Vernalization has proven to pivot around chromatin changes, so that the balance of antagonistic chromatin-modifying complexes provides a fine level of control that is a target for adaptation.
Abstract
Analysis of how seasonal cues influence the timing of the floral transition has revealed many important principles for how epigenetic regulation can integrate a variety of environmental cues with developmental signals. The study of the pathways that necessitate overwintering in plants and their ability to respond to prolonged cold (the vernalization requirement and response pathways) has elaborated different chromatin regulatory pathways and the involvement of noncoding RNAs. The major target of these vernalization pathways in Arabidopsis (Arabidopsis thaliana) is Flowering Locus C (FLC). A relatively simple picture of FLC regulation is emerging of a few core complexes and mechanisms that antagonize each other’s actions. This balance provides a fine degree of control that has nevertheless permitted evolution of a wide range of natural variation in vernalization in Arabidopsis. Similar simple routes of adaptation may underlie life history variation between species.
The time at which different species flower is an important marker of seasonal and climatic changes and is ecologically and economically important. The sessile nature of plants means that they experience the full range of environmental changes over the seasons. Flowering time control in many species is highly responsive to environmental cues and therefore very sensitive to local climate conditions. The impact of this on many ecosystem and agricultural processes has made understanding flowering time control an important objective.
Many genetic pathways influence flowering time, either as part of seasonal (photoperiod and past and present temperature), developmental (developmental phase and age), or stress response (overcrowding and nutrient stress). Despite the variety of competing inputs, these many and various mechanisms are integrated at the action of a small number of nodes in Arabidopsis (Arabidopsis thaliana) termed floral pathway integrators (Simpson and Dean, 2002). Analyses of the genes identified by flowering time mutants have shown many have roles as chromatin modifiers (Andrés and Coupland, 2012; Pajoro et al., 2014). Timing of flowering seems particularly sensitive to chromatin regulation, potentially due to the necessity for long-term storage of seasonal information.
In this review, we focus on vernalization in Arabidopsis and summarize our understanding of how chromatin modifiers interact with other proteins and noncoding RNAs to integrate developmental and temperature cues into chromatin changes at the key integrating locus Flowering Locus C (FLC). Further, we explore how changes in these mechanisms underlie different life history strategies and vernalization responses in different accessions and species. A key observation we wish to convey is that the complexity of these systems at the molecular level belies simplicity in balancing forces that enable a fine degree of control and adaptive responses at the phenotypic level.
FLOWERING TIME CONTROL
Analysis of the first genetic screens of Arabidopsis mutants affecting the transition to flowering specifically (as opposed to those that pleiotropically affect development or downstream steps) classed mutants into three principal floral transition pathways (Koornneef et al., 1991; Simpson and Dean, 2002). The three pathways are the photoperiod pathway (controlling response to changes in daylength), vernalization (accelerated flowering in response to prolonged winter-like cold), and the autonomous pathway, a group of mutants in which flowering remains prompted by vernalization but is otherwise severely delayed (Simpson and Dean, 2002). The expression of the floral integrator FT is influenced by all three pathways and, as such, is the major regulator of flowering in response to seasonal changes. The autonomous and vernalization pathways converge upstream of FT at the repression of the major floral regulator FLC (Simpson and Dean, 2002). FLC is a MADS-box transcription factor that directly represses the expression of FT, and its expression must normally be reduced before FT can be activated (Searle et al., 2006). The main photoperiod pathway integrator, CONSTANS (CO), is a transcription factor that acts antagonistically to FLC by activating the FT locus, and its protein level is only high when production coincides with certain circadian periods. Stabilization of the CO protein occurs only in the light, so that FT expression is only promoted both when winter has passed (FLC is low) and the day lengthens to be light in the evening (CO is high; for review, see Andrés and Coupland, 2012).
Other inputs that affect flowering often act via modulation of these canonical pathways; for example, both heat and short-term cold also affect FLC expression via effects at its chromatin (Jung et al., 2013; Gan et al., 2014; Bouché et al., 2015) or by changing the balance between them, such as the shade avoidance response, which allows the photoperiod pathway to override the repression of FT by FLC (Wollenberg et al., 2008). Redundancy is also present in these pathways: FLC has a clade of five homologs called the MADS AFFECTING FLOWERING1 (MAF1) to MAF5 genes, although they do not play exactly the same role as FLC (Ratcliffe et al., 2003). Chromatin modification in flowering time control acts at several loci, including the MAF genes, but it has been best characterized at the key integrators for control in response to seasonal cues, FT and especially FLC (Andrés and Coupland, 2012).
FLC AS A GATEWAY TO UNDERSTANDING CHROMATIN COMPLEXITY
The study of regulation at the FLC locus is shedding light on the different mechanisms that can be employed to program one locus with multiple commands, in part because the vernalization response allows the study of chromatin responses to environmental cues and partly due to the plethora of mechanisms acting at FLC. FLC represses flowering in a dose-dependent manner (Michaels and Amasino, 1999; Sheldon et al., 1999, 2000), and Arabidopsis accessions that have high initial FLC flower relatively late, while those with lower starting FLC flower proportionately earlier (Shindo et al., 2006). However, during cold exposure, FLC expression is reduced and the locus is quantitatively silenced in proportion to the duration of cold (Sheldon et al., 2000; Angel et al., 2011). On return to warm conditions the silencing is maintained epigenetically, enabling other environmental cues to promote the floral switch (Simpson and Dean, 2002). This epigenetic silencing is stable through development until the epigenetic state of FLC is reset and expression restored to high levels during embryogenesis, a process that involves both a histone demethylase and protein called FRIGIDA (FRI; Sheldon et al., 2008; Choi et al., 2009; Crevillén et al., 2014).
Balancing Up: High FLC
The ability to appropriately reset epigenetic states is a prerequisite for the marking of phase transitions and to prevent epigenetic information being carried over to the next generation. In the case of FLC, the silencing generated during vernalization leads to the accumulation of posttranslational modifications of the histones present at the FLC locus, in particular, trimethylation of lysine-27 of histone H3 (H3K27me3; Shindo et al., 2006), a mark associated with Polycomb gene repression in many organisms. This then needs to be removed to establish high levels of FLC expression at the beginning of every new generation. A mutagenesis screen aimed at identifying players required for FLC resetting implicated the Jumonji-C domain-containing (JMJ) demethylase EARLY FLOWERING6 in the removal of H3K27me3 at FLC during early embryogenesis (Crevillén et al., 2014). This hypomorphic allele leads to failure to completely remove H3K27me3 at FLC and thus to reset FLC expression, so resulting in inheritance of a partially silenced FLC into the next generation (Crevillén et al., 2014).
Other factors are also required to promote FLC expression to a high level in the young plant (Choi et al., 2009). FLC expression is reset during early embryo development, but it is then increased throughout embryogenesis to a maximum at final seed formation (Sheldon et al., 2008; Choi et al., 2009; Crevillén et al., 2014). Key to this increase is FRI, a coiled-coil protein that directly interacts with the nuclear cap-binding complex and increases the proportion of FLC mRNA carrying a 5′ cap (Fig. 1; Choi et al., 2009; Geraldo et al., 2009). FRI levels are limited by activity of the proteasome (Hu et al., 2014). FRI associates with other transcriptional regulators, including members of the FRIGIDA-LIKE and FLC EXPRESSOR families and SUPPRESSOR OF FRIGIDA4 to form a FRI-complex (Choi et al., 2011). The FRI-complex seems to act as a scaffold to recruit several more general regulators and chromatin modifiers, including a COMPASS-like H3K4 methyltransferase complex that includes ARABIDOPSIS TRITHORAX1, a H3K36me3 methyltransferase (EARLY FLOWERING IN SHORT DAYS), and the SWR1 complex, which is implicated in the remodelling of H2A histones to the activity-associated H2A.Z variant (Deal et al., 2007; Jiang et al., 2009; Choi et al., 2011). As well as increasing the proportion of capped FLC mRNA, the FRI complex promotes the levels of H3K4me3, H3K36me3, and acetylation on H3 and H4 within the FLC gene, all marks associated with high transcription. Thus, this complex acts to remodel the chromatin environment at FLC to promote high expression. In the absence of FRI, other components remain active, but FLC is not reactivated to such a high level and therefore flowering is not so significantly delayed (Ding et al., 2013).
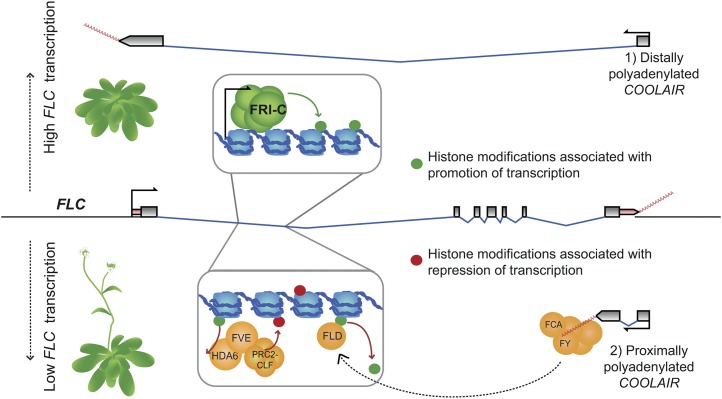
Figure 1.
FLC and COOLAIR circuitry: setting FLC expression in the warm. High expression of FLC is promoted by FRI, COMPASS-like, and SWR1 complexes (FRI-C). Their activity is associated with active histone modifications (green circles). This high-expression state promotes the distally polyadenylated form of COOLAIR (1). In opposition, FPA and the FCA/FY complex promote proximally polyadenylated COOLAIR (2) and, via FLD, remove the active histone modifications. FVE also acts, independently, to reduce FLC expression by removing active histone modifications (with HISTONE DEACETYLASE6 [HDA6]) and aiding deposition of repressive ones (red circles). FRI-C, FRI-complex.
FLC and COOLAIR Circuitry: Maintaining the Balance between Pathways in the Warm
The autonomous pathway consists of a group of mutants characterized by causing late flowering in rapid-cycling Columbia (Col-0) and Landsberg erecta ecotypes. Both Col-0 and Landsberg erecta have low FLC expression throughout their life cycle (due to lack of an active FRI allele; Johanson et al., 2000), so they do not require vernalization to flower rapidly. Mutants of FCA, FPA, FY, FLOWERING LOCUS D (FLD), FLOWERING LOCUS K, LUMINIDEPENDENS, and FVE up-regulate FLC despite the absence of FRI, and these higher levels are reversed by vernalization. These genes do not all act in the same molecular mechanism, so their activities are classified within the autonomous pathway through their common phenotypic effects; however, they all do counterbalance the activating complexes in the absence of vernalization, helping to establish the steady-state level of FLC expression (Fig. 1; Simpson and Dean, 2002).
Three of the classical loci, FCA, FPA, and FY, are RNA-processing factors, and their function involves a set of long noncoding antisense transcripts that cover the FLC locus, collectively termed COOLAIR (for review, see Ietswaart et al., 2012). The COOLAIR promoter is just downstream of the FLC polyadenylation [poly(A)] site and drives transcription of a number of capped, alternatively spliced, and polyadenylated transcripts (Hornyik et al., 2010; Liu et al., 2010). The two main classes of COOLAIR transcripts either terminate at a proximal poly(A) site cluster within intron 6 of the FLC gene body (class I) or read through the entire FLC coding sequence to a distal site cluster within the FLC promoter (class II; Fig. 1; Hornyik et al., 2010; Liu et al., 2010). FCA and FPA, both RNA recognition motif proteins, function partially redundantly to control alternative splicing and 3′-end processing of mRNAs (Hornyik et al., 2010). At COOLAIR, they act with FY, a cleavage and poly(A) specificity factor component, and the cleavage stimulation factors CstF64 and CstF77 to promote the choice of the proximal poly(A) site (Liu et al., 2010). The choice of this site is also increased by the activity of the core spliceosome component PRP8 and CYCLIN DEPENDENT KINASE GROUP C2, a Positive Transcription Elongation Factor b component (Marquardt et al., 2014; Wang et al., 2014b). The use of the proximal site decreases the level of H3K4me2 methylation in the FLC gene body via the action of the H3K4 demethylase FLD, reducing FLC transcription (Liu et al., 2007, 2010). The state of the chromatin in turn influences the choice of the COOLAIR poly(A) site, as fld mutants have higher ratios of class II to class I, possibly due to higher transcription rates at permissive H3K4me2-marked chromatin promoting read-through to the distal site (Liu et al., 2007; Marquardt et al., 2014). Such positive feedback loops help stabilize active or inactive chromatin states.
Two other features are present at the COOLAIR promoter. Firstly, this region is also targeted by the small interfering RNA pathway and marked by the heterochromatin-associated H3K9me2 modification, both of which act to repress FLC expression (Swiezewski et al., 2007). Secondly, an RNA-DNA hybrid forms over the COOLAIR promoter and much of the proximal COOLAIR species (Sun et al., 2013). This structure, known as an R-loop, is formed when antisense RNA invades double-stranded DNA to form a heteroduplex. The R-loop at FLC is stabilized by the binding of plant homeodomain-containing protein AtNODULIN HOMEOBOX and reduces transcription of COOLAIR (Sun et al., 2013).
Despite their balancing activities, only one autonomous pathway component has been reported to interact genetically with the FRI complex. Loss of FVE synergistically enhances FLC expression level in an active FRI background, suggesting it may directly antagonize the action of the FRI-complex (Lee and Amasino, 2013). FVE/MULTICOPY SUPPRESSOR OF IRA1 4 (MSI4) encodes a relative of retinoblastoma-associated proteins and has been reported to repress FLC by mediating histone deacetylation via association with HISTONE DEACETYLASE6 (Ausín et al., 2004; Gu et al., 2011). FVE has also been reported to interact with a Polycomb Repressive Complex2 (PRC2; Pazhouhandeh et al., 2011) but not with LIKE HETEROCHROMATIN PROTEIN1 (LHP1) nor as a core component of PRC2 (Derkacheva et al., 2013). Core components of PRC2 are Enhancer of zeste [E(z); a histone methyltransferase], Suppressor of zeste12 [Su(z)12], Extra Sex Combs, and MSI1. In plants, multiple semiredundant homologs exist that form at least three independent complexes differing in their targets (Xiao and Wagner, 2015). PRC2 complexes deposit H3K27me3 at target loci, and in warm conditions, a PRC2 containing the E(z) homolog CURLY LEAF (CLF) deposits H3K27me3 at FLC (Schubert et al., 2006). FVE, an MSI1 ortholog, is required for the CLF-PRC2 to induce a low level of H3K27me3-mediated repression in warm conditions (Pazhouhandeh et al., 2011).
The action of FVE in deacetylation also acts as a route for environmental inputs to FLC. Short-term intermittent cold stress (as opposed to long-term, vernalizing cold) causes the up-regulation of FLC via the interaction of FVE with a cold-induced E3 ubiquitin ligase HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1, delaying flowering by interfering with FVE-associated histone deacetylation in response to cold snaps (Jung et al., 2013). Similarly, high temperatures up-regulate FLC by the action of redundant JMJ demethylases JMJ30 and JMJ32, which remove the low prevernalization H3K27me3 level to prevent precocious flowering (Gan et al., 2014). Thus, the balance of autonomous pathway activities acts to integrate environmental information into flowering control.
FLC during Vernalization: Tipping the Balance in the Cold
During vernalization, FLC expression is down-regulated within 2 weeks of experiencing cold and is then epigenetically silenced by the activity of a plant-homeodomain zinc-finger (PHD)-PRC2 complex (Fig. 2; Song et al., 2013). COOLAIR transcripts do not seem to be absolutely required for vernalization (Helliwell et al., 2011), but replacement of the FLC terminator/COOLAIR promoter with an alternative promoter (terminator exchange FLCTEX) revealed that in the absence of COOLAIR, the rate of FLC down-regulation in the cold is reduced (Csorba et al., 2014). In wild-type plants, COOLAIR expression is normally up-regulated within 10 d of constant cold and reaches a maximum after 3 weeks. This is the earliest cold response among the known vernalization regulators; COOLAIR expression then reduces before plants return to the warmth (Swiezewski et al., 2009). Proximal COOLAIR increases significantly more than the distal class, suggesting the early phase of vernalization may be similar to that of the autonomous pathway in warm conditions (Csorba et al., 2014).
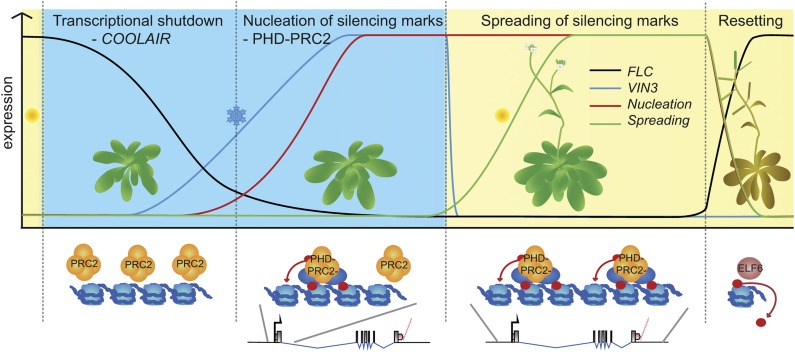
Figure 2.
FLC and vernalization. A series of stages during vernalization ensures the stable shutdown of the FLC locus. In the first phase of cold exposure, FLC reduction correlates with increased COOLAIR. Longer cold induces expression of VIN3 (blue line and ovals), which triggers nucleation of PHD-PRC2 (red line) and increased H3K27me3 at the nucleation region. On return to warmth, PHD-PRC2 spreads across the whole locus, leading to high H3K27me3 over the entire gene (green line). This maintains epigenetic silencing of FLC until resetting occurs in the embryo. Red circles show repressive histone modifications. ELF6, EARLY FLOWERING6.
Once FLC expression has been down-regulated, it is epigenetically silenced by a PHD-PRC2 complex that contains a constitutively expressed protein called VERNALIZATION2 (VRN2) as its Su(z)12 homolog, MSI1, and either CLF or, more usually, SWINGER as its E(z) member (Fig. 2; Gendall et al., 2001; Wood et al., 2006; De Lucia et al., 2008). VRN2 is present at the locus before cold, but its activity is significantly boosted through association with PHD proteins of the VERNALIZATION5/VIN3-Like family (Mylne et al., 2004; Sung et al., 2006b; Greb et al., 2007). This activation is independent of COOLAIR but has been proposed to involve another noncoding RNA named COLDAIR (Heo and Sung, 2011; Csorba et al., 2014). COLDAIR is a capped, but not polyadenylated, noncoding transcript that arises from the long intron I of FLC and is up-regulated by cold with a peak approximately 10 d after the COOLAIR maximum, albeit at a much lower level than COOLAIR (Heo and Sung, 2011). COLDAIR associates transiently with CLF in vivo and has been proposed to promote its recruitment to FLC (Heo and Sung, 2011). Epigenetic silencing is dependent on the cold-induced up-regulation of the PHD-encoding gene VERNALIZATION INSENSITIVE3 (VIN3), up-regulated only during long cold with expression maximal after approximately 4 to 6 weeks (Sung and Amasino, 2004). VIN3 up-regulation increases faster in a mutant defective in the extra-nuclear methyltransferase SET DOMAIN GROUP7 (Lee et al., 2015). VIN3 is thought to recruit its constitutively expressed homolog VRN5 to the first few nucleosomes of the FLC first exon and beginning of intron 1, called the nucleation region (Sung and Amasino, 2004; Greb et al., 2007; De Lucia et al., 2008). H3K27me3 levels increase at the nucleation region in proportion with the length of cold experienced. After return to the warmth, the PHD-PRC2 spreads to cover the whole FLC locus, leading to high H3K27me3 along its entire length. This is required to maintain the epigenetic silencing at FLC throughout the many cell divisions of subsequent development in the warmth (Finnegan and Dennis, 2007; De Lucia et al., 2008).
VRN2, VRN5, VIN3, and the genes VRN1 and LHP1 are all necessary for the maintenance of FLC silencing after the cold, and their respective mutants have reduced sensitivity to vernalization (Gendall et al., 2001; Levy et al., 2002; Sung and Amasino, 2004; Mylne et al., 2006; Sung et al., 2006a; Greb et al., 2007). LHP1 binds and maintains H3K27me3, with LHP1 and EMBRYONIC FLOWER1 proposed to form a PRC1-like complex. This is required to maintain repression at FT (Wang et al., 2014a) and could play a similar role at FLC (Sung et al., 2006a). VRN1 contains plant-specific B3 domains and binds double-stranded DNA nonspecifically along all the chromosomes. Unusually, it remains bound throughout mitosis, a feature that may relate to the mitotic stability of epigenetic silencing, but otherwise its mode of action is unknown (Mylne et al., 2006).
Calculating the Balance
As well as reduced H3K27me3, the vrn, vin3, and lhp1 mutants are defective in the cold-induced decrease of histone acetylation and increased H3K9me2 methylation at FLC (Mylne et al., 2006; Sung et al., 2006a; Greb et al., 2007; Kim and Sung, 2013). The interactions between different chromatin modifications is a key question in epigenetics, and at FLC, some of the answers to it are arising from answers to a very different problem: how to generate a memory of cold that is quantitative in proportion to the length of cold experienced.
Angel et al. (2011) tackled the question of quantitative silencing of FLC using a combination of mathematical modeling and experimentation. They showed that the quantitative accumulation of H3K27me3 was not a gradual accumulation at every FLC locus, but the result of a Polycomb-mediated cell-autonomous switch promoted by cold exposure. FLC is in either of two states, ON or OFF, the latter marked by H3K27me3, with the fraction of cells in the OFF state increasing with cold exposure duration. The model envisages two counteracting positive feedback loops that result in bistability: one involving H3K27me3 and the other involving an activating mark (Angel et al., 2011). Given the role of H3K4 and H3K36me3 methyltransferases in the activating FRI-complex, these modifications were good candidates for the activating mark and proved to match some but not all of the model’s predictions (Yang et al., 2014). H3K36me3 particularly fulfils two requirements of the model: H3K36me3 and H3K27me3 do not occur on the same histone tail, and the dynamics of H3K36me3 accumulation are broadly antagonistic to those of H3K27me3 at the nucleation region (Yang et al., 2014). However, the spatial patterns of activating versus repressing marks do not exactly counterbalance, and moreover, when histone dynamics were compared in the COOLAIR-disrupted FLCTEX line and in the vrn5 mutant, H3K36me3 dynamics were uncoupled from the tight pairing with H3K27me3 that the Angel et al. (2011) model predicts (Csorba et al., 2014). This emphasizes the roles of the other factors known to act at FLC and presents the challenge of incorporating their action into a global model of regulation at FLC.
The cell-autonomous ON/OFF nature of Polycomb silencing at FLC enabled a key question to be tackled: Are the chromatin modifications induced by the Polycomb system directing the epigenetic memory, or are they a consequence of the action of a trans-factor network? Epigenetic memory can be stored in the concentrations of diffusible regulatory factors (trans-memory; Ptashne, 2014). Alternatively, memory can be stored locally in the chromatin environment of individual genes (cis-memory; Moazed, 2011). In trans-memory, the chromatin responds to the transcriptional state defined by heritable concentrations of the transfactors, whereas in cis-memory, the local chromatin environment instructs its own inheritance and is therefore the key epigenetic memory element. By monitoring the expression of two copies of the Arabidopsis Polycomb target gene FLC in the same plants, Berry et al. (2015) showed that one copy can be repressed while the other is active. Furthermore, this mixed expression state is inherited through many cell divisions as plants develop; the local chromatin environment at FLC thus instructs its own epigenetic inheritance (Berry and Dean, 2015).
Temperature perception during vernalization is also likely to be registered in an all-or-nothing manner (Angel et al., 2015). Cold promotes a low-probability stochastic switch resulting in nucleation of H3K27me3 at the FLC nucleation region. Such a mechanism facilitates integration of the very noisy temperature signals normally experienced by plants over winter. How these digital switches influence nuclear organization of FLC has been extensively reviewed by Zhu et al. (2015). An early cold step, preceding H3K27me3 nucleation, is disruption of a gene loop between the FLC promoter and COOLAIR promoters (Crevillén et al., 2013). FLC alleles also cluster within the nucleus in the cold, with similar dynamics to the appearance of H3K27me3 nucleation. This clustering’s dependence on VRN2 and VRN5 suggests that it is tightly connected with the triggering of Polycomb silencing (Rosa et al., 2013).
FLC IN A COLD CLIMATE
The level of FLC expression and the responsiveness to vernalization largely defines the life history of a particular genotype. Arabidopsis accessions lacking a functional FRIGIDA, such as Col-0, do not require vernalization, permitting them to go through several life cycles within a year (Johanson et al., 2000; Le Corre et al., 2002). While the allele of FRI, the trans-activating complex, largely defines the requirement for vernalization (Johanson et al., 2000; Le Corre et al., 2002; Stinchcombe et al., 2004; Werner et al., 2005), cis-variation at the FLC allele defines the vernalization response profile (Werner et al., 2005; Shindo et al., 2006; Coustham et al., 2012; Sánchez-Bermejo et al., 2012; Li et al., 2014). Studies using Quantitative Trait Loci approaches to investigate flowering time have repeatedly identified the importance of FLC cis-variation to life history and seasonal response (Werner et al., 2005; Shindo et al., 2006; Strange et al., 2011; Sánchez-Bermejo et al., 2012). This is despite FLC rarely appearing in Genome Wide Association Studies, due to loss of statistical power caused by structurally distinct alleles conferring similar functions (Atwell et al., 2010; Li et al., 2010). In one accession, Lövvik-1, a distinctive requirement for a very long period of cold has been mapped to the presence of only four single nucleotide polymorphisms (SNPs) at the 5′ end of the locus (Coustham et al., 2012). These SNPs change the silencing profile of FLC so that longer vernalization is required to build up sufficient H3K27me3; without sufficient cold, silencing lapses and FLC expression reactivates (Coustham et al., 2012).
In a study of the structure of variation at FLC, Li et al. (2014) identified that Arabidopsis accessions worldwide cluster into only 20 haplotypes, which extend many kilobases either side of FLC. These SNPs do not affect the FLC protein and are largely in noncoding regions. The most common five haplotypes show different geographic distributions but can be grouped by their response to vernalization into two Rapid Vernalizer types that are fully vernalized after 4 weeks and three Slow Vernalizer types requiring 8 or more weeks. The cis-variation at FLC of each group is sufficient to generate their characteristic vernalization response when transgenically placed in the same genetic background (Li et al., 2014). For one haplotype, a single noncoding polymorphism influencing COOLAIR splicing accounts for its characteristic high FLC expression (Li et al., 2015). The suppression of recombination between haplotypes strongly suggests that these cis-variations have been selected for altered epigenetic regulation that results in life history variation. As FLC has other roles in development, this may involve more than just flowering time (Springthorpe and Penfield, 2015).
LESSONS FOR OTHER SPECIES
The study of FLC is proving to be highly transferable into other systems. Within the Brassicaceae, the FLC homologs of perennial species Arabidopsis halleri (AhFLC) and Arabis alpina (PERPETUAL FLOWERING1 [PEP1]) both show annual cycles of repression in response to vernalization in which FLC controls the timing of both onset of flowering via its repression and the reversion to vegetative growth via FLC reactivation (Wang et al., 2009; Aikawa et al., 2010). As in A. thaliana, much expression variation in A. alpina is due to cis-variation in PEP1, and the mechanisms at work, H3K27me3 methylation and COOLAIR, are also conserved (Albani et al., 2012; Castaings et al., 2014). COOLAIR is also conserved in Arabidopsis lyrata, but only the proximal (class I) is highly conserved between all three systems (Castaings et al., 2014). More broadly, FLC homologs have been identified as quantitative trait loci for flowering in the more distantly related genus Brassica, and homologs that respond to vernalization have even been found in monocots (Ruelens et al., 2013; Xiao et al., 2013).
Results from these systems suggest that the A. thaliana system represents a truncated version of an ancestral perennial system in which FLC reactivates, a phenomenon still seen in strongly reactivating accessions such as Lövvik-1 (Coustham et al., 2012). From this ancestral system, small changes such as increased epigenetic silencing could have allowed a radiation of life histories such as in Figure 3. Short cuts then arise to radically different phenologies via loss of key actors such as FRI, but there are also ample mechanisms that can be fine-tuned to provide appropriate responses to the very complex temperature profiles that plants experience.
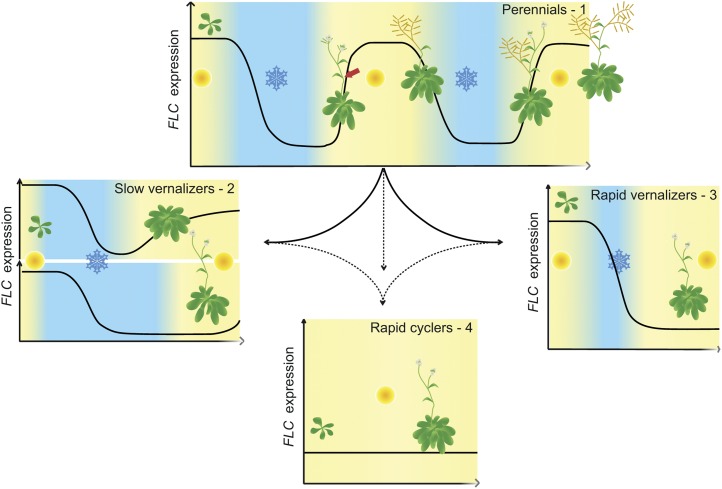
Figure 3.
A radiation of adaptations. In a generalized model of FLC homolog behavior in perennials (1) drawn from data from A. alpina and A. halleri, seasonal FLC repression promotes flowering in some meristems, whereas its programmed reactivation ensures some meristems remain vegetative (red arrow). In other models, such as the annual Arabidopsis, this reactivation still occurs after insufficient cold in slow-vernalizing accessions but is abolished by longer periods of cold (2) and has been lost completely in rapid-vernalizing haplotypes (3), with this variation in the vernalization response being defined by cis-variation at FLC. In rapid-cycling accessions, loss of the trans-factor FRI further changes life history (4).
This fine-tuning of response to climate has been demonstrated in A. halleri, in which, despite the complexity of diurnal temperature fluctuations, modeling has shown that the AhFLC profile responds to an accumulation of the low temperatures of the previous 6 weeks (Aikawa et al., 2010). However, in A. thaliana, instead of the local ecotype best suiting its original environment, ecotypes from warmer climes have shown higher fitness at given locations in recent years (Wilczek et al., 2014). One of the major challenges for the A. thaliana vernalization model now is to start moving mechanisms from experimental conditions into the understanding of responses to field temperatures. Although many components are known to be required for vernalization in the lab, which of these sense different elements of the field temperature profile, and how, remain largely unanswered. If most variation occurs at FLC itself, which mechanisms are being adapted to produce location-appropriate response? And how will vernalization of native plants and crops respond to global warming?
CONCLUSION
The scientific scrutiny applied to flowering time for many decades has provided examples of the complex mechanisms with which endogenous and environmental cues can be used to entrain developmental phase change, in plants and beyond. The two genes integrating a high number of environmental inputs, FT and FLC, are those with apparently the most complex regulation, much of it involving chromatin modification. In these complex external and internal environments, modeling has been a useful innovation in understanding this complexity in a single species and system, allowing reduction of multiprotein complexes and multiple inputs to understanding at the level of antagonizing forces.
Arabidopsis is a limited model for flowering control as a monocarpic annual, but conservation of mechanisms has allowed its lessons to be spread throughout Brassicaceae and taken as simple paradigms. The wide geographical spread of Arabidopsis, combined with comparison to other models, facilitates research into how modulation of chromatin and long noncoding RNAs can define adaptation over different scales. In this way, Arabidopsis FLC can be seen as translation software for researchers to understand adaptation of gene expression control. Just as FLC allows Arabidopsis to translate its environment, Arabidopsis research can help researchers to understand the impact of anthropogenic climate change on ecosystems.
Acknowledgments
We thank Michael Lenhard, Vinod Kumar, Julia Qüesta, Rebecca Bloomer, and Scott Berry for helpful comments on the article.
Glossary
- Col-0
ecotype Columbia
- poly(A)
polyadenylation
- SNP
single nucleotide polymorphism
Footnotes
This work was supported by the European Research Council (grant no. MEXTIM) and the Biotechnology and Biological Sciences Research Council Institute Strategic program (grant no. BB/J004588).
References
- Aikawa S, Kobayashi MJ, Satake A, Shimizu KK, Kudoh H (2010) Robust control of the seasonal expression of the Arabidopsis FLC gene in a fluctuating environment. Proc Natl Acad Sci USA 107: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani MC, Castaings L, Wötzel S, Mateos JL, Wunder J, Wang R, Reymond M, Coupland G (2012) PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering. PLoS Genet 8: e1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Angel A, Song J, Dean C, Howard M (2011) A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476: 105–108 [DOI] [PubMed] [Google Scholar]
- Angel A, Song J, Yang H, Questa JI, Dean C, Howard M (2015) Vernalizing cold is registered digitally at FLC. Proc Natl Acad Sci USA 112: 4146–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausín I, Alonso-Blanco C, Jarillo JA, Ruiz-García L, Martínez-Zapater JM (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36: 162–166 [DOI] [PubMed] [Google Scholar]
- Berry S, Dean C (2015) Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J 83: 133–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry S, Hartley M, Olsson TSG, Dean C, Howard M (2015) Local chromatin environment of a Polycomb target gene instructs its own epigenetic inheritance. eLife 4: 07205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché F, Detry N, Périlleux C (2015) Heat can erase epigenetic marks of vernalization in Arabidopsis. Plant Signal Behav 10: e990799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L, Bergonzi S, Albani MC, Kemi U, Savolainen O, Coupland G (2014) Evolutionary conservation of cold-induced antisense RNAs of FLOWERING LOCUS C in Arabidopsis thaliana perennial relatives. Nat Commun 5: 4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Hyun Y, Kang MJ, In Yun H, Yun JY, Lister C, Dean C, Amasino RM, Noh B, Noh YS, et al. (2009) Resetting and regulation of Flowering Locus C expression during Arabidopsis reproductive development. Plant J 57: 918–931 [DOI] [PubMed] [Google Scholar]
- Choi K, Kim J, Hwang HJ, Kim S, Park C, Kim SY, Lee I (2011) The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23: 289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J, Dean C (2012) Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science 337: 584–587 [DOI] [PubMed] [Google Scholar]
- Crevillén P, Sonmez C, Wu Z, Dean C (2013) A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J 32: 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crevillén P, Yang H, Cui X, Greeff C, Trick M, Qiu Q, Cao X, Dean C (2014) Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515: 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Questa JI, Sun Q, Dean C (2014) Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci USA 111: 16160–16165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AME, Greb T, Dean C (2008) A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA 105: 16831–16836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Topp CN, McKinney EC, Meagher RB (2007) Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkacheva M, Steinbach Y, Wildhaber T, Mozgová I, Mahrez W, Nanni P, Bischof S, Gruissem W, Hennig L (2013) Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J 32: 2073–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Kim SY, Michaels SD (2013) FLOWERING LOCUS C EXPRESSOR family proteins regulate FLOWERING LOCUS C expression in both winter-annual and rapid-cycling Arabidopsis. Plant Physiol 163: 243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES (2007) Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr Biol 17: 1978–1983 [DOI] [PubMed] [Google Scholar]
- Gan ES, Xu Y, Wong JY, Goh JG, Sun B, Wee WY, Huang J, Ito T (2014) Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat Commun 5: 5098. [DOI] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525–535 [DOI] [PubMed] [Google Scholar]
- Geraldo N, Bäurle I, Kidou S, Hu X, Dean C (2009) FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol 150: 1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Mylne JS, Crevillen P, Geraldo N, An H, Gendall AR, Dean C (2007) The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol 17: 73–78 [DOI] [PubMed] [Google Scholar]
- Gu X, Jiang D, Yang W, Jacob Y, Michaels SD, He Y (2011) Arabidopsis homologs of retinoblastoma-associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PLoS Genet 7: e1002366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Robertson M, Finnegan EJ, Buzas DM, Dennis ES (2011) Vernalization-repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. PLoS One 6: e21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hornyik C, Terzi LC, Simpson GG (2010) The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell 18: 203–213 [DOI] [PubMed] [Google Scholar]
- Hu X, Kong X, Wang C, Ma L, Zhao J, Wei J, Zhang X, Loake GJ, Zhang T, Huang J, et al. (2014) Proteasome-mediated degradation of FRIGIDA modulates flowering time in Arabidopsis during vernalization. Plant Cell 26: 4763–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ietswaart R, Wu Z, Dean C (2012) Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet 28: 445–453 [DOI] [PubMed] [Google Scholar]
- Jiang D, Gu X, He Y (2009) Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21: 1733–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Jung JH, Park JH, Lee S, To TK, Kim JM, Seki M, Park CM (2013) The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell 25: 4378–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sung S (2013) Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 25: 454–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Le Corre V, Roux F, Reboud X (2002) DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: Extensive nonsynonymous variation is consistent with local selection for flowering time. Mol Biol Evol 19: 1261–1271 [DOI] [PubMed] [Google Scholar]
- Lee J, Amasino RM (2013) Two FLX family members are non-redundantly required to establish the vernalization requirement in Arabidopsis. Nat Commun 4: 2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Yun JY, Zhao W, Shen WH, Amasino RM (2015) A methyltransferase required for proper timing of the vernalization response in Arabidopsis. Proc Natl Acad Sci USA 112: 2269–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C (2002) Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297: 243–246 [DOI] [PubMed] [Google Scholar]
- Li P, Filiault D, Box MS, Kerdaffrec E, van Oosterhout C, Wilczek AM, Schmitt J, McMullan M, Bergelson J, Nordborg M, et al. (2014) Multiple FLC haplotypes defined by independent cis-regulatory variation underpin life history diversity in Arabidopsis thaliana. Genes Dev 28: 1635–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Tao Z, Dean C (2015) Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev 29: 696–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO (2010) Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 21199–21204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Marquardt S, Lister C, Swiezewski S, Dean C (2010) Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327: 94–97 [DOI] [PubMed] [Google Scholar]
- Liu F, Quesada V, Crevillén P, Bäurle I, Swiezewski S, Dean C (2007) The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell 28: 398–407 [DOI] [PubMed] [Google Scholar]
- Marquardt S, Raitskin O, Wu Z, Liu F, Sun Q, Dean C (2014) Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol Cell 54: 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. (2011) Mechanisms for the inheritance of chromatin states. Cell 146: 510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne J, Greb T, Lister C, Dean C (2004) Epigenetic regulation in the control of flowering. Cold Spring Harb Symp Quant Biol 69: 457–464 [DOI] [PubMed] [Google Scholar]
- Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P, Dean C (2006) LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci USA 103: 5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajoro A, Biewers S, Dougali E, Leal Valentim F, Mendes MA, Porri A, Coupland G, Van de Peer Y, van Dijk ADJ, Colombo L, et al. (2014) The (r)evolution of gene regulatory networks controlling Arabidopsis plant reproduction: a two-decade history. J Exp Bot 65: 4731–4745 [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh M, Molinier J, Berr A, Genschik P (2011) MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc Natl Acad Sci USA 108: 3430–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. (2014) The chemistry of regulation of genes and other things. J Biol Chem 289: 5417–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S, De Lucia F, Mylne JS, Zhu D, Ohmido N, Pendle A, Kato N, Shaw P, Dean C (2013) Physical clustering of FLC alleles during Polycomb-mediated epigenetic silencing in vernalization. Genes Dev 27: 1845–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruelens P, de Maagd RA, Proost S, Theißen G, Geuten K, Kaufmann K (2013) FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat Commun 4: 2280. [DOI] [PubMed] [Google Scholar]
- Sánchez-Bermejo E, Méndez-Vigo B, Picó FX, Martínez-Zapater JM, Alonso-Blanco C (2012) Novel natural alleles at FLC and LVR loci account for enhanced vernalization responses in Arabidopsis thaliana. Plant Cell Environ 35: 1672–1684 [DOI] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J (2006) Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Kröber S, Amasino RA, Coupland G (2006) The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Hills MJ, Lister C, Dean C, Dennis ES, Peacock WJ (2008) Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc Natl Acad Sci USA 105: 2214–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES (2000) The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci USA 97: 3753–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Lister C, Crevillen P, Nordborg M, Dean C (2006) Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev 20: 3079–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dean C (2002) Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289 [DOI] [PubMed] [Google Scholar]
- Song J, Irwin J, Dean C (2013) Remembering the prolonged cold of winter. Curr Biol 23: R807–R811 [DOI] [PubMed] [Google Scholar]
- Springthorpe V, Penfield S (2015) Flowering time and seed dormancy control use external coincidence to generate life history strategy. eLife 4: e05557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, Purugganan MD, Schmitt J (2004) A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc Natl Acad Sci USA 101: 4712–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange A, Li P, Lister C, Anderson J, Warthmann N, Shindo C, Irwin J, Nordborg M, Dean C (2011) Major-effect alleles at relatively few loci underlie distinct vernalization and flowering variation in Arabidopsis accessions. PLoS One 6: e19949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C (2013) R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340: 619–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM (2006a) Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet 38: 706–710 [DOI] [PubMed] [Google Scholar]
- Sung S, Schmitz RJ, Amasino RM (2006b) A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev 20: 3244–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Crevillen P, Liu F, Ecker JR, Jerzmanowski A, Dean C (2007) Small RNA-mediated chromatin silencing directed to the 3′ region of the Arabidopsis gene encoding the developmental regulator, FLC. Proc Natl Acad Sci USA 104: 3633–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Wang R, Farrona S, Vincent C, Joecker A, Schoof H, Turck F, Alonso-Blanco C, Coupland G, Albani MC (2009) PEP1 regulates perennial flowering in Arabis alpina. Nature 459: 423–427 [DOI] [PubMed] [Google Scholar]
- Wang Y, Gu X, Yuan W, Schmitz RJ, He Y (2014a) Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev Cell 28: 727–736 [DOI] [PubMed] [Google Scholar]
- Wang ZW, Wu Z, Raitskin O, Sun Q, Dean C (2014b) Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc Natl Acad Sci USA 111: 7468–7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JD, Borevitz JO, Uhlenhaut NH, Ecker JR, Chory J, Weigel D (2005) FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170: 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczek AM, Cooper MD, Korves TM, Schmitt J (2014) Lagging adaptation to warming climate in Arabidopsis thaliana. Proc Natl Acad Sci USA 111: 7906–7913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg AC, Strasser B, Cerdán PD, Amasino RM (2008) Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol 148: 1681–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA (2006) The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA 103: 14631–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D, Zhao JJ, Hou XL, Basnet RK, Carpio DPD, Zhang NW, Bucher J, Lin K, Cheng F, Wang XW, et al. (2013) The Brassica rapa FLC homologue FLC2 is a key regulator of flowering time, identified through transcriptional co-expression networks. J Exp Bot 64: 4503–4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Wagner D (2015) Polycomb repression in the regulation of growth and development in Arabidopsis. Curr Opin Plant Biol 23: 15–24 [DOI] [PubMed] [Google Scholar]
- Yang H, Howard M, Dean C (2014) Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr Biol 24: 1793–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Rosa S, Dean C (2015) Nuclear organization changes and the epigenetic silencing of FLC during vernalization. J Mol Biol 427: 659–669 [DOI] [PubMed] [Google Scholar]