Abstract
Many insects harbour facultative symbiotic bacteria, some of which have been shown to provide resistance against natural enemies. One of the best-known protective symbionts is Hamiltonella defensa, which in pea aphid (Acyrthosiphon pisum) confers resistance against attack by parasitoid wasps in the genus Aphidius (Braconidae). We asked (i) whether this symbiont also confers protection against a phylogenetically distant group of parasitoids (Aphelinidae) and (ii) whether there are consistent differences in the effects of bacteria found in pea aphid biotypes adapted to different host plants. We found that some H. defensa strains do provide protection against an aphelinid parasitoid Aphelinus abdominalis. Hamiltonella defensa from the Lotus biotype provided high resistance to A. abdominalis and moderate to low resistance to Aphidius ervi, while the reverse was seen from Medicago biotype isolates. Aphids from Ononis showed no evidence of symbiont-mediated protection against either wasp species and were relatively vulnerable to both. Our results may reflect the different selection pressures exerted by the parasitoid community on aphids feeding on different host plants, and could help explain the maintenance of genetic diversity in bacterial symbionts.
Keywords: aphid, facultative symbiosis, parasitoid, resistance, symbiont
1. Introduction
Facultative bacterial symbionts are found commonly in many groups of insects [1]. Unlike obligate symbionts, they are not essential for successful growth and reproduction and they persist either by manipulating host reproduction for their own benefit [2] or because they confer some fitness advantage on their host. Symbionts have been shown to help their hosts resist abiotic challenges and attack by natural enemies [3–5]. The importance of facultative symbionts has become increasingly apparent in the last two decades yet we still know relatively little about how they affect their hosts' interactions in complex food webs.
The pea aphid, Acyrthosiphon pisum (Hemiptera: Aphididae), is widely used as a model for studying insect facultative symbiosis. Seven species of facultative bacteria are found commonly in pea aphids [6–10] and have been shown to affect resistance to parasitoids [11,12] or fungal pathogens [13–15], as well as having a variety of other effects on host plant use and life history [16–18]. Aphids are attacked by several groups of primary parasitoids, all solitary koinobiont endoparasitoids [19]. Parasitoids typically attack pre-adult aphids but their larvae do not destructively consume the host until late in development [20,21]. The full-grown parasitoid larva pupates inside the mummified husk of its host. The symbiont Hamiltonella defensa (Gammaproteobacteria: Enterobacteriaceae) has been shown to protect aphids against a number of parasitoids within the subfamily Aphidiinae (Hymenoptera: Braconidae) [11,12,22,23], although the extent of this protection varies considerably among bacterial isolates [11]. In aphids infected with a protective symbiont, the parasitoid dies during the egg or larval stage. Protection by H. defensa is dependent on the presence of a bacteriophage, termed APSE (A. pisum Secondary Endosymbiont) [24], which carries putative toxin genes affecting eukaryotes [25,26]. Variation in toxin genes is correlated with variation in the strength of protection [11,27,28], while strains lacking the phage [25,28], or which carry phages with apparently inactivated toxin genes [29], provide no protection.
What processes might allow these multiple strains of the same symbiont to coexist in aphid populations? Symbiont carriage can carry costs for the aphid [30–32] and one possibility is that more protective symbionts have higher costs. Alternatively, different symbiont strains may provide protection against different parasitoid genotypes or species. In addition to aphidiine braconids, aphids are attacked by a second major group of primary parasitoids: wasps in the genus Aphelinus from the family Aphelinidae (superfamily Chalcidoidea, rather than the Ichneumonoidea which include the Braconidae). Only one recent study has investigated whether the symbionts that protect against Aphidiinae have the same effects against Aphelinidae. Working with the black bean aphid (Aphis fabae), Cayetano & Vorburger [22] found that a strain of H. defensa that provided protection against the aphidiine Lysiphlebus fabarum provided no protection against the aphelinid Aphelinus chaonia. There are substantial differences in the biology of aphelinids and aphidiine braconids [33–35] that may affect interactions with symbionts. The eggs of aphelinids are much larger, which has been suggested to be an explanation for their immunity to symbiont toxins [22]. In addition, aphidiines produce teratocytes (cells derived from egg membranes that circulate in the host haemocoel and influence host physiology) which have never been recorded from Aphelinidae [21,36]. Teratocytes may be a target for symbiont toxins [37,38], and such a defence mechanism would thus affect aphidiines but not aphelinids.
This study set out to answer two questions. First, we asked whether the symbiont H. defensa confers protection against both major groups of aphid primary parasitoid, or only to aphidiine braconids. Second, we asked whether there are consistent differences in different pea aphid biotypes in their resistance to parasitoid attack. The pea aphid taxon comprises a set of genetically differentiated populations or biotypes which feed on different host plants [39] and which show consistent differences in their symbiont communities [40]. This will help us address whether biotype differentiation is influenced by selection pressures from higher trophic levels in addition to selection for host plant utilization.
We addressed these questions using two relatively generalist species of aphid parasitoid, representing the two major groups attacking aphids: the aphidiine braconid Aphidius ervi and the aphelinid Aphelinus abdominalis. We took 10 clonal lines of pea aphids, belonging to three different biotypes, all of which had natural H. defensa infections. Using antibiotics, sub-lines were created from which the H. defensa had been removed and resistance to the two parasitoid species was measured with or without the symbiont. We also investigated the presence of APSE in each H. defensa strain.
2. Material and methods
(a). Experimental organisms
Aphids were collected from three host plant species, Ononis spinosa, Lotus pedunculatus and Medicago sativa, in the south of England, UK, between 2003 and 2012 (hereafter referred to as the Ononis, Lotus and Medicago biotypes; table 1). In each case, a single adult individual was used to establish a line of aphids that were subsequently maintained in laboratory culture on broad bean (Vicia faba). Diagnostic PCR was used to confirm that each clone carried the symbiont H. defensa and to find out whether any other known facultative symbionts of pea aphids were present, a procedure that was repeated immediately prior to our experiments (see Henry et al. [41] for details of primers and PCR conditions used). Microsatellite typing was used to establish that the clones are distinct from one another and belong to biotypes known to be associated with the different plant species [39]. Hamiltonella defensa infections were removed from 10 aphid clones by using antibiotics which do not affect the primary symbiont, Buchnera aphidicola. To do this, first instar aphids were placed for 3 days on a leaf of V. faba with the stem inserted into a solution of 50 µg ml−1 cefotaxime, 50 µg ml−1 gentamicin and 100 µg ml−1 ampicillin. For the aphids specialized on Medicago (table 1), it was found necessary to use a solution of double this concentration and to apply a second dose of antibiotics for a further 3 days. Successful removal was confirmed by repeated diagnostic PCR for H. defensa, with appropriate positive and negative controls. All cured lines consistently tested negative for six generations before being used in experiments. Two experimental aphid clones carried a co-infection, in both cases of the bacterium known as ‘X-type’ (Gammaproteobacteria: Enterobacteriaceae), which was retained in the ‘cured’ lines (table 1).
Table 1.
Details of pea aphid clones used in phylogenetic analysis; indication is given where the clones were also included in the experimental assays.
| clone code | biotype | colour | used in experiment? | collection year | additional symbionts carried |
|---|---|---|---|---|---|
| 11 | Lotus pedunculatus | green | yes | 2010 | |
| 74 | L. pedunculatus | green | yes | 2010 | |
| 132 | L. pedunculatus | green | yes | 2003 | |
| 159 | L. pedunculatus | green | yes | 2010 | |
| 208 | L. pedunculatus | red | no | 2003 | Rickettsiella |
| 224 | L. pedunculatus | green | no | 2003 | |
| 123 | Ononis spinosa | red | yes | 2003 | |
| 256 | O. spinosa | green | yes | 2010 | |
| 257 | O. spinosa | red | yes | 2010 | |
| 101 | O. spinosa | green | no | 2003 | |
| 302 | Medicago sativa | green | yes | 2012 | X-type |
| 308 | M. sativa | red | yes | 2012 | |
| 312 | M. sativa | red | yes | 2012 | X-type |
| 161 | M. sativa | red | no | 2003 | Spiroplasma |
| 304 | M. sativa | green | no | 2012 | X-type |
| 305 | M. sativa | red | no | 2012 | X-type |
| 217 | M. sativa | red | no | 2012 |
Aphids were routinely maintained at 14°C with a 16 L : 8 D cycle in 9 cm Petri dishes containing a single leaf of V. faba with the petiole inserted in 2% agar gel. The aphids were transferred to a fresh dish once a week. Two generations before the experiments were carried out, the aphids were moved to a 20°C room, and this was the temperature at which the experiments were conducted.
Individuals of both species of parasitoid wasp to be used in the experiment were taken from inbred stocks maintained in the laboratory at 20°C and a 16 L : 8 D cycle (for over 5 years in the case of A. ervi and for a minimum of nine months for A. abdominalis). The wasps were reared on a highly susceptible pea aphid clone which lacks any described facultative endosymbionts. Female A. abdominalis mature eggs throughout their lifetime and have the capacity to resorb eggs in the absence of aphids (a synovarial life history [42]); the adults also feed on aphids, which A. ervi does not [43]. We therefore decided that A. abdominalis females should be exposed to aphids prior to their use in experiments, both to ensure that they possessed mature eggs [35] and to minimize host-feeding during the experiment [33]. Aphidius ervi females were also allowed access to aphids, because previous exposure has been shown to improve later oviposition success in the closely related Aphidius colemani [44]. A 30% honey solution was provided to ensure adequate levels of nutrition. Both wasp species were kept in mixed-sex cages and were therefore presumed to be mated. Aphidius ervi females were between 24 h and 5 days old when used in experiments; A. abdominalis females, which are longer lived, were between 24 h and 8 days old.
(b). Exposure of aphids to parasitoids
In each replicate, 15 third-instar (4–5 days old) aphids were exposed to a single female wasp. We chose this age class because previous findings in A. fabae [45] and our own observations in pea aphid suggest that symbiont-conferred resistance may not be effective until aphids are at least 3–4 days old. Aphids were placed in a Petri dish containing half a leaf of V. faba, split down the petiole, with the remaining stalk inserted into 2% agar. Splitting the leaves ensured that the search area was as simple as possible and reduced variation between dishes. Leaves of approximately equal size were used in each dish to ensure that the search area was similar between dishes.
The two wasp species have very different parasitism techniques [46]. Aphidius species attack quickly, laying each egg in less than a second but causing considerable disruption to the aphid colony; Aphelinus are furtive and cause minimal disturbance to their hosts, but oviposition takes several minutes [33,34,46]. The goal of the experimental design was to allow enough time for all aphids in the dish to be parasitized but to minimize superparasitism. Based on pilot experiments, A. abdominalis females were given access to aphids for 24 h and A. ervi for 3 h. In each temporal block, experiments with A. abdominalis were initiated 21 h ahead of those with A. ervi, and the two species were removed at the same time. At the end of the exposure period, the aphids were transferred to fresh Petri dishes with leaves of V. faba.
Both wasps form distinctive mummies: Aphelinus mummies are black and appear approximately one week after oviposition, while Aphidius mummies are swollen and golden, and appear approximately 10 days after oviposition. All mummies were assumed to have been formed after 10 days for A. abdominalis and after 14 days for A. ervi, at which point the number of live aphids and mummies in each dish were recorded. Experiments were carried out over 11 temporal blocks, each containing a subset of aphid clones, with all clones represented in more than one block. For every block in which a particular clone was included, both cured and infected lines were exposed to at least one parasitoid species.
(c). Statistical analysis of results
Data from the parasitism success rate assays were analysed using generalized linear mixed modelling techniques implemented in the ‘lme4’ [47] and ‘car’ [48] packages written for the software R v. 3.0.2 [49]. The full model included aphid clone and experimental block as random factors and aphid symbiont status (cured or infected) and biotype as fixed factors. Binomial error variances were assumed and an observation-level term included as a random factor to account for any overdispersion in the data. Contrasts responsible for significant results were identified using Tukey's adjusted pairwise method.
(d). Comparison of Hamiltonella defensa and Acyrthosiphon pisum secondary endosymbiont sequences
DNA was extracted from adult aphids using the DNeasy Blood and Tissue kit (Qiagen). Partial sequences from two housekeeping genes, murE and hrpA (855 bp and 812 bp, respectively), were used to compare the genetic distance between the H. defensa isolates from the experimental aphids. These genes were selected because previous studies have shown that they display greater between than within biotype variation [41]. Primers and PCR conditions are given in Henry et al. [41]. In addition to the aphid clones used in the experiment, we also included seven additional samples to obtain a better understanding of genetic variation within biotypes (table 1). The samples were tested for the presence of APSE phage using specific primers [25] and reaction conditions (given in the electronic supplementary material) designed to amplify two conserved phage genes, P3 and P51, which are known to exhibit considerable variation between phage strains [25]. Successfully amplified products were Sanger sequenced in one direction by Source Bioscience (Nottingham, UK). Sequence traces were assembled and edited in CodonCode Aligner (CodonCode Corporation), before alignment using Clustal X [50] and BioEdit Sequence Alignment Editor [51].
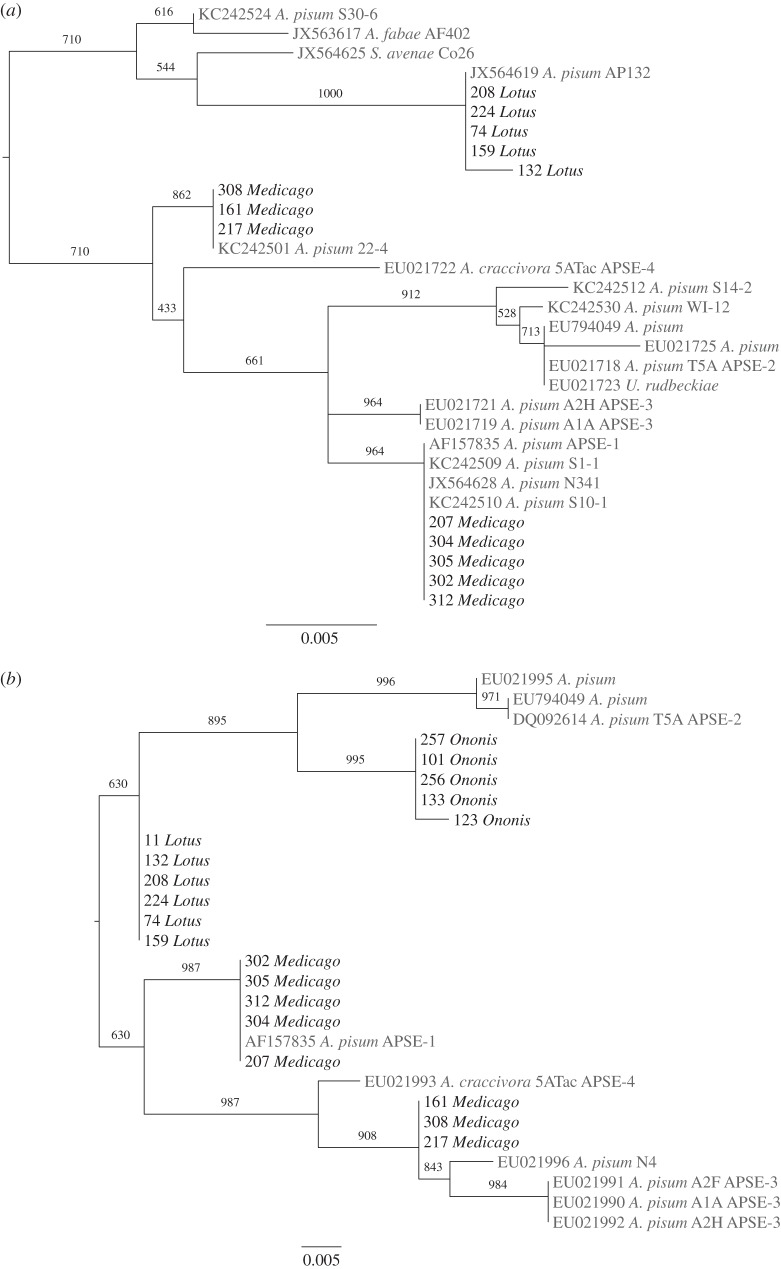
The partial sequences for P3 and P51 (989 bp and 497–813 bp, respectively) were then compared to published APSE sequences available in GenBank (see electronic supplementary material, tables S1 and S2, for details of comparison strains). A number of different phage strain types have been previously identified, only some of which have associated phenotypic data: strain APSE-3 has been found associated with strong protection against A. ervi and APSE-2 with partial protection [28]. Phylogenetic trees were constructed for P3 and P51 using maximum-likelihood methods implemented in PhyML v. 3.0 [52] using HKY and K80 substitution models, respectively. These were identified as the most suitable models using FindModel (http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html). Tree topologies were improved using nearest neighbour interchange and support assessed using 1000 bootstrap replicates.
3. Results
(a). Resistance to Aphidius ervi
In agreement with previous studies [11,12,24,53], we found that the overall proportion of aphids forming mummies after parasitism by A. ervi was significantly reduced by the presence of the symbiont H. defensa (table 2a). However, we identified significant differences between the biotypes in both the probability of parasitism and the effects of the symbiont (table 2a and figure 1a). We carried out multiple comparisons with Tukey adjustments to identify the source of these differences (electronic supplementary material, tables S3 and S4). We found no evidence that H. defensa improves survival in aphids from Ononis (z = 0.376, p = 0.999), whereas aphids from both Lotus and Medicago were significantly protected (Tukey comparison: Lotus z = −3.327, p < 0.001; Medicago z = −4.376, p < 0.001). Medicago aphids have greater resistance than Lotus aphids both with (z = −4.588, p < 0.001) and without (z = −2.976, p = 0.032) H. defensa. Model comparison showed that the removal of neither block (χ2 = 1.136, p = 0.286) nor clone (χ2 = 0.225, p = 0.635) significantly impaired the explanatory power of the model.
Table 2.
Results of analysis of deviance for generalized linear mixed models investigating effects of symbiont presence and aphid biotype on the parasitism success of (a) A. ervi and (b) A. abdominalis.
| d.f. | Wald χ2 | p | |
|---|---|---|---|
| (a) A. ervi | |||
| symbiont | 1 | 13.448 | <0.001 |
| biotype | 2 | 16.555 | <0.001 |
| symbiont × biotype | 2 | 16.905 | <0.001 |
| (b) A. abdominalis | |||
| symbiont | 1 | 7.2233 | <0.001 |
| biotype | 2 | 9.0624 | 0.011 |
| symbiont × biotype | 2 | 51.8976 | <0.001 |
Figure 1.
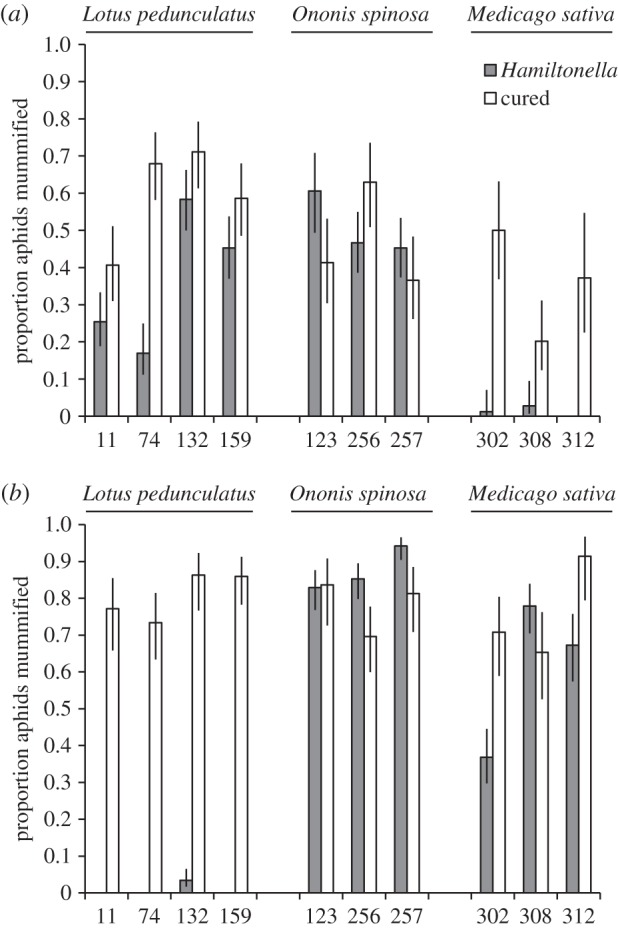
Mean proportion of aphids from the three different host plants forming mummies for (a) A. ervi and (b) A. abdominalis. Error bars denote standard error.
(b). Resistance to Aphelinus abdominalis
Our results demonstrate that certain strains of H. defensa can significantly improve aphid resistance to A. abdominalis, the first time we believe that this effect has been shown against a parasitoid outside the Braconidae (table 2b). As with A. ervi, this protection was found to vary significantly among biotypes (table 2b and figure 1b). Again, aphids from Ononis showed no evidence of symbiont-conferred protection (Tukey comparison: z = 1.278, p = 0.779). Medicago aphids also showed no significant protection (z = −1.824, p = 0.424) but Lotus aphids succumbed to parasitoid attack less frequently when they carried H. defensa (z = −7.361, p < 0.001). Lotus aphids have greater symbiont-conferred resistance than aphids from Medicago (z = 5.270, p < 0.001), the reverse of the pattern with A. ervi attack. Model comparison found that removing block significantly reduced the model's explanatory power (χ2 = 5.058, p = 0.025) but the effect of removing clone was not significant (χ2 = 0.108, p = 0.743).
(c). Symbiont and phage sequence data
Analysis of the sequence data for the H. defensa hrpA and murE genes confirm that clones collected from the different host plants have genotypes characteristic of the H. defensa strains previously associated with these plants [41]. There were two different H. defensa types associated with Medicago, one of which is extremely similar to the single H. defensa clade associated with Lotus; there was also a single H. defensa clade associated with Ononis (see electronic supplementary material, figure S1).
Maximum-likelihood phylogenies for the phage loci P3 and P51 are shown in figure 2; however, we caution that high levels of recombination among APSE phage [26] mean that phylogenetic trees may not reliably show relationships between phage strains. We were unsuccessful in amplifying gene P3 from Ononis aphids despite repeated attempts. As has been reported previously, we find considerable variation among APSE sequences [26] and some of the clades are poorly supported. Lotus clones cluster together and are not clearly associated with any previously described phage strain. The results from the P51 gene suggest that phage hosted by H. defensa from Ononis aphids are most similar to APSE-2. Both genes in five Medicago clones show high similarity to previously published APSE-1 sequences (for which no phenotypic data were previously available), while the other three Medicago clones showed some sequence similarity to isolates ascribed to APSE-3 (previously associated with strong protection against A. ervi). Each of the two types of phage in Medicago aphids is associated with one of the two different H. defensa genotypes in these insects (electronic supplementary material, figure S1).
Figure 2.
Maximum-likelihood phylogenies of phage genes (a) P3 and (b) P51. Clones from this study (in black) are indicated by a number and host plant genus affiliation (table 1). Accession number, aphid species, isolate name and phage type are given for previously published sequences (in grey), where available. For all accession numbers and references, see electronic supplementary material, tables S1 and S2. Bootstrap values (1000 replicates) are shown on branches for major clades; scale bar indicates substitutions per site.
4. Discussion
We investigated whether aphids carrying the facultative symbiont H. defensa can benefit from symbiont-conferred resistance against aphelinid as well as distantly related braconid parasitoids. We found that some H. defensa strains protect against the aphelinid A. abdominalis. However, different symbiont isolates varied in the protection they afforded against the two wasp species and some provided no significant protection against one or both parasitoids. Our second aim was to determine whether there were any consistent differences between aphid biotypes carrying their natural isolates of H. defensa in their resistance phenotype. Aphids from Lotus, when naturally infected with H. defensa, showed high resistance to A. abdominalis and moderate to low resistance to A. ervi; aphids from Medicago, when naturally infected with H. defensa, showed high resistance to A. ervi and little resistance to A. abdominalis. Aphids from Ononis showed no evidence of symbiont-mediated protection against either wasp species and were relatively vulnerable to both.
Our results may help explain why some H. defensa strains that provide apparently poor protection against A. ervi persist in aphid populations: they may in fact provide high levels of protection, but against a different parasitoid species. Degnan & Moran [26] have speculated that different phage-encoded toxins may act specifically against different parasitoid species, and there is some evidence that Aphis craccivora symbionts show variation in specificity against different aphidiine braconid species [54]. As yet the hypothesis that APSE toxins are responsible for parasitoid protection has not been confirmed, but a comparison of toxin proteins in the different strains and potential targets in the two families of wasp may help resolve this question. As mentioned in the Introduction, it has been suggested that phage toxins target teratocytes [37,38] but the absence of teratocytes in aphelinids [21] shows this cannot be a universal model of action. Previous work has shown that Regiella insecticola can confer resistance against parasitoids even though it does not carry APSE (or other recognized phages) [55], and it is possible that non-phage mechanisms may also operate in H. defensa.
Our second aim was to examine patterns of resistance across aphid biotypes. Clones from different biotypes, with their naturally associated symbionts, showed consistent patterns of resistance to the two parasitoid species in our study (figure 3). However, symbionts from the same biotype are phylogenetically more closely related, which means we have few degrees of freedom to explore whether strong protection against one parasitoid type is associated with weak protection against a second. A larger study, with more clones from different biotypes, will be needed to test this hypothesis. There is considerable genotypic and phenotypic diversity in H. defensa and their associated phage (APSE), though no correlations with aphid species or geography have been discovered so far, admittedly with relatively restrictive sampling (figure 2; see also [56]). The similarity of the phages found within each of the three biotypes, despite their collection at different times, suggests that there may be some phage population structure at the biotype scale, just as there is for the host symbiont [41]. The lack of protection in Ononis clones could conceivably result from decay of the trait over time while being cultured in the laboratory, but we consider this unlikely because there is no correlation between phenotype and the length of time in culture (table 1), and clones from all plants have been maintained in the same way since initial sampling.
Figure 3.
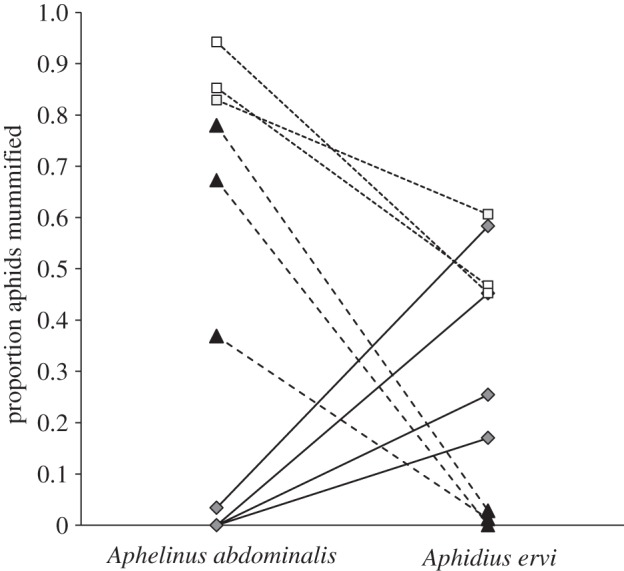
Comparison of proportion parasitism seen for the two wasps against the 10 aphid clones (all with natural H. defensa infections). White squares, Ononis aphids; black triangles, Medicago aphids; grey diamonds, Lotus aphids.
Both aphelinids and aphidiine braconids are important and common natural enemies of aphids in the field [57] and it is quite likely that the selection pressure exerted by the different parasitoid groups varies across host plants. Existing data are not extensive enough to explore this question but our results lead us to predict that aphidiines would be more important than aphelinids on Medicago and the reverse on Lotus. However, the situation in the field may be more complex. Our experiments were carried out using only single inbred lines of parasitoids and so we could not explore genotype-by-genotype specificity between symbionts and parasitoids in the effectiveness of protection, something that has previously been observed in interactions between A. fabae and L. fabarum [23,58]. It is possible that similar genotypic variation could exist within the parasitoid species attacking A. pisum. In addition, it would be desirable to confirm that our results also obtain when the aphids feed on their natural host plant rather than the ‘universal’ food plant V. faba. There is a need for field studies of the benefits and costs of symbiosis in different biotypes if we are to understand fully the dynamics of H. defensa and its bacteriophage.
Although secondary symbionts are an important component of aphid resistance to parasitoids [11,12], some pea aphid clones have been found to display considerable intrinsic resistance in the absence of symbionts [29,59]. We found little evidence for intrinsic resistance to A. abdominalis in any clone (figure 1b). However, there was greater variation in the case of intrinsic A. ervi resistance, with aphids from Ononis and Medicago showing the least innate susceptibility (figure 1a). Our assays did not distinguish between physiological (immune) defence mechanisms and physical resistance such as kicking. The fact that both symbiont and intrinsic resistance was high in Medicago clones suggest that the two mechanisms are responding to similar selection pressures rather than one substituting for the other (although data from other biotypes are needed to confirm this). In addition, the X-type symbiont which remains present in the ‘cured’ lines of two Medicago clones could also provide some resistance; the role of X-type is not well understood [10], but aphid symbionts other than H. defensa have previously been shown to improve resistance, albeit to a lesser extent [11,60].
Symbiotic bacteria play important roles in protecting their insect hosts from various threats, including viruses [61,62], fungal pathogens [13,15,63], parasites [64] and predators [65,66], as well as abiotic hazards such as heat shock [67,68]. However, these protective effects can differ markedly within a single symbiont ‘species' [5,11,15]. Our study suggests that different strains of the protective symbiont H. defensa may be adapted to defend their host against different parasitoid families. These results add to an increasingly complex and fascinating picture of phenotypic variation in the ecological effects of facultative symbionts.
Acknowledgements
We are grateful to Enric Frago, Lee Henry, Jan Hrček and Ben Parker for useful discussions; to Julia Ferrari for providing aphid clones; to Jean-Christophe Simon, Jean Peccoud and Jan Hrček for advice and assistance with the microsatellite typing; to Yang Wang and Ciara Mann for assistance with practical work; and to three anonymous reviewers for their constructive comments on earlier versions of this manuscript.
Data accessibility
Data on parasitoid resistance are available in Dryad (doi:10.5061/dryad.9n36s). DNA sequences are available in GenBank (accession numbers KT028603–KT028671).
Authors' contributions
A.H.C.M. initiated the study and carried out the experimental work and phylogenetic analysis. A.H.C.M. and H.C.J.G. designed the study, conducted statistical analysis and drafted the manuscript.
Competing interests
The authors declare no competing interests.
Funding
This work was funded by the University of Oxford and the Natural Environment Research Council.
References
- 1.Duron O, Hurst GDD. 2013. Arthropods and inherited bacteria: from counting the symbionts to understanding how symbionts count. BMC Biol. 11, 45 ( 10.1186/1741-7007-11-45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engelstädter J, Hurst GDD. 2009. The ecology and evolution of microbes that manipulate host reproduction. Annu. Rev. Ecol. Evol. Syst. 40, 127–149. ( 10.1146/annurev.ecolsys.110308.120206) [DOI] [Google Scholar]
- 3.Duron O, Bouchon D, Boutin S, Bellamy L, Zhou LQ, Engelstadter J, Hurst GD. 2008. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 6, 12 ( 10.1186/1741-7007-6-27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brownlie JC, Johnson KN. 2009. Symbiont-mediated protection in insect hosts. Trends Microbiol. 17, 348–354. ( 10.1016/j.tim.2009.05.005) [DOI] [PubMed] [Google Scholar]
- 5.Hurst GDD, Hutchence KJ. 2010. Host defence: getting by with a little help from our friends. Curr. Biol. 20, R806–R808. ( 10.1016/j.cub.2010.07.038) [DOI] [PubMed] [Google Scholar]
- 6.Chen DQ, Campbell BC, Purcell AH. 1996. A new Rickettsia from a herbivorous insect, the pea aphid Acyrthosiphon pisum (Harris). Curr. Microbiol. 33, 123–128. ( 10.1007/s002849900086) [DOI] [PubMed] [Google Scholar]
- 7.Fukatsu T, Tsuchida T, Nikoh N, Koga R. 2001. Spiroplasma symbiont of the pea aphid, Acyrthosiphon pisum (Insecta: Homoptera). Appl. Environ. Microbiol. 67, 1284–1291. ( 10.1128/AEM.67.3.1284-1291.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsuchida T, Koga R, Shibao H, Matsumoto T, Fukatsu T. 2002. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol. Ecol. 11, 2123–2135. ( 10.1046/j.1365-294X.2002.01606.x) [DOI] [PubMed] [Google Scholar]
- 9.Moran NA, Russell JA, Koga R, Fukatsu T. 2005. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 71, 3302–3310. ( 10.1128/AEM.71.6.3302-3310.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guay J-F, Boudreault S, Michaud D, Cloutier C. 2009. Impact of environmental stress on aphid clonal resistance to parasitoids: role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. J. Insect Physiol. 55, 919–926. ( 10.1016/j.jinsphys.2009.06.006) [DOI] [PubMed] [Google Scholar]
- 11.Oliver KM, Moran NA, Hunter MS. 2005. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc. Natl Acad. Sci. USA 102, 12 795–12 800. ( 10.1073/pnas.0506131102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver KM, Russell JA, Moran NA, Hunter MS. 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl Acad. Sci. USA 100, 1803–1807. ( 10.1073/pnas.0335320100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarborough CL, Ferrari J, Godfray HCJ. 2005. Aphid protected from pathogen by endosymbiont. Science 310, 1781 ( 10.1126/science.1120180) [DOI] [PubMed] [Google Scholar]
- 14.Parker BJ, Spragg CJ, Altincicek B, Gerardo NM. 2013. Symbiont-mediated protection against fungal pathogens in pea aphids: a role for pathogen specificity? Appl. Environ. Microbiol. 79, 2455–2458. ( 10.1128/aem.03193-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Łukasik P, van Asch M, Guo H, Ferrari J, Charles J, Godfray H. 2013. Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol. Lett. 16, 214–218. ( 10.1111/ele.12031) [DOI] [PubMed] [Google Scholar]
- 16.Leonardo TE, Mondor EB. 2006. Symbiont modifies host life-history traits that affect gene flow. Proc. R. Soc. B 273, 1079–1084. ( 10.1098/rspb.2005.3408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuchida T, Koga R, Fukatsu T. 2004. Host plant specialization governed by facultative symbiont. Science 303, 1989 ( 10.1126/science.1094611) [DOI] [PubMed] [Google Scholar]
- 18.Tsuchida T, Koga R, Horikawa M, Tsunoda T, Maoka T, Matsumoto S, Simon J-C, Fukatsu T. 2010. Symbiotic bacterium modifies aphid body color. Science 330, 1102–1104. ( 10.1126/science.1195463) [DOI] [PubMed] [Google Scholar]
- 19.Dixon AFG. 1998. Aphid ecology: an optimization approach, 2nd edn London, UK: Chapman and Hall. [Google Scholar]
- 20.Sequeira R, Mackauer M. 1992. Nutritional ecology of an insect host–parasitoid association: the pea aphid–Aphidius ervi system. Ecology 73, 183–189. ( 10.2307/1938730) [DOI] [Google Scholar]
- 21.Christiansen-Weniger P. 1994. Morphological observations on the preimaginal stages of Aphelinus varipes (Hym, Aphelinidae) and the effects of this parasitoid on the aphid Rhopalosiphum padi (Hom, Aphididae). Entomophaga 39, 267–274. ( 10.1007/bf02373031) [DOI] [Google Scholar]
- 22.Cayetano L, Vorburger C. 2015. Symbiont-conferred protection against hymenopteran parasitoids in aphids: how general is it? Ecol. Entomol. 40, 85–93. ( 10.1111/een.12161) [DOI] [Google Scholar]
- 23.Vorburger C, Sandrock C, Gouskov A, Castaneda LE, Ferrari J. 2009. Genotypic variation and the role of defensive endosymbionts in an all-parthenogenetic host–parasitoid interaction. Evolution 63, 1439–1450. ( 10.1111/j.1558-5646.2009.00660.x) [DOI] [PubMed] [Google Scholar]
- 24.Oliver KM, Degnan PH, Hunter MS, Moran NA. 2009. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325, 992–994. ( 10.1126/science.1174463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Degnan PH, Moran NA. 2008. Evolutionary genetics of a defensive facultative symbiont of insects: exchange of toxin-encoding bacteriophage. Mol. Ecol. 17, 916–929. ( 10.1111/j.1365-294X.2007.03616.x) [DOI] [PubMed] [Google Scholar]
- 26.Degnan PH, Moran NA. 2008. Diverse phage-encoded toxins in a protective insect endosymbiont. Appl. Environ. Microbiol. 74, 6782–6791. ( 10.1128/aem.01285-08) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moran NA, Degnan PH, Santos SR, Dunbar HE, Ochman H. 2005. The players in a mutualistic symbiosis: insects, bacteria, viruses, and virulence genes. Proc. Natl Acad. Sci. USA 102, 16 919–16 926. ( 10.1073/pnas.0507029102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weldon SR, Strand MR, Oliver KM. 2013. Phage loss and the breakdown of a defensive symbiosis in aphids. Proc. R. Soc. B 280, 20122103 ( 10.1098/rspb.2012.2103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez AJ, Weldon SR, Oliver KM. 2013. Effects of parasitism on aphid nutritional and protective symbioses. Mol. Ecol. 23, 1594–1607. ( 10.1111/mec.12550) [DOI] [PubMed] [Google Scholar]
- 30.Herzog J, Muller CB, Vorburger C. 2007. Strong parasitoid-mediated selection in experimental populations of aphids. Biol. Lett. 3, 667–669. ( 10.1098/rsbl.2007.0362) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vorburger C, Gouskov A. 2011. Only helpful when required: a longevity cost of harbouring defensive symbionts. J. Evol. Biol. 24, 1611–1617. ( 10.1111/j.1420-9101.2011.02292.x) [DOI] [PubMed] [Google Scholar]
- 32.Oliver KM, Campos J, Moran NA, Hunter MS. 2008. Population dynamics of defensive symbionts in aphids. Proc. R. Soc. B 275, 293–299. ( 10.1098/rspb.2007.1192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai B, Mackauer M. 1990. Oviposition and host-feeding patterns in Aphelinus asychis (Hymenoptera: Aphelinidae) at different aphid densities. Ecol. Entomol. 15, 9–16. ( 10.1111/j.1365-2311.1990.tb00778.x) [DOI] [Google Scholar]
- 34.Gerling D, Roitberg BD, Mackauer M. 1990. Instar-specific defense of the pea aphid Acyrthosiphon pisum: influence on oviposition success of the parasite Aphelinus asychis (Hymenoptera: Aphelmidae). J. Insect Behav. 3, 501–514. ( 10.1007/bf01052014) [DOI] [Google Scholar]
- 35.Wu Z, Heimpel GE. 2007. Dynamic egg maturation strategies in an aphid parasitoid. Physiol. Entomol. 32, 143–149. ( 10.1111/j.1365-3032.2007.00560.x) [DOI] [Google Scholar]
- 36.Pedata PA, Garonna AP, Zabatta A, Zeppa P, Romani R, Isidoro N. 2003. Development and morphology of teratocytes in Encarsia berlesei and Encarsia citrina: first record for Chalcidoidea. J. Insect Physiol. 49, 1063–1071. ( 10.1016/j.jinsphys.2003.08.003) [DOI] [PubMed] [Google Scholar]
- 37.Li S, et al. 2002. Pea aphid clonal resistance to the endophagous parasitoid Aphidius ervi. J. Insect Physiol. 48, 971–980. ( 10.1016/S0022-1910(02)00176-2) [DOI] [PubMed] [Google Scholar]
- 38.Nyabuga FN, Outreman Y, Simon JC, Heckel DG, Weisser WW. 2010. Effects of pea aphid secondary endosymbionts on aphid resistance and development of the aphid parasitoid Aphidius ervi: a correlative study. Entomol. Exp. Appl. 136, 243–253. ( 10.1111/j.1570-7458.2010.01021.x) [DOI] [Google Scholar]
- 39.Peccoud J, Ollivier A, Plantegenest M, Simon JC. 2009. A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc. Natl Acad. Sci. USA 106, 7495–7500. ( 10.1073/pnas.0811117106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrari J, West JA, Via S, Godfray HCJ. 2012. Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution 66, 375–390. ( 10.1111/j.1558-5646.2011.01436.x) [DOI] [PubMed] [Google Scholar]
- 41.Henry LM, et al. 2013. Horizontally transmitted symbionts and host colonization of ecological niches. Curr. Biol. 23, 1713–1717. ( 10.1016/j.cub.2013.07.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Godfray HCJ. 1994. Parasitoids: behavioural and evolutionary ecology. Princeton, NJ: Princeton University Press. [Google Scholar]
- 43.Bai B, Mackauer M. 1991. Recognition of heterospecific parasitism: competition between Aphidiid (Aphidius ervi) and Aphelinid (Aphelinus asychis) parasitoids of aphids (Hymenoptera: Aphidiidae; Aphelinidae). J. Insect Behav. 4, 333–345. ( 10.1007/BF01048282) [DOI] [Google Scholar]
- 44.Grasswitz TR. 1998. Effect of adult experience on the host-location behavior of the aphid parasitoid Aphidius colemani Viereck (Hymenoptera: Aphidiidae). Biol. Control 12, 177–181. ( 10.1006/bcon.1998.0633) [DOI] [Google Scholar]
- 45.Schmid M, Sieber R, Zimmermann YS, Vorburger C. 2012. Development, specificity and sublethal effects of symbiont-conferred resistance to parasitoids in aphids. Funct. Ecol. 26, 207–215. ( 10.1111/j.1365-2435.2011.01904.x) [DOI] [Google Scholar]
- 46.Henry LM, Bannerman JA, Gillespie DR, Roitberg BD. 2010. Predator identity and the nature and strength of food web interactions. J. Anim. Ecol. 79, 1164–1171. ( 10.1111/j.1365-2656.2010.01723.x) [DOI] [PubMed] [Google Scholar]
- 47.Bates D, Maechler M, Bolker B, Walker S. 2014. lme4: Linear mixed-effects models using Eigen and S4. R package v. 1.0–6 See http://CRAN.R-project.org/package=lme4. [Google Scholar]
- 48.Fox J, Weisberg S. 2011. An R companion to applied regression, 2nd edn Thousand Oaks, CA: Sage; (http://socserv.socsci.mcmaster.ca/jfox/Books/Companion) [Google Scholar]
- 49.R Development Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; (http://www.R-project.org/) [Google Scholar]
- 50.Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. ( 10.1093/bioinformatics/btm404) [DOI] [PubMed] [Google Scholar]
- 51.Hall TA. 1999. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- 52.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. ( 10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 53.Ferrari J, Darby AC, Daniell TJ, Godfray HCJ, Douglas AE. 2004. Linking the bacterial community in pea aphids with host-plant use and natural enemy resistance. Ecol. Entomol. 29, 60–65. ( 10.1111/j.1365-2311.2004.00574.x) [DOI] [Google Scholar]
- 54.Asplen MK, Bano N, Brady CM, Desneux N, Hopper KR, Malouines C, Oliver KM, White JA, Heimpel GE. 2014. Specialisation of bacterial endosymbionts that protect aphids from parasitoids. Ecol. Entomol. 39, 736–739. ( 10.1111/een.12153) [DOI] [Google Scholar]
- 55.Hansen AK, Vorburger C, Moran NA. 2012. Genomic basis of endosymbiont-conferred protection against an insect parasitoid. Genome Res. 22, 106–114. ( 10.1101/gr.125351.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russell JA, et al. 2013. Uncovering symbiont-driven genetic diversity across North American pea aphids. Mol. Ecol. 22, 2045–2059. ( 10.1111/mec.12211) [DOI] [PubMed] [Google Scholar]
- 57.Müller CB, Adriaanse ICT, Belshaw R, Godfray HCJ. 1999. The structure of an aphid–parasitoid community. J. Anim. Ecol. 68, 346–370. ( 10.1046/j.1365-2656.1999.00288.x) [DOI] [Google Scholar]
- 58.Rouchet R, Vorburger C. 2012. Strong specificity in the interaction between parasitoids and symbiont-protected hosts. J. Evol Biol. 25, 2369–2375. ( 10.1111/j.1420-9101.2012.02608.x) [DOI] [PubMed] [Google Scholar]
- 59.Martinez AJ, Ritter SG, Doremus MR, Russell JA, Oliver KM. 2014. Aphid-encoded variability in susceptibility to a parasitoid. BMC Evol. Biol. 14, 127 ( 10.1186/1471-2148-14-127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vorburger C, Gehrer L, Rodriguez P. 2010. A strain of the bacterial symbiont Regiella insecticola protects aphids against parasitoids. Biol. Lett. 6, 109–111. ( 10.1098/rsbl.2009.0642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hedges LM, Brownlie JC, O'Neill SL, Johnson KN. 2008. Wolbachia and virus protection in insects. Science 322, 702 ( 10.1126/science.1162418) [DOI] [PubMed] [Google Scholar]
- 62.Teixeira L, Ferreira A, Ashburner M. 2008. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6, 2753–2763. ( 10.1371/journal.pbio.1000002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaltenpoth M, Gottler W, Herzner G, Strohm E. 2005. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr. Biol. 15, 475–479. ( 10.1016/j.cub.2004.12.084) [DOI] [PubMed] [Google Scholar]
- 64.Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. 2010. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329, 212–215. ( 10.1126/science.1188235) [DOI] [PubMed] [Google Scholar]
- 65.Kellner RLL. 1999. What is the basis of pederin polymorphism in Paederus riparius rove beetles? The endosymbiotic hypothesis. Entomol. Exp. Appl. 93, 41–49. ( 10.1046/j.1570-7458.1999.00560.x) [DOI] [Google Scholar]
- 66.Piel J, Höfer I, Hui D. 2004. Evidence for a symbiosis island involved in horizontal acquisition of pederin biosynthetic capabilities by the bacterial symbiont of Paederus fuscipes beetles. J. Bacteriol. 186, 1280–1286. ( 10.1128/jb.186.5.1280-1286.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Montllor CB, Maxmen A, Purcell AH. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27, 189–195. ( 10.1046/j.1365-2311.2002.00393.x) [DOI] [Google Scholar]
- 68.Chen DQ, Montllor CB, Purcell AH. 2000. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid A. kondoi. Entomol. Exp. Appl. 95, 315–323. ( 10.1046/j.1570-7458.2000.00670.x) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data on parasitoid resistance are available in Dryad (doi:10.5061/dryad.9n36s). DNA sequences are available in GenBank (accession numbers KT028603–KT028671).