Abstract
Following the two rounds of whole-genome duplication that occurred during deuterostome evolution, a third genome duplication event occurred in the stem lineage of ray-finned fishes. This teleost-specific genome duplication is thought to be responsible for the biological diversification of ray-finned fishes. DEAD-box polypeptide 3 (DDX3) belongs to the DEAD-box RNA helicase family. Although their functions in humans have been well studied, limited information is available regarding their function in teleosts. In this study, two teleost Ddx3 genes were first identified in the transcriptome of Japanese flounder (Paralichthys olivaceus). We confirmed that the two genes originated from teleost-specific genome duplication through synteny and phylogenetic analysis. Additionally, comparative analysis of genome structure, molecular evolution rate, and expression pattern of the two genes in Japanese flounder revealed evidence of subfunctionalization of the duplicated Ddx3 genes in teleosts. Thus, the results of this study reveal novel insights into the evolution of the teleost Ddx3 genes and constitute important groundwork for further research on this gene family.
Keywords: DDX3, genome duplication, teleost, evolution, subfunctionalization
Helicase proteins are classified into three superfamilies and two families (SF1−SF5) based on their conserved motifs (Gorbalenya and Koonin 1993), of which motif II and the Walker B motif are found in all helicases (Walker et al. 1982). DEAD-box RNA helicases are members of superfamily 2 (SF2) and are characterized by 12 conserved motifs (Linder and Fuller-Pace 2013). The DEAD-box helicase family is named after the amino acid sequence Asp-Glu-Ala-Asp (D-E-A-D) of its motif II (Linder et al. 1989). These proteins have important functions in RNA metabolism and are associated with processes involving RNA, such as transcription and degradation (Rocak and Linder 2004). On the basis of their sequences and functions, DEAD-box RNA helicases consist of three subfamilies, including the DEAD-box polypeptide 3 (Ddx3) gene family.
The human genome contains two functional genes of the Ddx3 family: Ddx3X located in chromosome X (Park et al. 1998) and Ddx3Y located in chromosome Y (Lahn and Page 1997). DDX3X is implicated in nucleocytoplasmic shuttling with RNA-dependent ATPase/helicase activity (You et al. 1999; Yedavalli et al. 2004). DDX3X also reportedly participates in various mRNA and protein biogenesis steps, including transcription (Chao et al. 2006), mRNA migration (Kanai et al. 2004), pre-mRNA splicing (Zhou et al. 2002), and translation (Lai et al. 2008), which suggests that DDX3X has a regulatory role in gene expression. In contrast, the human DDX3Y gene, which lies in the azoospermia factor a region, is involved in male fertility. Deletion of Ddx3Y results in oligozoospermia, azoospermia, and male sertoli-cell only syndrome (Foresta et al. 2001; Kuo et al. 2004). Furthermore, the human Ddx3Y gene is considered to be one of the genes essential for human spermatogenesis and male fertility. Although several studies have contributed to our understanding of the function of DDX3 in development, studies have been rarely conducted to elucidate the role of the Ddx3 gene family in teleosts.
Studies have provided evidence of multiple rounds of whole-genome duplication in vertebrate lineages. In particular, two successive rounds of whole-genome duplication are thought to have occurred near the base of vertebrate lineages and played a significant role in vertebrate evolution (Meyer 1998; Hoegg and Meyer 2005; Van De Peer et al. 2009; Hoffmann et al. 2012). At ∼350Ma, a third-round whole-genome duplication event occurred in the common ancestor of ray-finned fishes (Amores et al. 1998; Christoffels et al. 2004; Jaillon et al. 2004; Vandepoele et al. 2004; Meyer and Van De Peer 2005; Brunet et al. 2006; Amores et al. 2011). Teleost-specific genome duplication (TGD) likely provided gene copies that contributed to evolutionary radiation and phenotypic diversification of teleost fishes. Studies on TGD-derived gene duplicates that evolved distinct physiological or developmental functions in various teleost lineages provide evidence, supporting a cause−effect relationship between gene copy number and species diversity (Braasch et al. 2006, 2009; Mulley et al. 2006; Hoegg and Meyer 2007; Siegel et al. 2007; Liu et al. 2009; Liu et al. 2013; Li et al. 2014). This finding shows that duplicated genes may have diverged from the roles of their ancestors, and this divergence could be demonstrated by changes in evolutionary rates, expression patterns, and regulatory mechanisms. On the basis of a duplication−degeneration−complementation model, duplicated genes may have three main fates, including nonfunctionalization (i.e., one of the two copies is lost or inactivated by mutation), subfunctionalization (i.e., both copies assume more specialized functions than those of their ancestor), and neofunctionalization (i.e., one copy evolves a new function) (Ohno 1970; Hughes 1994; Force et al. 1999; Lynch and Conery 2000; Lynch and Force 2000). Studies have revealed a combination of the last two fates, termed subneofunctionalization, when rapid subfunctionalization is accompanied by prolonged and substantial rates of neofunctionalization in duplicated gene evolution (He and Zhang 2005).
In this study, two homologous Ddx3 genes initially were identified from the transcriptome of Japanese flounder and other teleost genomes. We conducted synteny and phylogenetic analyses of vertebrate Ddx3 genes. Then, comparisons of genomic structure, molecular evolutionary rate, and expression pattern of the two Ddx3 genes in Japanese flounder were performed; results revealed probable subfunctionalization of teleost duplicated Ddx3 genes. This study lays the foundation for evolutionary and functional studies of the Ddx3 gene family in teleosts.
Materials and Methods
Ethics statement
Japanese flounder (Paralichthys olivaceus) samples were collected from local aquatic farms. Permission to collect samples was obtained from the local government of Yantai, Shandong, China. All of the procedures complied with and were approved by the Institutional Animal Care and Use Committee of the Ocean University of China.
Fish
Randomly selected 2-yr-old healthy adult Japanese flounder (five females and five males) were dissected. Muscle, gill, heart, intestine, brain, kidney, liver, spleen, and gonad tissues were collected. Each sample was collected in triplicate. All of the samples were immediately frozen using liquid nitrogen and stored at −80° for total RNA or genomic DNA preparation.
RNA and genomic DNA extraction
Total RNA was extracted using Trizol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The extracted total RNA was treated with RNase-free DNase I (TaKaRa, Dalian, China) to degrade genomic DNA and then frozen at −80°. cDNA was synthesized using 1 µg of total RNA and random hexamer primers with reverse transcriptase M-MLV (RNase H−) kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. Genomic DNA was isolated from muscle tissues by the traditional phenol/chloroform extraction method. The quality and the quantity of RNA and DNA were evaluated by 1.5% agarose gel electrophoresis and spectrophotometry using NanoPhotometer Pearl (Implen, Munich, Germany).
Data collection
The human Ddx3X/Y coding sequences were used as queries for local TBLASTX searches against the transcriptome (SRA, Accession number SRX500343) and genome (unpublished) of Japanese flounder to manually predict putative Ddx3 genes. Other Ddx3 genes were identified from TBLASTX search against the genome database at the National Center for Biotechnology Information or Ensembl over the internet. The Assembly ID and Accession numbers are provided in Supporting Information, Table S1.
Sex-specific amplification of Ddx3 in Japanese flounder
We designed specific primers (Table S2) based on the genomic sequence of each Ddx3 gene from Japanese flounder. Polymerase chain reaction (PCR) amplification was performed using the isolated genomic DNA as templates under the following conditions: initial denaturation at 95° for 5 min, followed by 30 cycles at 95° for 30 sec, at 60° for 30 sec, and at 72° for 1 min, and a final extension at 72° for 10 min. PCR products were examined by 1.5% agarose gel electrophoresis.
Phylogenetic analysis
The coding sequences of vertebrate Ddx3 genes were aligned in MAFFT v7 (Katoh and Standley 2013). Alignment was carefully checked and gaps and ambiguous sites were removed before phylogenetic analyses were performed. We used ModelGenerator (Keane et al. 2006) to choose an appropriate model of sequence evolution for the alignment. The Bayesian method was used as implemented in MrBayes 3.2 (Huelsenbeck et al. 2001; Ronquist et al. 2012). Maximum likelihood (ML) phylogeny was reconstructed by MEGA6 (Tamura et al. 2013), and the branching reliability was tested via bootstrap resampling with 1000 replicates.
Synteny analysis
Annotated genes surrounding Ddx3 genes were extracted from the genome databases at the National Center for Biotechnology Information or Ensembl. These genes were mapped according to their relative locations in the chromosome to perform synteny analysis. Then, the Synteny Database was used to detect conserved synteny between chromosomes containing Ddx3 genes (Catchen et al. 2009).
Genomic structure analysis
The coding sequences of teleost Ddx3 genes were used as queries for BLASTn searches against the corresponding genomic sequences to find exon−intron boundaries. The genomic structure of each gene was illustrated according to the size and position of exons and introns.
Alignments of the deduced amino acid sequences of teleost DDX3a/b proteins were conducted by MAFFT v7 (Katoh and Standley 2013). Locations of the conserved motifs were marked based on the genomic structure.
Tests for positive selection of teleost Ddx3 genes
Alignments, along with Bayesian trees and ML trees, were constructed as described. These alignments were then analyzed with the CODEML package from PAML (Pathology Associated Medical Laboratory) v4.7 (Yang 2007) to estimate the strength and the form of selection among teleost Ddx3 genes. Nonsynonymous (dN) and synonymous (dS) substitution rate ratios (dN/dS or ω) were estimated with various site models.
The model M0 assumes a constant ω ratio whereas M3 assumes three classes of ω. The null model M1a assumes two classes of codon sites for ω (0 < ω < 1 and ω = 1), whereas the alternative model M2a assumes one additional class of site with ω > 1. The M7 model assumes that ω follows a beta distribution with 10 categories, each corresponding to a distinctive ω value that is always less than 1, whereas the M8a model allows for an extra class of codons with ω = 1. The alternative model M8 has an extra category with ω > 1 (Yang et al. 2000; Swanson et al. 2001; Wong et al. 2004). Comparisons between the two nested site models were performed to evaluate the variation in ω (M3 vs. M0) and to determine the presence of a positively selected class of sites (M2a vs. M1a; M8 vs. M7 and M8 vs. M8a). The M2a−M1a comparison appears to be less powerful than the M8−M7 comparison according to the author (Yang 2007).
Analyses were run starting with branch lengths estimated from the Ddx3 gene tree and repeated thrice with varying initial starting points of κ (transition to transversion rate ratio) and ω (0.4, 1, and 4, respectively) as recommended to check multiple local optima. The model pairs were compared using a likelihood ratio test with χ2 distribution.
Tissue distribution pattern of Ddx3 genes in Japanese flounder
Two specific primer pairs (Table S2) were designed for the Japanese flounder Ddx3a and Ddx3b genes. A pre-experiment was conducted to confirm single cDNA PCR product and avoid genomic DNA amplification. Specific PCR products were verified by sequencing. Five biological replicates of each sample were analyzed, and each sample was run in triplicate. Quantitative real-time PCR was performed with SYBR Premix Ex Taq II (TaKaRa, Dalian, China) by using LightCycler480 (Roche Applied Science, Mannheim, Germany) at 95° (5 min) for preincubation followed by 40 cycles at 95° (15 sec) and 60° (45 sec). The melting curve was analyzed to detect single amplification. Accumulation of fluorescent signal from SYBR Green I was recorded at the 60° (45 sec) phase during each cycle in LightCycler480 Software 1.5. Negative control (no-template reaction) was included.
Relative expression was determined using 18S rRNA as the reference gene, as previously described (Zhong et al. 2008; Gao et al. 2013). The target gene was relatively quantified and expressed as fold variation of the reference gene 18S rRNA by the 2-ΔΔCq comparative Cq method. Data were statistically analyzed by one-way analysis of variance followed by a Tukey’s post-hoc test using SPSS 20.0 (SPSS, Chicago, IL). P < 0.05 was considered to indicate statistically significant difference.
Results
Identification of Ddx3 genes
Using TBLASTX searches with E-value at or effectively 0, we identified two Ddx3 genes from Japanese flounder transcritptome and genome. Two Ddx3 genes also were identified from the genomes of other teleost species such as zebrafish, torafugu, and Nile tilapia. Spotted gar was the exception, with only a single Ddx3 gene. In addition to humans, Ddx3X/Y could also be found in other eutherians, such as house mouse, cattle, and chimpanzee. Only one Ddx3 gene was extracted from the genomes of other species, such as elephant shark, coelacanth, and chicken.
Using genomic DNA from male or female Japanese flounder as templates, we found that the amplification products of Japanese flounder Ddx3 genes showed no sexual specificity (Figure S1).
Phylogenetic analysis of vertebrate Ddx3 genes
According to the Akaike Information Criterion and Bayesian Information Criterion from ModelGenerator (Keane et al. 2006), both the Bayesian method and the ML method chose the General Time Reversible model and Gamma distributed with Invariant sites (G + I) as the optimal model for Ddx3 coding sequence alignments.
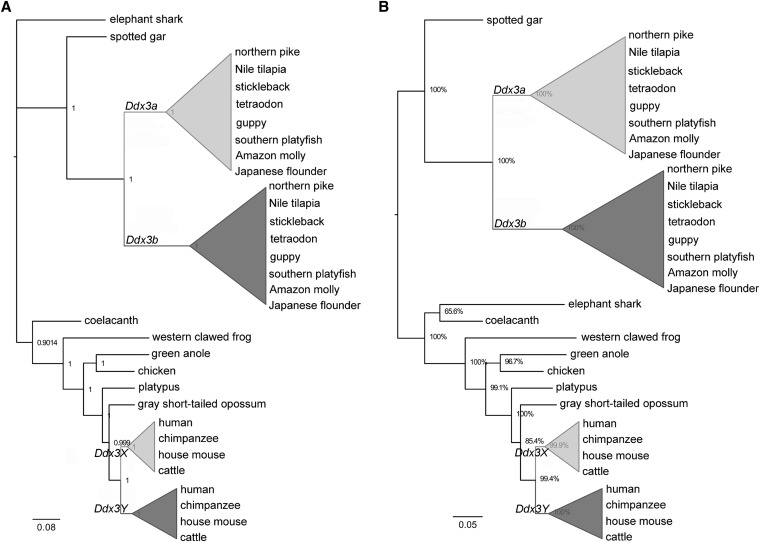
The result of Bayesian analysis was congruent with accepted species relationships (Figure 1A), whereas the out-group elephant shark was grouped with coelacanth in an analysis based on the ML method (Figure 1B). Nevertheless, both phylogenetic trees were consistent in terms of the topological structure of other species, especially the teleost clade and the eutherian clade. The teleost clade was organized into two clades. The spotted gar Ddx3 occupied a clade, and the teleost Ddx3a genes were clearly separated from Ddx3b genes in the other clade. Eutherian Ddx3 genes were also arranged into two distinct clades: the Ddx3X clade and the Ddx3Y clade.
Figure 1.
Phylogenetic analyses of vertebrate Ddx3 genes. (A) Bayesian method was used to construct the gene tree. Numbers at the nodes are Bayesian posterior probabilities. Scale bar = 0.08 substitutions per site. (B) The gene tree was built by the maximum likelihood method. Numbers at the nodes are bootstrap support values with a percentage based on 1000 replicates. Scale bar = 0.05 substitutions per site. Phylogenetic reconstructions were based on the coding sequences of Ddx3 genes. The accession numbers of these genes at GenBank or Ensembl database are provided in Table S1. Elephant shark Ddx3 was used as the out-group. Teleost Ddx3a or Ddx3b genes that clustered together are marked as a cartoon clade, as are the eutherian Ddx3X/Y genes.
Synteny analyses of Ddx3 genes
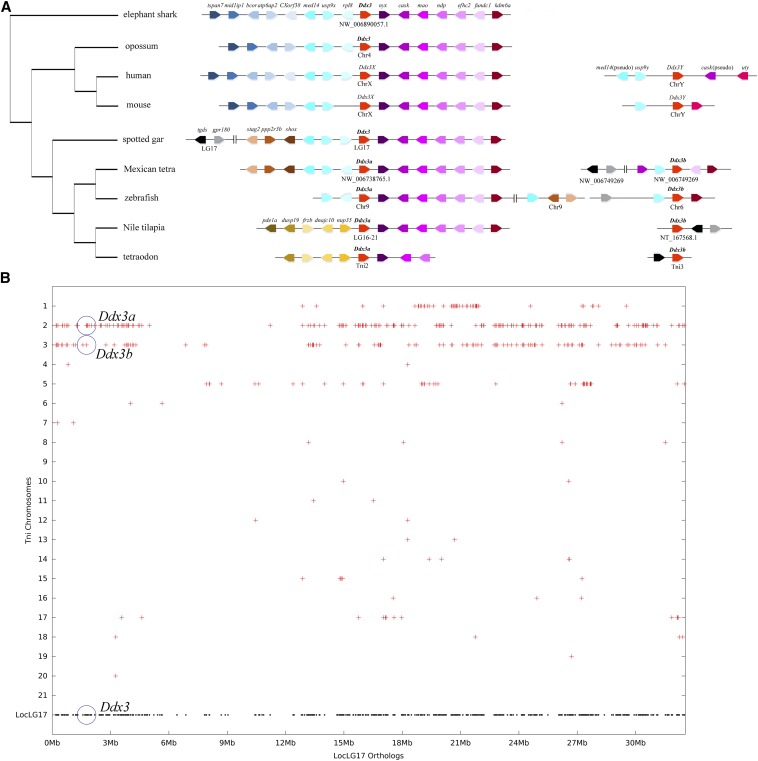
As shown in Figure 2A, the Ddx3 gene and the surrounding genes were mapped according to their relative locations on the same chromosome or scaffold; vertical lines in the figure indicate noncontiguous chromosomal regions. A conserved syntenic relationship was detected among the Ddx3 genes from elephant shark, opossum, and Ddx3X genes from eutherian. In contrast, the surrounding genes of eutherian Ddx3Y changed a lot at the Y chromosome compared with the syntenic genes of Ddx3X at the X chromosome. Only the paralogous gene of usp9x was retained and others were pseudogenized or deleted.
Figure 2.
Synteny analyses of vertebrate Ddx3 genes. (A) Illustration of the syntenic relationship among several Ddx3 genes, eutherian DDX3X/Y genes, and teleost Ddx3a and Ddx3b genes. The arrows in different colors stand for different genes and the arrowheads point in the direction of the corresponding gene. Gene order was determined according to their relative positions in the same chromosome or scaffold. Vertical lines indicate noncontiguous chromosomal regions. The phylogenetic tree was constructed using the coding sequences of Ddx3, Ddx3a, and Ddx3X from these species by Neighbor Joining method in MEGA6. The Assembly ID at NCBI or Ensembl database was provided in Table S1. (B) Synteny analysis of spotted gar LG17 and tetraodon chromosomes. Spotted gar Ddx3 gene is at LG17.Tetraodon Ddx3a is at Tni2, and Ddx3b is at Tni3.
In teleosts, the downstream genes of Ddx3 or Ddx3a genes also were conserved compared with those in elephant shark, although several downstream genes were lost or shifted in tetraodon (Figure 2A). In contrast, the upstream genes were found to have changed through evolution. From elephant shark to spotted gar, several genes were found to have moved and new genes were found upstream of spotted gar Ddx3. The upstream genes of Mexican tetra and zebrafish Ddx3a were conserved with those of spotted gar Ddx3 to some extent, whereas some new genes were translocated upstream of Ddx3a in Nile tilapia and tetraodon. Although the syntenic relationship of Ddx3b was not consistent among those species, regularities also could be discovered. First, the genes gpr180 and tgds were located in the same chromosome as Ddx3b and they often were situated close to Ddx3b. Second, paralogous genes of some Ddx3a syntenic genes also could be found surrounding Ddx3b or away from Ddx3b in the same chromosome.
The spotted gar Ddx3 was located at the chromosome LG17, whereas tetraodon Ddx3a and Ddx3b were located at chromosomes Tni2 and Tni3, respectively. Using the Synteny Database to detect conserved synteny between spotted gar LG17 and tetraodon chromosomes revealed double conserved synteny among the spotted gar LG17 and the tetraodon chromosomes Tni2 and Tni3 (Figure 2B). In addition, the Tni2 and Tni3 chromosomes also exhibited conserved synteny with each other.
Genomic structure analyses of teleost Ddx3 genes
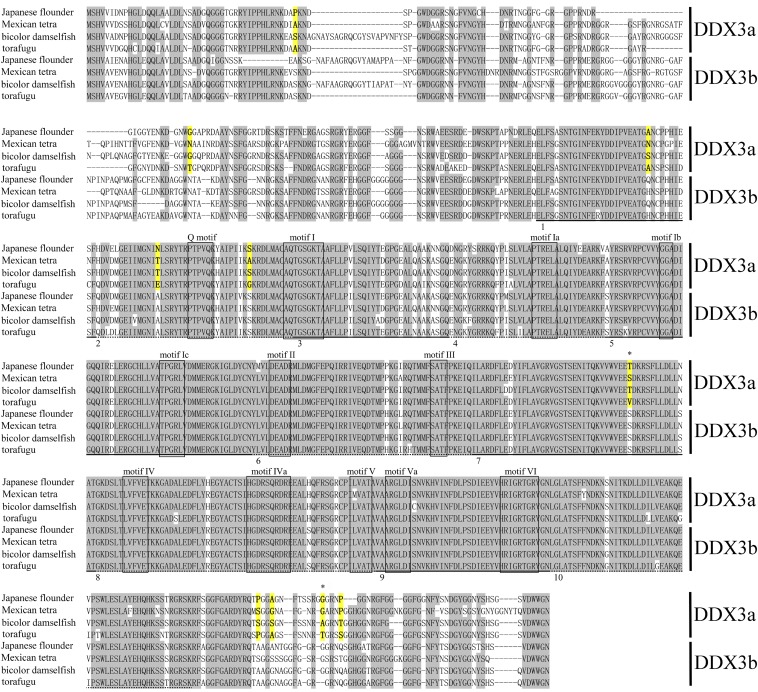
Multiple sequence alignment of deduced full-length DDX3a/b proteins revealed that teleost DDX3a proteins shared higher identity with homologous genes in different species than with DDX3b within the same species (Figure 3). Figure S2 illustrates the genomic structure of Ddx3 genes in four teleost species. The teleost Ddx3 genes contained 18−19 exons. In addition, deep analysis of these exons revealed the presence of 10 exons of uniform size and position in both Ddx3a and Ddx3b genes (Figure 3). Notably, these 10 exons encoded highly identical amino acids containing all the motifs characteristic of DEAD-box proteins (Figure 3).
Figure 3.
Alignment of the deduced amino acid sequences of teleost Ddx3 genes. Identical amino acids are in gray background. Amino acid sequences with identical underline are encoded by the same exon. Numbers 1−10 stand for the ten exons of uniform size and position in both Ddx3a and Ddx3b genes. The 12 conserved motifs characteristic of DEAD-box proteins are boxed. Amino acid sites under positive selection are in yellow background. Asterisk stand for posterior probability > 0.95.
Molecular evolutionary analysis of teleost Ddx3 genes
To explore variation in selective pressure between Ddx3a and Ddx3b genes in teleosts, we used codon-based models of evolution, as implemented in PAML (Yang 2007). For Bayesian trees of teleost Ddx3a and Ddx3b genes (Figure S3A and Figure S4A), the results are shown in Table S3. For Ddx3a and Ddx3b, M0 was rejected in favor of the alternative model M3 (M3 vs. M0, P < 0.0001), indicating variable selection pressure among sites across these genes. Comparison of M2a vs. M1a showed no evidence for positive selection (P = 1). For Ddx3a, M8 vs. M7 and M8 vs. M8a comparisons were statistically significant and the model M8 permitting positive selection showed a better fit to the data for Ddx3a (P < 0.05). For site models, when the likelihood ratio test was significant, the Bayes Empirical Bayes (BEB) tool implemented in PAML (Yang et al. 2005) was used to calculate posterior probabilities to identify sites under positive selection (ω > 1) in the M2a and M8 models. As a result, 12 sites under positive selection among Ddx3a were analyzed by BEB (Table S4). However, neither comparison (M7 vs. M8 and M8a vs. M8) indicated any site classes as being significantly favored for teleost Ddx3b genes (Table S3). This indicated that no amino acid sites were subjected to positive evolution.
For ML trees (Figure S3B and Figure S4B), the results are shown in Table S5. Notably, these results are consistent with the results of Bayesian gene trees. A total of 13 sites in Ddx3a genes were found to be under positive selection by BEB (Table S6).
Ten positively selected sites in Ddx3a were identified not only from Bayesian tree but also from ML tree; for example, site 429 and site 616 (posterior probability >0.95). All such sites are marked in the amino acid sequence of teleost DDX3a protein (yellow background) in Figure 3. Eight sites including site 616 were at the C- or N-termini, one site was located between motif Q and motif I, and site 429 was located between motif III and motif IV.
Tissue distribution pattern of Japanese flounder Ddx3 genes
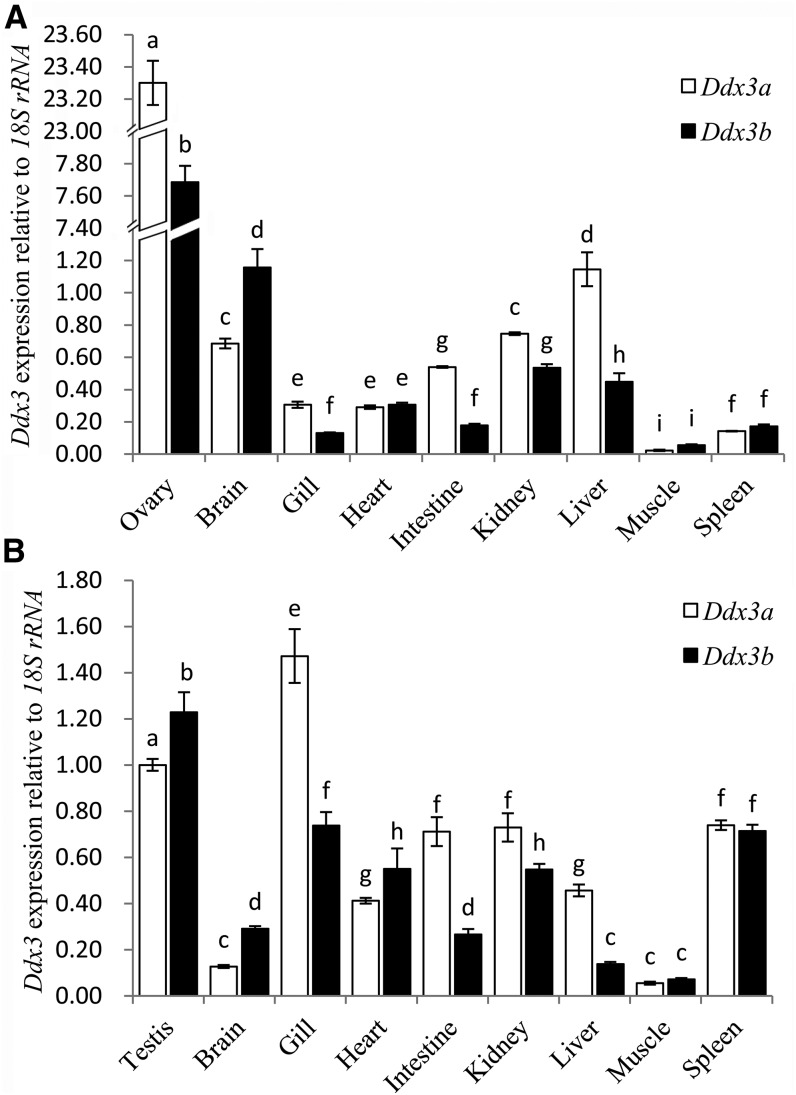
Quantitative real-time PCR results indicated that Ddx3a and Ddx3b showed differential tissue-specific expression pattern in Japanese flounder. In females (Figure 4A), they both showed very high relative expression levels in the ovary, with the level of Ddx3a being much greater than that of Ddx3b. In addition, the relative expression level of Ddx3a was also greater than that of Ddx3b in gills, intestine, kidney, and liver, but lower in the brain. In males (Figure 4B), Ddx3a had the greatest relative expression level in gills, whereas Ddx3b expression was the greatest in testes. The relative expression level of Ddx3b was greater than that of Ddx3a in testes and the brain, whereas that of Ddx3a was higher in gills, intestine, kidney, and liver.
Figure 4.
Tissue distribution patterns of Japanese flounder Ddx3 genes. (A) Relative expression levels of Ddx3 genes in female tissues. (B) Relative expression levels of Ddx3 genes in male tissues. Relative expression level of Ddx3a in the testis was used as calibrator. Data are shown as mean ± SD (n = 5). Values with different superscripts indicate statistical significance (P < 0.05), which was calculated by one-way analysis of variance.
Discussion
In this study, Japanese flounder as well as other teleost species were each found to contain two Ddx3 genes. We therefore thought to explore the evolutionary relationship between teleost Ddx3a/b genes.
Are Ddx3a/b genes sex-linked in teleosts?
In eutherians, there were also two Ddx3 genes (Ddx3X/Y) found to be separately located on the sex chromosomes. Sexual dimorphism of Japanese flounder has been observed, and its gender is genetically determined on the basis of XX (female)–XY (male) type. However, this fish does not present morphologically differentiated sex chromosomes (Tabata 1991; Mei and Gui 2015). Therefore, we first tested whether the Ddx3 gene is X- or Y-linked in Japanese flounder. Surprisingly, the PCR-amplification products showed no sexual specificity (Figure S1), demonstrating that neither gene is located on the Y chromosome. Synteny analysis of other teleost Ddx3a/b genes also revealed that neither is located in the sex chromosomes (Figure 2A). These results clearly indicated that the teleost Ddx3a/b genes are not sex-linked genes as they are in eutherian Ddx3X/Y.
Phylogenetic analysis of teleost Ddx3a/b genes
The out-group elephant shark was grouped with coelacanth in our analysis based on the ML method (Figure 1B). This result can be explained by studying the elephant shark genome. Comparison of syntenic genes in elephant shark and their orthologs in human and zebrafish genomes showed that the level of conserved synteny between the elephant shark and humans is higher (Venkatesh et al. 2007). Furthermore, in this study, we found a conserved syntenic relationship among the Ddx3 genes from elephant shark and opossum, as well as Ddx3X genes from eutherian. Therefore, it is possible that the elephant shark Ddx3 gene was clustered with the tetrapod Ddx3 genes in the phylogenetic analysis. Then, in the phylogenetic analysis based on Bayesian and ML methods, the Ddx3a clade was clearly separated from the Ddx3b clade under the teleost clade (Figure 1), indicating that Ddx3a and Ddx3b diverged from a common ancestral gene.
The origin of teleost Ddx3a and Ddx3b genes
Eutherians contain another autosomal Ddx3 gene besides Ddx3X/Y (Chang and Liu 2010). This gene is intronless and is thought to derive from retroposition of the Ddx3X genes (Mazeyrat et al. 1998; Emerson et al. 2004; Vong et al. 2006). Genomic analysis of the teleost Ddx3a/b genes revealed that both contained exon−intron structures (Figure S2). Furthermore, analysis of deduced amino acid sequences suggested that both genes maintained their protein-coding potential (Figure 3), whereas the coding potential of the autosomal Ddx3 gene in eutherians, except in mice, was diminished (Chang and Liu 2010). On the basis of these results, we conclude that neither of the two teleost Ddx3 genes evolved from retroposition.
Considering that we could find two Ddx3 genes only in teleost species that underwent TGD, we hypothesized that the two Ddx3 genes originated from genome duplication. This could explain why paralogous genes of several Ddx3a syntenic genes are located around Ddx3b in the same chromosome and why the genes gpr180 and tgds are located near Ddx3b despite being far away from the ancestral Ddx3 gene, as they still are in spotted gar (Figure 2A). For a long period after the chromosome was fully duplicated, recombination must have taken place between the two copies of the duplicated chromosomes and one copy of syntenic genes of Ddx3a and Ddx3b may have been pseudogenized, deleted, or translocated. It is possible that the chromosome corresponding to the Ddx3a gene retained more of the ancestral genes, although the extent of this is clearly lineage-dependent. Similarly, one copy each of gpr180 and tgds appear to have been lost, and the other copies of gpr180 and tgds were likely translocated closer to post-duplication Ddx3b.
Only one Ddx3 gene is located in chromosome LG17 of spotted gar, which did not undergo TGD, whereas tetraodon Ddx3a is located in chromosome Tni2 and Ddx3b is located in chromosome Tni3. Synteny analysis revealed that the tetraodon chromosomes Tni2 and Tni3 share conserved synteny with each other and with the spotted gar LG17 (Figure 2B). It is noteworthy here that Tni2 and Tni3 were previously shown to be derived from the ancestral chromosome c and duplicated during TGD (Kasahara et al. 2007). Therefore, our evidence showing conserved synteny between these chromosomes further confirms that the ancestral Ddx3 gene was duplicated during TGD. Our result also rules out the possibility that Ddx3a/b just originated from large-scale segmental duplication through evolution.
To further confirm these findings, dot plots of the spotted gar LG17 with stickleback were performed. Our results showed another double conserved synteny between GacgroupI and GacgroupXVI (Figure S5A). Ddx3a is found in groupXVI in stickleback, and strong synteny was observed between Tni2 and GacgroupXVI (Figure S5B). Ddx3b is found in groupI in stickleback and unequivocal evidence of synteny between Tni3 and GacgroupI was observed (Figure S5C). These results present strong evidence of whole-genome duplication that occurred specifically in teleosts.
Subfunctionalization of the duplicated teleost Ddx3 genes
The 10 uniform exons of Ddx3 genes encode highly homologous amino acids that span the entire conserved motifs and domains characteristic of DEAD-box protein (Figure 3), suggesting conserved genomic structure and protein function between teleost Ddx3a genes and Ddx3b genes.
No models of selection were statistically significant for the teleost Ddx3b genes (Table S3 and Table S5), suggesting probable purifying selection of Ddx3b during evolution. In contrast, 10 positively selected sites (Figure 3) can be identified in DDX3a, suggesting divergent evolutionary fates between Ddx3a and Ddx3b. In addition, the C- or N-termini of DEAD box helicases can facilitate RNA binding (Mohr et al. 2008). The Q motif and motif I regulate ATP binding and hydrolysis (Tanner et al. 2003; Cordin et al. 2004). The linker between motif III and motif IV connects the two core domains of DEAD box helicase and can regulate their orientation (Andreou and Klostermeier 2012). Therefore, mutation of these positively selected sites may have affected the RNA helicase and ATPase activities of DDX3a, resulting in functional divergence between DDX3a and DDX3b.
The function of DDX3 proteins in gametogenesis has been well described in other metazoans (Leroy et al. 1989; Mazeyrat et al. 1998; Mochizuki et al. 2001; Session et al. 2001; Johnstone et al. 2005). In this study, we found differential tissue-specific expression patterns of Ddx3a and Ddx3b, especially in the gonads, in Japanese flounder (Figure 4). The relative expression level of Ddx3a was much greater than that of Ddx3b in the ovary, whereas that of Ddx3b was greater than that of Ddx3a in the testis. These results provide evidence for divergent functions between DDX3a and DDX3b in teleost gametogenesis. However, this remains to be directly addressed through in vitro and in vivo analysis of the exact functions of DDX3a and DDX3b in suitable fish models.
Considering the aforementioned results, we conclude that the teleost DDX3a and DDX3b proteins retained the conserved function of DEAD-box RNA helicases, whereas their divergent evolutionary fates resulted in their functional differences. Thus, there is sufficient evidence for subfunctionalization of the duplicated Ddx3 genes in teleosts after TGD.
In summary, we investigated the origin of teleost Ddx3a/b genes in the current study. It is the first to report that two Ddx3 genes are present in teleosts as a result of TGD. In addition, our findings suggest probable subfunctionalization of the duplicated Ddx3 genes in teleost through evolution. Therefore, this study provides novel insights into the teleost Ddx3 gene family and open doors to further functional studies in suitable fish models.
Supplementary Material
Acknowledgments
This work was supported by the National High-Tech Research and Development Program (2012AA10A402) and the National Natural Science Foundation of China (31172385).
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.018911/-/DC1
Communicating editor: W. S. Davidson
Literature Cited
- Amores A., Force A., Yan Y. L., Joly L., Amemiya C., et al. , 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711–1714. [DOI] [PubMed] [Google Scholar]
- Amores A., Catchen J., Ferrara A., Fontenot Q., Postlethwait J. H., 2011. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics 188: 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreou A. Z., Klostermeier D., 2012. Conformational changes of DEAD-box helicases monitored by single molecule fluorescence resonance energy transfer. Methods Enzymol. 511: 75–109. [DOI] [PubMed] [Google Scholar]
- Braasch I., Salzburger W., Meyer A., 2006. Asymmetric evolution in two fish-specifically duplicated receptor tyrosine kinase paralogons involved in teleost coloration. Mol. Biol. Evol. 23: 1192–1202. [DOI] [PubMed] [Google Scholar]
- Braasch I., Volff J. N., Schartl M., 2009. The endothelin system: evolution of vertebrate-specific ligand-receptor interactions by three rounds of genome duplication. Mol. Biol. Evol. 26: 783–799. [DOI] [PubMed] [Google Scholar]
- Brunet F. G., Roest Crollius H., Paris M., Aury J. M., Gibert P., et al. , 2006. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol. Biol. Evol. 23: 1808–1816. [DOI] [PubMed] [Google Scholar]
- Catchen J. M., Conery J. S., Postlethwait J. H., 2009. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 19: 1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T. C., Liu W. S., 2010. The molecular evolution of PL10 homologs. BMC Evol. Biol. 10: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao C. H., Chen C. M., Cheng P. L., Shih J. W., Tsou A. P., et al. , 2006. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res. 66: 6579–6588. [DOI] [PubMed] [Google Scholar]
- Christoffels A., Koh E. G., Chia J. M., Brenner S., Aparicio S., et al. , 2004. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol. Biol. Evol. 21: 1146–1151. [DOI] [PubMed] [Google Scholar]
- Cordin O., Tanner N. K., Doere M., Linder P., Banroques J., 2004. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 23: 2478–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson J. J., Kaessmann H., Betrán E., Long M., 2004. Extensive gene traffic on the mammalian X chromosome. Science 303: 537–540. [DOI] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., et al. , 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresta C., Moro E., Ferlin A., 2001. Prognostic value of Y deletion analysis The role of current methods. Hum. Reprod. 16: 1543–1547. [DOI] [PubMed] [Google Scholar]
- Gao J., Wang J., Jiang J., Fan L., Wang W., et al. , 2013. Identification and characterization of a nanog homolog in Japanese flounder (Paralichthys olivaceus). Gene 531: 411–421. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V., 1993. Helicases: amino acid sequence comparisons and structure–function relationships. Curr. Opin. Struct. Biol. 3: 419–429. [Google Scholar]
- He X., Zhang J., 2005. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169: 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegg S., Meyer A., 2005. Hox clusters as models for vertebrate genome evolution. Trends Genet. 21: 421–424. [DOI] [PubMed] [Google Scholar]
- Hoegg S., Meyer A., 2007. Phylogenomic analyses of KCNA gene clusters in vertebrates: why do gene clusters stay intact? BMC Evol. Biol. 7: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann F. G., Opazo J. C., Storz J. F., 2012. Whole-genome duplications spurred the functional diversification of the globin gene superfamily in vertebrates. Mol. Biol. Evol. 29: 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Ronquist F., Nielsen R., Bollback J. P., 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294: 2310–2314. [DOI] [PubMed] [Google Scholar]
- Hughes A. L., 1994. The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 256: 119–124. [DOI] [PubMed] [Google Scholar]
- Jaillon O., Aury J. M., Brunet F., Petit J. L., Stange-Thomann N., et al. , 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431: 946–957. [DOI] [PubMed] [Google Scholar]
- Johnstone O., Deuring R., Bock R., Linder P., Fuller M. T., et al. , 2005. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev. Biol. 277: 92–101. [DOI] [PubMed] [Google Scholar]
- Kanai Y., Dohmae N., Hirokawa N., 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43: 513–525. [DOI] [PubMed] [Google Scholar]
- Kasahara M., Naruse K., Sasaki S., Nakatani Y., Qu W., et al. , 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447: 714–719. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T. M., Creevey C. J., Pentony M. M., Naughton T. J., McLnerney J. O., 2006. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo P. L., Lin Y. H., Teng Y. N., Hsu C. C., Lin J. S., et al. , 2004. Transcriptional levels of four Y chromosome-linked AZF genes in azoospermic men and their association with successful sperm retrieval. Urology 63: 131–136. [DOI] [PubMed] [Google Scholar]
- Lahn B. T., Page D. C., 1997. Functional coherence of the human Y chromosome. Science 278: 675–680. [DOI] [PubMed] [Google Scholar]
- Lai M. C., Lee Y. H. W., Tarn W. Y., 2008. The DEAD-Box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well asTip-associated protein and participates in translational control. Mol. Biol. Cell 19: 3847–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy P., Alzari P., Sassoon D., Wolgemuth D., Fellous M., 1989. The protein encoded by a murine male germ cell-specific transcript is a putative ATP-dependent RNA helicase. Cell 57: 549–559. [DOI] [PubMed] [Google Scholar]
- Li X. Y., Zhang X. J., Li Z., Hong W., Liu W., et al. , 2014. Evolutionary history of two divergent Dmrt1 genes reveals two rounds of polyploidy origins in gibel carp. Mol. Phylogenet. Evol. 78: 96–104. [DOI] [PubMed] [Google Scholar]
- Linder P., Fuller-Pace F. V., 2013. Looking back on the birth of DEAD-box RNA helicases. Biochim. Biophys. Acta. Gene Regul. Mech. 1829: 750–755. [DOI] [PubMed] [Google Scholar]
- Linder P., Lasko P. F., Ashburner M., Leroy P., Nielsen P. J., et al. , 1989. Birth of the D-E-A-D box. Nature 337: 121–122. [DOI] [PubMed] [Google Scholar]
- Liu S., Li Z., Gui J. F., 2009. Fish-specific duplicated dmrt2b contributes to a divergent function through Hedgehog pathway and maintains left-right asymmetry establishment function. PLoS One 4: e7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang Y. B., Liu T. K., Gui J. F., 2013. Lineage-specific expansion of IFIT gene family: an insight into coevolution with IFN gene family. PLoS One 8: e66859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J. S., 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch M., Force A., 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazeyrat S., Saut N., Sargent C. A., Grimmond S., Longepied G., et al. , 1998. The mouse Y chromosome interval necessary for spermatogonial proliferation is gene dense with syntenic homology to the human AZFa region. Hum. Mol. Genet. 7: 1713–1724. [DOI] [PubMed] [Google Scholar]
- Mei J., Gui J. F., 2015. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 58: 124–136. [DOI] [PubMed] [Google Scholar]
- Meyer A., 1998. Hox gene variation and evolution. Nature 391: 225–228. [DOI] [PubMed] [Google Scholar]
- Meyer A., Van de Peer Y., 2005. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). BioEssays 27: 937–945. [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Nishimiya-Fujisawa C., Fujisawa T., 2001. Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev. Genes Evol. 211: 299–308. [DOI] [PubMed] [Google Scholar]
- Mohr G., Del Campo M., Mohr S., Yang Q., Jia H., et al. , 2008. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J. Mol. Biol. 375: 1344–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulley J. F., Chiu C. H., Holland P. W., 2006. Breakup of a homeobox cluster after genome duplication in teleosts. Proc. Natl. Acad. Sci. USA 103: 10369–10372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., 1970. Evolution by Gene Duplication. Springer-Verlag, New York. [Google Scholar]
- Park S. H., Lee S. G., Kim Y., Song K., 1998. Assignment of a human putative RNA helicase gene, DDX3, to human X chromosome bands p11.3→p11.23. Cytogenet. Cell Genet. 81: 178–179. [DOI] [PubMed] [Google Scholar]
- Rocak S., Linder P., 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 5: 232–241. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., et al. , 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session D. R., Lee G. S., Wolgemuth D. J., 2001. Characterization of D1Pas1, a mouse autosomal homologue of the human AZFa region DBY, as a nuclear protein in spermatogenic cells. Fertil. Steril. 76: 804–811. [DOI] [PubMed] [Google Scholar]
- Siegel N., Hoegg S., Salzburger W., Braasch I., Meyer A., 2007. Comparative genomics of ParaHox clusters of teleost fishes: gene cluster breakup and the retention of gene sets following whole genome duplications. BMC Genomics 8: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W. J., Yang Z., Wolfner M. F., Aquadro C. F., 2001. Positive Darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc. Natl. Acad. Sci. USA 98: 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata K., 1991. Induction of gynogenetic diploid males and presumption of sex determination mechanisms in the hirame Paralichthys olivaceus. Bull. Jpn. Soc. Sci. Fish. 57: 845–850. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner N. K., Cordin O., Banroques J., Doere M., Linder P., 2003. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell 11: 127–138. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., Maere S., Meyer A., 2009. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10: 725–732. [DOI] [PubMed] [Google Scholar]
- Vandepoele K., De Vos W., Taylor J. S., Meyer A., Van de Peer Y., 2004. Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc. Natl. Acad. Sci. USA 101: 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B., Kirkness E. F., Loh Y.H., Halpern A. L., Lee A. P., et al. , 2007. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 5: e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vong Q. P., Li Y., Lau Y. F. C., Dym M., Rennert O. M., et al. , 2006. Structural characterization and expression studies of Dby and its homologs in the mouse. J. Androl. 27(5): 653–661. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J., 1982. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1: 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W. S., Yang Z., Goldman N., Nielsen R., 2004. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168: 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., Goldman N., Pedersen A. M. K., 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155: 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Wong W. S., Nielsen R., 2005. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22: 1107–1118. [DOI] [PubMed] [Google Scholar]
- Yedavalli V. S., Neuveut C., Chi Y. H., Kleiman L., Jeang K. T., 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119: 381–392. [DOI] [PubMed] [Google Scholar]
- You L. R., Chen C. M., Yeh T. S., Tsai T. Y., Mai R. T., et al. , 1999. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J. Virol. 73: 2841–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q., Zhang Q., Wang Z., Qi J., Chen Y., et al. , 2008. Expression profiling and validation of potential reference genes during Paralichthys olivaceus embryogenesis. Mar. Biotechnol. (NY) 10: 310–318. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Licklider L. J., Gygi S. P., Reed R., 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.