-NAGEL). For sqt-1(sc1), lin-12(n950), and dpy-10(cn64) phenocopy, injection mixes contained 50 ng/μL pIK155 Peft-3::Cas9::tbb-2 3′ UTR, 100 ng/μL gene-specific sgRNA construct in pIK111 (sqt-1, lin-12) or pIK198 (dpy-10) PU6::sgRNA plasmid, 20 ng/μL Pmyo-3::gfp, and 50 ng/μL template oligonucleotide, or 100 ng/μL double-stranded circular template, or 50 ng/μL single-stranded circular template. Injections were performed in N2 animals. For comparison with the R07E5.16 promoter, sqt-1(sc1) sgRNA #2 was cloned in pIK155 Peft-3::Cas9::tbb-2 3′UTR, PU6::sgRNA plasmid. The injection mix contained 50 ng/μL of the resulting plasmid pIK177, 50 ng/μL template oligonucleotide, and 20 ng/μL Pmyo-3::gfp.
For unc-22 mutagenesis, injection mixes contained 50 ng/μL pIK155 Peft-3::Cas9::tbb-2 3′ UTR, 100 ng/μL gene-specific sgRNA construct in pIK198 PU6::sgRNA plasmid, and 20 ng/μL Pmyo-3::gfp. Injections were performed in N2 animals.
For inserting Peft-3::gfp::h2b::tbb-2 3′UTR into unc-22, the mix contained 5 ng/μL pCFJ104, 5 ng/μL pGH8, 2.5 ng/μL pCFJ90 (Frøkjær-Jensen et al. 2012), 100 ng/μL pIK155 Peft-3::Cas9::tbb-2 3′ UTR, 100 ng/μL pIK199 (5′ GCTCCATTGGTATGGTACCG 3′ sgRNA in F+E backbone pIK198 plasmid), 100 ng/μL pIK206 (5′ GACAAGCCGAAACCACCAAA 3′ sgRNA in F+E backbone pIK198 plasmid), and 100 ng/μL pIK211 (5′ TAAGGACAAGCCGAAACCACCAAAGGGTCC 3′ cloned in pIK137 [Peft-3::gfp::h2b::tbb-2 3′UTR, C. briggsae unc-119]). The mix was injected into HT1593 unc-119(ed3) animals.
For inserting Peft-3::gfp::h2b::tbb-2 3′UTR into lin-41, the mix contained 5 ng/μL pCFJ104, 5 ng/μL pGF8, 2.5 ng/μL pCFJ90 (Frøkjær-Jensen et al. 2012), 100 ng/μL pIK155 Peft-3::Cas9::tbb-2 3′ UTR, 100 ng/μL pIK123 (5′ GCTTCAAACTGAGATCGACG 3′ sgRNA in backbone pIK111 plasmid), and 100 ng/μL pIK204 (5′ GTCGGCTTCAAACTGAGATCGACGTGGACG 3′ cloned in pIK127 [Peft-3::gfp::h2b::tbb-2 3′UTR]). The mix was injected into N2 animals. In both cases, F2 progeny were screened for individuals exhibiting ubiquitous green fluorescence and no red fluorescence.
Single-stranded circular DNA generation
The double-stranded DNA in pBluescript SK+ phagemid vector was transformed into an F´ containing an Escherichia coli strain (XL1 Blue; Agilent Technologies). Transformants were infected with M13K07 helper phage as described in the manufacturer’s protocol (New England Biolabs). After harvesting cells by centrifugation, single-stranded DNA was purified by the PureLink HiPure Plasmid midiprep kit (Invitrogen; protocol version for single-stranded DNA purification).
PCR identification of NHEJ-mediated insertions in unc-22 and lin-41
The unc-22(bch26) insertion was identified by the following PCRs:
oIK803 (5′-AGAGAAGACCGTTCAACAACAGG-3′, in the unc-22 locus) –oIK777 (5′- TGCACATCTAACTCCTAGCACG-3′, in Peft-3 on the repair plasmid) and
oIK808 (5′-AGCCTCGTTCATCTCGATCTTTC-3′, in the unc-22 locus) – oIK818 (5′- TATGCGGCATCAGAGCAGATTG-3′, in the repair plasmid backbone).
oIK807 (5′-AGGAGCCACGGCATACATTC-3′, in the unc-22 locus) - oIK810 (5′-TGCCAATCTTTCTCGGACTTCTC-3′, in the unc-22 locus) PCR was performed to confirm that the unc-22 genomic locus was not modified through a sgRNA #1-mediated cut.
The lin-41(bch28) insertion was identified by the following PCR:
oIK796 (5′-CTTGTCATCAGCAGCCCTCG-3′, in the lin-41 locus) – oIK777 (5′- TGCACATCTAACTCCTAGCACG-3′, in Peft-3 on the repair plasmid)
oIK817 (5′- GTCCCGAGCCAAGATGATTATCC-3′, in the lin-41 locus) – oIK819 (5′- CTGCACATCTAACTCCTAGCACG-3′, in Peft-3 on the repair plasmid)
Results and Discussion
Oligonucleotides in the sense orientation with respect to the target gene are better repair templates
Repair of CRISPR/Cas9-induced lesions by oligonucleotides has been reported in C. elegans (Arribere et al. 2014; Zhao et al. 2014; Ward 2015). To compare relative repair frequencies by different templates, we focused on phenocopying three dominant mutations, sqt-1(sc1) (Cox et al. 1980), lin-12(n950) (Greenwald et al. 1983), and dpy-10(cn64) (Arribere et al. 2014), as phenotypic screening for recombinants can be performed in the first filial generation: lin-12(n950)/+ animals have a Multivulva phenotype, whereas sqt-1(sc1)/+ and dpy-10(cn64)/+ animals roll. We designed three sgRNAs to target sqt-1, one to target lin-12 (Figure 1A), and used a previously published sgRNA targeting dpy-10 (Arribere et al. 2014). In each experiment, wild-type (N2) animals were microinjected with a mix containing ubiquitously expressed Cas9, an sgRNA, a repair oligonucleotide in the sense or antisense direction with respect to the transcription of the gene, and a fluorescent marker. The oligonucleotides encode desired changes, which result in dominant phenotypes, and a silent change. Some of the silent changes disrupt sgRNA binding and prevent recutting of the recombinant genomic locus by Cas9. Each experiment was repeated three times, by two independent experimenters, and apparent F1 recombinants—roller animals or animals with multivulva phenotype, respectively—were scored for heritable phenotype segregation and presence of fluorescent marker expression (Table 1). The yield of recombinants resulting from each injection was normalized to the number of transgenic animals resulting from the injection; this normalization accounts for the efficiency of the microinjection procedure and any toxicity of the DNA mix.
Table 1. Heritability of dominant mutant phenotypes in F1 animals and their cosegregation with a fluorescent transformation marker.
| sgRNA and Recombination Template | Number of Mutant F1s | Heritable (%) | Of Heritable, Fluorescent (%) |
|---|---|---|---|
| Oligonucleotide templates | |||
| sqt-1 sgRNA #1 sense | 68 | 62 (91) | 2 (4)a |
| sqt-1 sgRNA #1 antisense | 2 | 2 (100) | 0 |
| sqt-1 sgRNA #2 sense | 25 | 19 (76) | 0 |
| sqt-1 sgRNA #2 antisense | 3 | 1 (33) | 0 |
| sqt-1 sgRNA #3 sense | 36 | 31 (86) | 4 (13) |
| sqt-1 sgRNA #3 antisense | 16 | 7 (44) | 1 (14) |
| lin-12 sense | 125 | 42 (34) | 12 (29) |
| lin-12 antisense | 73 | 4 (5) | 3 (75) |
| dpy-10 sense | 273 | 143 (52) | 53 (37) |
| dpy-10 antisense | 198 | 118 (60) | 36 (31) |
| Total oligonucleotide | 819 | 429 (52) | 111 (27)a |
| Double-stranded circular templates | |||
| sqt-1 sgRNA #2 | 99 | 9 (9) | 0 |
| lin-12 | 125 | 35 (28) | 14 (40) |
| Total double-stranded DNA | 224 | 44 (20) | 14 (32) |
| Single-stranded circular templates | |||
| sqt-1 sgRNA #2 | 105 | 15 (14) | 0 |
| lin-12 | 118 | 27 (23) | 9 (33) |
| Total single-stranded DNA | 223 | 42 (19) | 9 (21) |
The table shows the total number of mutant (roller or multivulva) animals resulting from microinjections with three sgRNAs targeting sqt-1, one sgRNA targeting lin-12, one sgRNA targeting dpy-10, and the Cas9 driven by the ubiquitous promoter of the eft-3 gene. Each mix was injected into 20-30 P0 animals in triplicate, except for dpy-10 sgRNA containing mixes, which were injected into 10-15 P0 animals in triplicate. Heritable changes are those that mutant animals segregate in the F2 generation (roller, dumpy, multivulva, and egg-laying deficient, respectively). The last column shows the proportion of F1 animals with heritable mutations which were also positive for the transgenic array.
The “Total oligonucleotide” row includes results of 30 separate microinjections with sense or antisense oligonucleotides as recombination templates.
Fluorescence status was known for just 412 animals. The “Total double-stranded DNA” and “Total single-stranded DNA” rows include results of 6 separate injections each.
In each case, we found that the oligonucleotide sense to the direction of gene transcription was a better recombination template, regardless of the DNA strand the sgRNA was complementary to (Figure 1B); observations consistent with ours also were made by Ward (2015) from a small number of co-conversion experiments. From our two studies, we have evidence that oligonucleotides sense to the direction of gene transcription are better recombination templates for seven pairs of oligonucleotides, targeting five genes, and irrespective of the DNA strand the sgRNA targets (Supporting Information, Table S1).
Similarly to Zhao et al. (2014) and Arribere et al. (2014), we find that recombinants are preferentially nontransgenic, i.e., they do not inherit the DNA array containing fluorescent markers, Cas9, sgRNAs, and oligonucleotide templates (Table 1), which has implications for experimental design. We also find that, using the Cas9 protein expressed from a ubiquitous eft-3 promoter, we obtain a proportion of apparent recombinant animals (rollers and animals with multivulvae, respectively) segregating only wild-type progeny (Table 1); it is likely that in these animals, recombination occurs in one or more somatic tissues during development.
In all experiments described here, we express sgRNAs from the K09B11.12 U6 promoter (Friedland et al. 2013). Farboud and Meyer (2015) reported failure to obtain rol-6(su1006) mutants using the K09B11.12 promoter, but were successful using the R07E5.16 promoter originally described by Dickinson et al. (2013). To compare these two promoters, we scored yields of heritable roller animals obtained from driving sqt-1(sc1) sgRNA #2 expression from the R07E5.16 promoter in the pDD162 Cas9 plasmid (Dickinson et al. 2013) with those obtained using the K09B11.12 promoter, in the context of the gene conversion mix with the oligonucleotide sense to sqt-1 transcription. The 95% confidence interval using 100 ng/μl of the K09B11.12-driven sgRNA was 5.0 ± 3.4% recombinants; for 50 ng/μL of the R07E5.16 promoter-driven sgRNA, it was 0.5 ± 0.1% recombinants. Thus, we continued to use the K09B11.12 promoter, which is also effective in co-conversion experiments (I. Katic, unpublished data).
Single-stranded oligonucleotides and double-stranded or single-stranded circular DNA with extensive homology to the target locus are comparable templates for homologous recombination (HR)
It had been reported that single-stranded DNA might be a better template than double-stranded DNA in fission yeast (Simon and Moore 1987) and that it can act as a HR template in mammalian cells (Fujioka et al. 1993). We tested whether this might be the case in C. elegans.
We again assayed phenocopy of dominant mutations in sqt-1 and lin-12, now using double-stranded phagemid DNA with >1 kb homology arms and corresponding circular single-stranded DNAs. We do not observe significant differences between sense oligonucleotides, double- and single-stranded circular DNA (Figure 1C), so for ease of experimental design, we recommend oligonucleotides as templates for nucleotide changes and short tag insertions. With all kinds of templates, we found that recombinants were found more often among nontransgenic animals: more than 70% of all recombination events happened in animals that did not inherit the fluorescent array (Table 1). If HR events are desired, therefore, co-conversion approaches (Arribere et al. 2014; Ward 2015) where an oligonucleotide-templated HR event at one locus is used to enrich for a desired modification of a different locus, appear to be preferable to cloning transgenic F1 animals and analyzing their progeny. We also attempted to generate single-stranded linear DNA templates by restriction digest of single-stranded circular DNA, but we observed widespread toxicity (I. Katic, unpublished data).
sgRNAs 2ith target site homology of >20 nt can also guide Cas9
We and others have previously shown that mismatches at the 5′ end of an sgRNA can be tolerated (Jinek et al. 2012; Cong et al. 2013; Fu et al. 2013; Hwang et al. 2013; Katic and Großhans 2013; Paix et al. 2014; Ward 2015; Farboud and Meyer 2015). DNA-based CRISPR approaches in C. elegans use U6 promoters to express sgRNAs (Friedland et al. 2013; Dickinson et al. 2013; Chen et al. 2013b). Although the exact sequence requirements of C. elegans U6 promoters have not been studied, transcription from a mouse U6 promoter initiates at the first A or G nucleotide starting from the annotated −1 position (Ma et al. 2014). The apparent requirement of a starting A or G reduces the choices of available sgRNAs with perfect complementarity to the target.
We compared a series of sgRNAs targeting the same sequence within sqt-1 but differing in length. All three sgRNAs are perfectly complementary to the target sequence and all have an A as the 5′-most nucleotide. They are 20, 26, and 30 nucleotides long. Although microinjection of the 20-nt sgRNA results in the greatest proportion of recombinants with respect to transgenic animals obtained (32%), the remaining sgRNAs also can form functional complexes with Cas9 (13% and 4% recombinants, respectively; Table S2), as was shown in mammalian systems (Qi et al. 2013; Ran et al. 2013). This flexibility in choosing an sgRNA of different length, in addition to the possibility of adding 5′ A or G nucleotides noncomplementary to the template (Katic and Großhans 2013; Paix et al. 2014; Ward 2015; Farboud and Meyer 2015) increases the pool of sgRNAs available to the C. elegans experimenter.
sgRNA effects on mutation rate
Efficiency of Cas9-mediated genome engineering crucially depends on sgRNA efficacy, which varies widely (summarized by Farboud and Meyer (2015) for C. elegans). To improve the stability of the sgRNA backbone, we synthesized a “flipped and extended” version (Chen et al. 2013a), where an A-U basepair is flipped within the sgRNA stem-loop to disrupt a potential Pol III terminator and the stem-loop is extended by 5 bp. We placed this new backbone behind the K09B11.12 U6 promoter (Friedland et al. 2013) to create a universal sgRNA cloning vector, pIK198, which will be available through Addgene. The identical backbone was used by Ward (2015), but the snRNA promoter in that study, R07E5.16, is different.
To test a model for prediction of sgRNA activity based on experimental data in mammalian systems (Doench et al. 2014), we designed four sgRNAs targeting the long 20th exon of the unc-22 gene and expressed them from the improved backbone. Two were predicted to be efficient (score >0.7) and two were poor (score <0.1). We tested the mutagenicity of each sgRNA separately and found that the two sgRNAs predicted to be efficient indeed resulted in more Unc-22 animals in total than the two predicted to be inefficient (102 vs. 15 mutant animals obtained; Table S3). In addition, we analyzed C. elegans sgRNAs with published mutagenesis efficiencies according to the Doench et al. (2014) algorithm. Of the 46 sgRNAs that met our criteria, 34 showed a range of levels of activity, while 12 did not result in any mutants. Of the 12 inactive sgRNAs, 10 have scores of <0.2 in the Doench et al. (2014) algorithm, one has a score of 0.25, and one a high score of 0.67 (Table S4). Furthermore, of the 23 sgRNAs with scores of <0.2, 10 are inactive. Recently, Farboud and Meyer (2015) proposed and validated a model for designing efficient sgRNAs targeting C. elegans genes whose requirements are 5′ N17NGG(NGG)3′, where the protospacer adjacent motif sequence is in parentheses. When these sequence requirements are difficult to meet for the desired modification, our analysis suggest that low scoring sgRNAs from the Doench et al. (2014) model should be used with caution, and high scoring ones considered.
NHEJ-mediated knock-ins
Precise genome modification by homologous recombination is crucial in many cases, such as when precisely tagging a protein. However, despite continued improvements in screening techniques (Kim et al. 2014; Arribere et al. 2014; Ward 2015) and template optimization (Paix et al. 2014), we and others have had difficulties achieving consistent HR-mediated modification events (Farboud and Meyer (2015), and our unpublished results). On the other hand, NHEJ is clearly active in the germline of C. elegans, or in very early embryos, as heritable mutations resulting from Cas9 when a recombination template is not provided are repaired by that mechanism. NHEJ approaches have been used to achieve knock-ins upon ZFN and TALEN cleavage in cell lines (Maresca et al. 2013), as well as Cas9-mediated cleavage in zebrafish (Auer et al. 2014).
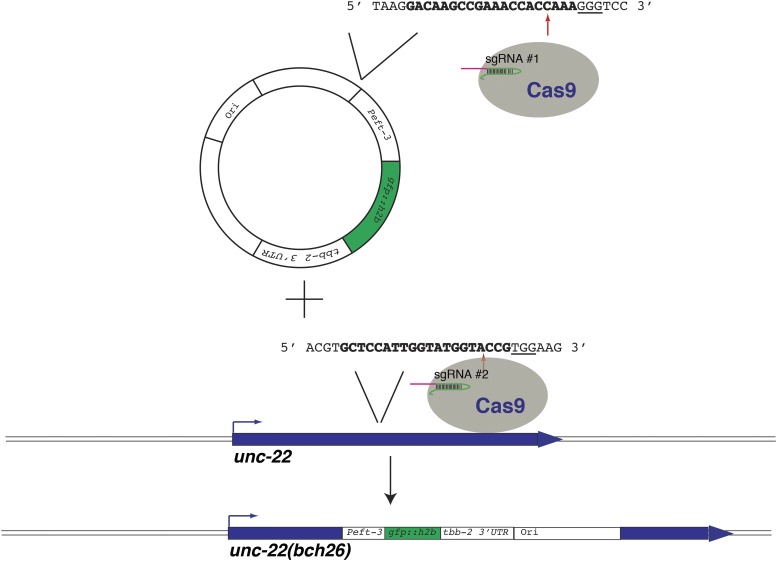
We explored the possibility of targeting insertion of a bright Peft-3::gfp::h2b::tbb-2 3′UTR construct into a genomic locus through NHEJ (Figure 2). We cloned a previously tested sgRNA targeting site for unc-22, whose corresponding sgRNA cuts efficiently, and its immediate genomic context, into a plasmid containing Peft-3::gfp::h2b::tbb-2 3′UTR and a C. briggsae unc-119 rescuing fragment, upstream of the transgenes. The 30 nucleotide length was chosen as the immediate context was shown to affect efficiency of sgRNA cutting the genomic site in mammalian cells (Doench et al. 2014). We reasoned that concomitant cutting of the plasmid and the genomic target might, in some cases, result in NHEJ-mediated knock-in of the plasmid into the genomic locus, as was shown by Auer et al. (2014) in zebrafish. We microinjected 30 unc-119(ed3) animals with a mixture containing Cas9, two sgRNAs targeting unc-22, the NHEJ targeting construct, and a mixture of red fluorescent markers (Frøkjær-Jensen et al. 2012) and screened them analogously to MosSCI (Frøkjaer-Jensen et al. 2008; Frøkjær-Jensen et al. 2012): integration of the transgene results in loss of red fluorescent markers, but non-Unc-119 animals are present on plates. One F2 animal expressed bright, ubiquitous green fluorescent protein (GFP) and no red markers, and twitched when moving, which is a phenotype of unc-22(lof) alleles (Moerman et al. 1988). The resulting strain was termed unc-22(bch26). We backcrossed this mutant to the wild-type parent strain twice and performed genetic linkage analysis. 189/189 Unc-22 animals segregating from unc-22(bch26)/+ parents give rise to only GFP positive progeny, showing that Peft-3::gfp::h2b::tbb-2 3′UTR insertion is closely linked to the unc-22 locus. Subsequent PCR analysis and sequencing of the mutant revealed that the insertion disrupts the unc-22 locus, as designed (Figure S1 and Figure S2).
Figure 2.
Knock-in of a plasmid into a genomic locus upon nonhomologous end joining-mediated repair of a Cas9/CRISPR lesion. Knock-in of a Peft-3::gfp::h2b::tbb-2 3′UTR –containing plasmid into the unc-22 locus. The 30-bp genomic sequence from the unc-22 locus was cloned into the Peft-3::gfp::h2b::tbb-2 3′UTR, C. briggsae unc-119 rescue fragment –containing plasmid, including the 20-bp sgRNA #1 target site (bold) and the protospacer adjacent motif sequence (underlined). Cas9 guided by two unc-22 sgRNAs concomitantly cuts the plasmid and the genomic locus, which can then lead to insertion of the cut plasmid into the genomic lesion. In the case of the insertion allele unc-22(bch26), sgRNA #1 guided Cas9 to the plasmid, resulting in a cut, but there was no evidence of a cut in the target site of the sgRNA #1 in the genomic locus. sgRNA #2 guided Cas9 to cut the genomic locus, which was repaired by insertion of the linearized plasmid, as shown.
On the basis of the brightness of green fluorescence in the unc-22(bch26) animals, we repeated the injection into 30 N2 animals, reasoning that screening by Unc-119 phenotype rescue was not essential. In this experiment, we also identified one GFP- but not mCherry-expressing, twitcher animal upon chunking starved P0 plates. However, this integration was not genetically linked to unc-22—perhaps inserting into an off-target site of one of the sgRNAs used—and was not analyzed further.
To show that this method could be used to target a different gene—one we have previously unsuccessfully tried to modify through homologous recombination—we cloned an sgRNA targeting site for lin-41 and its immediate genomic context into the Peft-3::gfp::h2b::tbb-2 3′UTR plasmid, 5′ to the transgene. This plasmid did not contain unc-119 rescuing sequences. As described previously, we injected the mix, this time containing just a single sgRNA, into gonads of 30 N2 animals and obtained one candidate GFP-expressing, but not mCherry-expressing, animal at the F2 stage. This animal segregated two kinds of GFP-positive progeny: sickly, dumpy animals that often died in early adulthood or were sterile (Figure S1B)—lin-41 loss-of-function candidates (Slack et al. 2000)—and apparent wild-type animals. It also segregated nonfluorescent, wild-type animals, so it appeared to have been heterozygous for the insert. We termed it lin-41(bch28), backcrossed it to the wild-type parent strain and then mated it with lin-41(xe8) mutants, which have a partial 3′ UTR deletion, resulting in inability of lin-41 to be down-regulated by let-7 (Ecsedi et al. 2015). Heterozygous lin-41(xe8)/lin-41(bch28 [Peft-3::gfp::h2b::tbb-2 3′ UTR]) animals are fertile and segregate GFP and non−GFP-expressing animals. All 44 of the non-GFP-expressing animals burst after vulva eversion and are homozygous for the xe8 deletion (M. Rausch, unpublished data). On sequencing analysis of the lin-41 locus in the lin-41(bch28) animals, we observe a complex insert, containing sequences from at least two copies of the Peft-3::gfp::h2b::tbb-2 3′UTR plasmid, one of them truncated (Figure S1 and Figure S3). The insertion occurred in the targeted sgRNA binding site, as designed.
This approach potentially can be generalized to targeting any site in the C. elegans genome. It requires very simple cloning of a 30-bp sgRNA sequence by oligonucleotide hybridization and Gibson assembly into a linearized vector, followed by microinjection and screening, analogous to MosSCI. Frøkjær-Jensen et al. (2014) have shown that Peft-3::tdTomato::h2b single-copy inserts are visible under a dissecting microscope, so a similar approach to a balancer labeled with red fluorescence should also be feasible. As there are still regions in the C. elegans genome without a balancer, and conventional balancers can break (Edgley et al. 2006), this method might simplify and accelerate progress for researchers whose experiments depend on successful balancing. We note that Frøkjær-Jensen et al. (2014) have recently made available a large set of fluorescent insertions. Alone or in combination with genome editing, they can serve as balancers. However, our NHEJ-based method can be used when a very stable, fluorescently-labeled balancer is desired, as recombination between the balanced locus and the fluorescent marker should be virtually absent.
In conclusion, we show that oligonucleotides, double- and single-stranded circular DNA are comparable templates for homologous recombination-based changes of a few nucleotides upon Cas9 cleavage, and that oligonucleotide-templated repair appears to depend on polarity of the oligonucleotides. Changes in three genes occurred more efficiently when the template oligonucleotide was oriented in the sense direction to the gene transcription, through an unknown mechanism. We also recommend screening for recombination events in the F2 generation, because the most widely used constructs at this time, where the nuclease is expressed from the ubiquitous eft-3 promoter, also lead to somatic recombination in the F1 generation. For all kinds of templates, we observe preferential recombinational repair in those progeny that will not inherit the DNA transgene array. Finally, we show that exogenous DNA can be inserted into precise loci of the genome by NHEJ through a simple method.
Supplementary Material
Acknowledgments
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). I.K. and L.X. are members of the Friedrich Miescher Institute (FMI) C. elegans facility, which is supported by FMI core funding. We are grateful to Marcello Maresca for bringing nonhomologous end joining (NHEJ)-based knock-ins in other model systems to our attention. We thank Helge Großhans for helpful discussions and critical reading of the manuscript. We are grateful to Magdalene Rausch for help with experiments involving lin-41(xe8). We thank the Calarco laboratory for the gift of U6::unc-119 sgRNA plasmid (through Addgene, #46169) and the Goldstein laboratory for the gift of pDD162 (through Addgene, #47549).
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.019273/-/DC1
Communicating editor: B. J. Andrews
Literature Cited
- Arribere J. A., Bell R. T., Fu B. X. H., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer T. O., Duroure K., De Cian A., Concordet J.-P., Del Bene F., 2014. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 24: 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Gilbert L. A., Cimini B. A., Schnitzbauer J., Zhang W., et al. , 2013a Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155: 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Fenk L. A., de Bono M., 2013b Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acids Res. 41: e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G. N., Laufer J. S., Kusch M., Edgar R. S., 1980. Genetic and phenotypic characterization of roller mutants of Caenorhabditis elegans. Genetics 95: 317–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J. G., Hartenian E., Graham D. B., Tothova Z., Hegde M., et al. , 2014. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecsedi M., Rausch M., Großhans H., 2015. The let-7 microRNA directs vulval development through a single target. Dev. Cell 32: 335–344. [DOI] [PubMed] [Google Scholar]
- Edgley M. L., Baillie D. L., Riddle D. L., Rose A. M., 2006 Genetic balancers. (April 6, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.89.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Farboud B., Meyer B. J., 2015. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics 199: 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiácovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Sarov M., Taylor J., Flibotte S., et al. , 2014. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11: 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., et al. , 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka K., Aratani Y., Kusano K., Koyama H., 1993. Targeted recombination with single-stranded DNA vectors in mammalian cells. Nucleic Acids Res. 21: 407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A., et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Greenwald I. S., Sternberg P. W., Horvitz H. R., 1983. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34: 435–444. [DOI] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Kaini P., et al. , 2013. Heritable and precise Zebrafish genome editing using a CRISPR-Cas system. PLoS ONE 8: e68708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E., et al. , 2013. RNA-programmed genome editing in human cells. eLife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic I., Großhans H., 2013. Targeted heritable mutation and gene conversion by Cas9-CRISPR in Caenorhabditis elegans. Genetics 195: 1173–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ishidate T., Ghanta K. S., Seth M., Conte D., et al. , 2014. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics 197: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Wu Y., Dang Y., Choi J.-G., Zhang J., et al. , 2014. Pol III promoters to express small RNAs: Delineation of Transcription Initiation. Mol Ther Nucleic Acids 3: e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. , 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca M., Lin V. G., Guo N., Yang Y., 2013. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 23: 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman D. G., Benian G. M., Barstead R. J., Schriefer L. A., Waterston R. H., 1988. Identification and intracellular localization of the unc-22 gene product of Caenorhabditis elegans. Genes Dev. 2: 93–105. [DOI] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H. E., Lee C.-Y. S., Calidas D., et al. , 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 Sites in Caenorhabditis elegans. Genetics 198: 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., et al. , 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Lin C.-Y., Gootenberg J. S., Konermann S., et al. , 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. R., Moore P. D., 1987. Homologous recombination between single-stranded DNA and chromosomal genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 2329–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack F. J., Basson M., Liu Z., Ambros V., Horvitz H. R., et al. , 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. [DOI] [PubMed] [Google Scholar]
- Waaijers S., Boxem M., 2014. Engineering the Caenorhabditis elegans genome with CRISPR/Cas9. Methods 68: 381–388. [DOI] [PubMed] [Google Scholar]
- Ward J. D., 2015. Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair. Genetics 199: 363–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Zhang Z., Ke H., Yue Y., Xue D., 2014. Oligonucleotide-based targeted gene editing in C. elegans via the CRISPR/Cas9 system. Cell Res. 24: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.