Respiratory chain complex I acts as a negative regulator of fluxes through glycolysis and the TCA cycle.
Abstract
Complex I (NADH:ubiquinone oxidoreductase) is central to cellular NAD+ recycling and accounts for approximately 40% of mitochondrial ATP production. To understand how complex I function impacts respiration and plant development, we isolated Arabidopsis (Arabidopsis thaliana) lines that lack complex I activity due to the absence of the catalytic subunit NDUFV1 (for NADH:ubiquinone oxidoreductase flavoprotein1) and compared these plants with ndufs4 (for NADH:ubiquinone oxidoreductase Fe-S protein4) mutants possessing trace amounts of complex I. Unlike ndufs4 plants, ndufv1 lines were largely unable to establish seedlings in the absence of externally supplied sucrose. Measurements of mitochondrial respiration and ATP synthesis revealed that compared with ndufv1, the complex I amounts retained by ndufs4 did not increase mitochondrial respiration and oxidative phosphorylation capacities. No major differences were seen in the mitochondrial proteomes, cellular metabolomes, or transcriptomes between ndufv1 and ndufs4. The analysis of fluxes through the respiratory pathway revealed that in ndufv1, fluxes through glycolysis and the tricarboxylic acid cycle were dramatically increased compared with ndufs4, which showed near wild-type-like fluxes. This indicates that the strong growth defects seen for plants lacking complex I originate from a switch in the metabolic mode of mitochondria and an up-regulation of respiratory fluxes. Partial reversion of these phenotypes when traces of active complex I are present suggests that complex I is essential for plant development and likely acts as a negative regulator of respiratory fluxes.
In most eukaryotic organisms, energy is mainly provided by cellular respiration, which is composed of three main pathways. Glycolysis in the cytosol, and additionally in the plastids of plants, degrades sugars into pyruvate. The tricarboxylic acid (TCA) cycle, which largely resides in the mitochondrial matrix, further dissimilates pyruvate into CO2. These two pathways generate reduced cofactors, mostly NADH. The oxidative phosphorylation (OXPHOS) system couples cofactor recycling with ATP production. The electron transfer chain (ETC) located in the mitochondrial inner membrane (IM) uses the redox energy of the reduced cofactors to create an electrochemical gradient across the IM. This gradient is then used by the ATP synthase to convert ADP to ATP, which is subsequently exported from the mitochondria to fuel cellular metabolism and sustain housekeeping functions and growth.
The ETC is composed of four large multiprotein complexes. The first of these complexes is the NADH-ubiquinone oxidoreductase, also called complex I. It plays a crucial role in recycling NAD+ for the TCA cycle, and its activity is responsible for about 40% of the total proton pumping across the IM (Wikström, 1984; Galkin et al., 2006). In plants, additional NADH dehydrogenases located on both sides of the IM are present (for review, see Rasmusson et al., 2008). These enzymes can recycle NAD+ for use in glycolysis and the TCA cycle and transfer the resulting electrons onto ubiquinone, but they do not participate in the formation of the proton gradient. These alternative NADH dehydrogenases can, therefore, be considered as bypasses for complex I.
Complex I is the largest complex of the ETC (for review, see Brandt, 2006; Hirst, 2013). It is present from bacteria to eukaryotes. In bacteria, it is composed of 14 subunits that are conserved in all complex I-containing organisms and are referred to as core subunits. The number of subunits increased to more than 40 in most eukaryotes with the addition of so-called accessory subunits. The roles of these accessory subunits have mostly remained unknown (Hirst, 2011). Overall, the subunit composition of complex I is well conserved in eukaryotes; however, a few lineage-specific subunits are present in plants, fungi, and mammals (Cardol, 2011). In eukaryotes, most complex I subunits are encoded by the nuclear genome, but five to nine subunits are encoded by mitochondrial genes. The three-dimensional structure of complex I has been solved in bacteria (Baradaran et al., 2013) and is conserved in eukaryotes (Hunte et al., 2010; Shimada et al., 2014; Vinothkumar et al., 2014; Zickermann et al., 2015). Complex I displays the shape of an L, with a membrane arm embedded in the IM and a matrix arm protruding into the mitochondrial matrix. The membrane arm is the site of proton pumping across the membrane and contains four antiporter-like domains (Efremov and Sazanov, 2011). The matrix arm contains several cofactors, a flavin mononucleotide (FMN) at the site of NADH oxidation and a chain of Fe-S clusters. The latter are responsible for the transfer of electrons from the tip of the matrix arm, where the NADH-binding site is located, to the ubiquinone-binding site situated at the level of the membrane (Efremov et al., 2010). The molecular mode of action of complex I, in particular the mechanism involved in the coupling of electron transfer and proton pumping, is still largely unknown (Friedrich, 2014; Sazanov, 2014).
Complex I deficiencies have dramatic effects on cellular physiology, and numerous plant growth defects and human diseases have been linked with mutations in complex I. Because of its intricacy (large number of subunits, dual genetic origin, and biogenesis depending on a complex assembly pathway), complex I is the main origin of human diseases resulting from mitochondrial dysfunction (Scaglia et al., 2004; Nouws et al., 2012). Interestingly, some subunits have been described as mutation hot spots in Homo sapiens (Tucker et al., 2011). In particular, the catalytic core subunit NDUFV1 (for NADH:ubiquinone oxidoreductase flavoprotein1) and the accessory subunit NDUFS4 (for NADH:ubiquinone oxidoreductase Fe-S protein4) are affected in approximately 15% of the mutations described in nucleus-encoded complex I subunits (Pagniez-Mammeri et al., 2012). Although the origin of the complex I deficiency is generally well characterized, the consequences of this deficiency at the mitochondrial and cellular levels remain poorly understood. A comprehensive analysis of physiological parameters of complex I-deficient individuals identified common features present in all patients (i.e. lactate and reactive oxygen species accumulation; Distelmaier et al., 2009). In general, the absence of complex I is particularly detrimental to organs with high energy demand and usually causes death within a few months or years after birth.
In plants, several complex I mutants have been reported (Gutierres et al., 1999; Karpova and Newton, 1999; Brangeon et al., 2000; Perales et al., 2005; de Longevialle et al., 2007; Juszczuk et al., 2007; Meyer et al., 2009; Haïli et al., 2013). Most of these show delayed growth, but unlike in H. sapiens, the absence of complex I does not cause premature death, possibly due to the presence of bypasses for complex I. The best-characterized mutants are cytoplasmic male sterile II (CMSII) in Nicotiana sylvestris, which has no detectable complex I due to a deletion in the mitochondrial NADH dehydrogenase subunit7 (nad7) gene encoding a complex I subunit (Pla et al., 1995; Gutierres et al., 1997; Sabar et al., 2000; Pineau et al., 2005), and Arabidopsis (Arabidopsis thaliana) ndufs4 that is impaired in complex I biogenesis due to loss of the accessory NDUFS4 subunit (Meyer et al., 2009). These studies reported delayed growth and modifications of the metabolite and transcript pools (Meyer et al., 2009), as well as potential secondary effects on other metabolic pathways, such as photosynthesis (Dutilleul et al., 2003; Priault et al., 2006; Meyer et al., 2009). However, the mitochondrial phenotypes observed were restricted to subtle changes of respiratory parameters (Sabar et al., 2000; Meyer et al., 2009). Defects in complex I biogenesis in plants have been associated with strikingly different degrees of growth inhibition. The most severe phenotype in Arabidopsis has been reported for the mutant organelle transcript processing defect43 (otp43; de Longevialle et al., 2007). otp43 plants are unable to produce mature nad1 mRNA due to a mitochondrial splicing defect and contain no detectable complex I; they exhibit strongly delayed development and produce a few malformed seeds that fail to germinate (de Longevialle et al., 2007). Similarly, in maize (Zea mays) nonchromosomal stripe2 (NCS2) mutants, a 3′ end deletion of the mitochondrial nad4 gene conditions the presence of only partially but no fully assembled complex I, which leads to lethality during kernel development (Newton and Coe, 1986; Marienfeld and Newton, 1994; Karpova and Newton, 1999). By contrast, Arabidopsis mutants affected in mitochondrial nad4 mRNA maturation and accumulating a subcomplex I similar in size to that detected in NCS2 produce seeds of which 20% germinate (Haïli et al., 2013). Several other complex I mutants including ndufs4 (Meyer et al., 2009) are apparently not impaired in seed development or germination. Conditional male sterility as reported for CMSII (De Paepe et al., 1990) has not been observed for severely affected mutants such as otp43 (de Longevialle et al., 2007). Thus, the importance of complex I activity for plant development has remained unclear.
To understand how complex I activity impacts respiration and development in plants, we isolated Arabidopsis mutants that completely lack complex I activity due to inactivation of the nuclear gene encoding the catalytic subunit NDUFV1. In contrast to ndufs4 and other previously described complex I mutants, NDUFV1-deficient seedlings could not survive under normal growth conditions. While this resembled complex I deficiencies in H. sapiens, Arabidopsis ndufv1 lines could be maintained under specific growth conditions on sugar-containing synthetic media. We performed a comparative systems analysis of Arabidopsis ndufv1 and ndufs4 mutants, the later showing a milder growth phenotype. We identified the uncoupling of glycolysis and TCA cycle from the ETC as the only major difference between these mutants, suggesting that the strong growth defects of ndufv1 originated from a switch in the metabolic mode of mitochondria. These data provide, to our knowledge, new fundamental insights into the metabolic rearrangements occurring in plants lacking complex I activity.
RESULTS
Complex I Mutants Show Diverse Growth Phenotypes
Several mutants impaired in complex I have been characterized in the model plant Arabidopsis. They include mutants in genes encoding complex I subunits (Perales et al., 2005; Meyer et al., 2009) and mutants in genes encoding proteins involved in the expression of the mitochondrial complex I genes (for review, see Colas des Francs-Small and Small, 2014). Most of these mutants showed delayed growth. We compared the growth phenotypes of several complex I mutants grown in vitro on synthetic media and found that the growth delay was variable among the different mutants, ranging from wild-type-like to extremely delayed growth (Supplemental Fig. S1). These diverse phenotypes could be due to (1) variable residual levels of complex I or (2) accumulation of different complex I fragments or assembly intermediates that affect mitochondrial metabolism or could reflect additional functions of complex I or specific complex I subunits. To distinguish between these possibilities, we isolated mutants that completely lacked complex I activity. This was achieved by inactivating the gene encoding the NADH-binding subunit NDUFV1. This protein, also named 51-kD subunit, is located at the tip of the matrix arm of complex I and carries the FMN cofactor that initiates electron transfer (Fig. 1A). NDUFV1 is encoded by a single gene in Arabidopsis, At5g08530. We used two independent transfer DNA (T-DNA) insertion lines (Fig. 1B) to isolate the mutant lines ndufv1-1 and ndufv1-2 (Fig. 1C), which were homozygous for the loss-of-function allele. Using anti-51-kD antibodies (Peters et al., 2012), we could not detect the NDUFV1 protein in mitochondrial fractions prepared from ndufv1 mutants (Fig. 1D). Compared with wild-type plants, both ndufv1-1 and ndufv1-2 showed strongly delayed growth and distorted leaves. This macroscopic phenotype was considerably more severe than that of the previously characterized ndufs4 mutant (Meyer et al., 2009), which was included as a reference (Fig. 1C).
Figure 1.
Isolation of ndufv1 mutants. A, Schematic representation of Arabidopsis complex I according to Meyer (2012). The position of NDUFV1 is shown in black, the FMN cofactor in yellow, and the Fe-S clusters in red. B, Representation of the structure of the gene At5g08530 that encodes NDUFV1. Untranslated regions (white boxes) and regions encoding the predicted mitochondrial transit peptide (black) and mature protein (gray) are indicated. Gaps between boxes represent introns. The T-DNA insertions are represented as triangles. Both mutant alleles contain two copies of the T-DNA present as inverted repeats. C, Growth phenotype of the ndufv1 mutants grown for 8 weeks on soil. The previously characterized ndufs4 mutant is shown for comparison. D, Western-blot analysis of extracted mitochondrial protein samples using anti-NDUFV1 antibodies. The asterisk indicates a nonspecific signal. A western blot with anti-ATPB antibodies was performed as control for equal loading. Col-0, Ecotype Columbia.
Severely Impaired Growth of ndufv1 Mutants
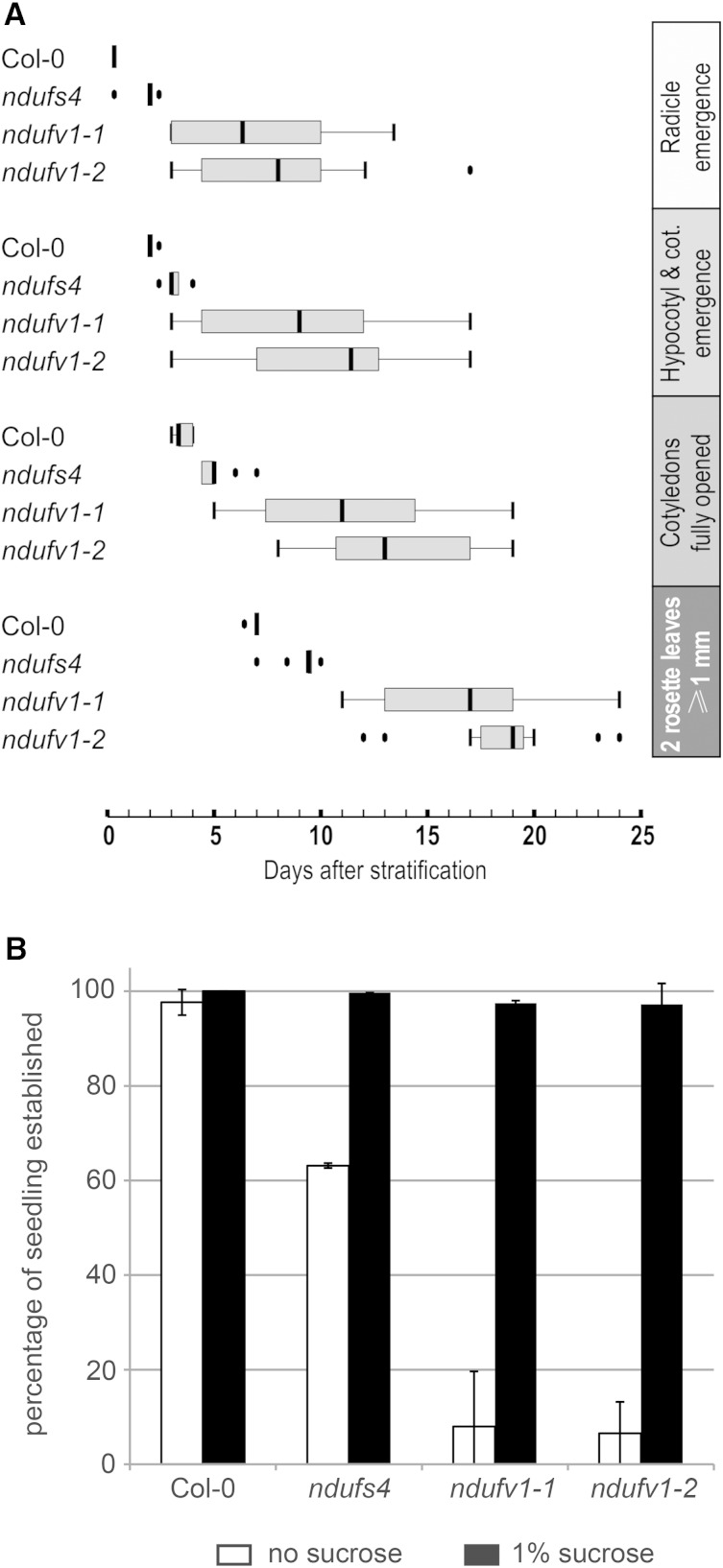
We quantified the growth retardation of ndufv1-1 and ndufv1-2 by monitoring the time required by these mutants and the wild type to reach different growth stages defined by Boyes et al. (2001); the ndufs4 mutant was included in the analysis (Fig. 2A). Growth and development at all stages were severely delayed for ndufv1-1 and ndufv1-2 compared with the wild type or ndufs4. For example, germination of ndufv1 seeds took, on average, 7 and 5 d longer than germination of wild-type and ndufs4 seeds, respectively (Fig. 2A). In addition, we observed substantial variation between individuals within the ndufv1-1 and ndufv1-2 lines. Interestingly, it was nearly impossible to recover seedlings when ndufv1 seeds were sown directly on soil. We thus speculated that the presence of sugar in the media was required for the establishment of viable ndufv1 seedlings. We tested this hypothesis by growth assays in vitro. While we did not observe any effect of sugar on germination efficiencies, only about 5% of the germinated ndufv1 seeds produced a seedling with true leaves in the absence of sugar (Fig. 2B). Most of the germinated seedlings of the ndufv1 mutants died at the cotyledon stage in the absence of sugar, whereas normal seedling development occurred in the presence of sugar (Supplemental Fig. S2). A similarly strong sugar effect was not observed for ndufs4. We will subsequently refer to mutants that are not affected in seedling establishment as mild mutants, in contrast to severe mutants that are similar to ndufv1. We isolated further severe mutants lacking the subunits NDUFS1 (Wydro et al., 2013), NDUFS7, or NDUFV2 (Supplemental Fig. S3). These three subunits are all located in the complex I matrix arm, contain Fe-S cofactors, and are involved in the electron transfer function of the complex. Isolation of several independent mutants in the electron transfer domain of complex I that all show similarly severe phenotypes suggests that the latter are due to a complete lack of complex I activity. Consequently, mild mutants, which are impaired in accessory subunits such as NDUFS4, might possess trace amounts of complex I activity that cannot easily be detected by activity assays.
Figure 2.
Analysis of the early growth stages of ndufv1. A, Box plot of the time required by several complex I mutants to reach the following growth stages: radicle emergence, cotyledon emergence, full opening of cotyledons, and two rosette leaves greater than 1 mm (seedling establishment). B, Quantification of the sugar dependence of seedling establishment in the ndufv1 and ndufs4 mutants and wild-type plants. The bars represent the percentage of germinated seeds that form a seedling in the absence (white bars) and presence (black bars) of 1% Suc in the media. Bars represent the means ± se (n = 4). Col-0, Ecotype Columbia; cot., cotyledons.
Impaired Complex I Biogenesis in ndufv1 Mutants
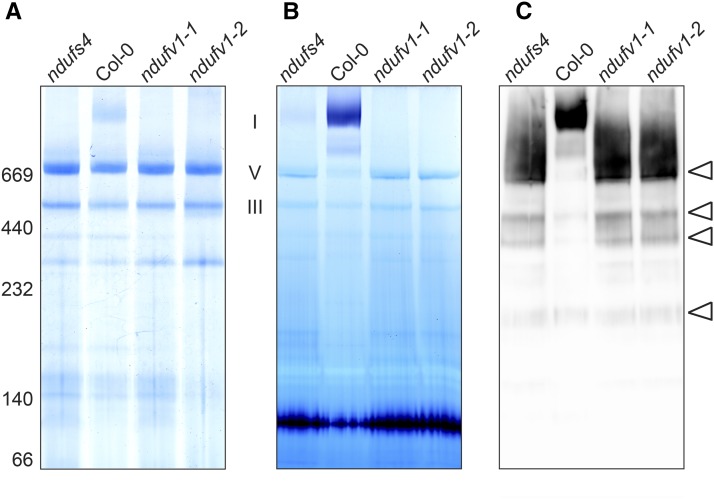
Our earlier study of ndufs4 had not detected functional complex I in this mutant (Meyer et al., 2009). To reexamine ndufs4 for complex I activity and compare complex I levels between this mutant and ndufv1-1 and ndufv1-2, we purified mitochondria from rosette leaves of wild-type and mutant plants and separated the solubilized mitochondrial membrane complexes by blue-native (BN) PAGE (Fig. 3). Colloidal Coomassie staining of the gel detected complex I in the wild type but not in ndufv1-1, ndufv1-2, or ndufs4 (Fig. 3A). By contrast, all mutant lines displayed wild-type-like levels of complexes III and V. A similar BN-PAGE gel was stained for NADH oxidase activity. While we were unable to detect complex I activity in ndufv1 mitochondria, ndufs4 exhibited trace amounts of activity (Fig. 3B). We next tested for the presence of complex I and its subcomplexes through immunodetection with antibodies against carbonic anhydrase (CA)-related subunits (Fig. 3C). The latter are incorporated into the complex during the initial stage of assembly (Kühn et al., 2011; Meyer et al., 2011). Their immunolabeling thus detects complex I intermediates at the early as well as the late assembly steps. CA immunodetection revealed identical subcomplex patterns for ndufv1 and ndufs4. Moreover, ndufv1 mutants accumulated similar amounts of the assembly intermediates as ndufs4 (Fig. 3C). Therefore, we concluded that the only difference between the respiratory chains of ndufs4 and ndufv1 mutants was the presence of trace amounts of active complex I in ndufs4, as opposed to the complete lack of complex I activity in ndufv1. Previous attempts to measure residual complex I activity in ndufs4 were unsuccessful (Meyer et al., 2009), most likely due to the low sensitivity of the enzymatic assays existing to measure complex I activity. The quantification of the residual complex I activity in ndufs4 would require the development of a unique highly sensitive assay.
Figure 3.
Analysis of complex I in ndufv1. A, Mitochondrial complexes were solubilized with 1% dodecylmaltoside and separated by BN-PAGE, followed by Coomassie staining. The positions of molecular weight markers are indicated on the left, and the migration of OXPHOS complexes I, V, and III is indicated on the right. B, Staining of BN-PAGE-resolved complex I for NADH oxidase activity. The signal detected in the bottom part of the gel corresponds to the activity of the dihydrolipoamide dehydrogenase (Meyer et al., 2009). C, Western-blot analysis of complex I and its subcomplexes. Antibodies against CAs were used for immunolabeling. The bands corresponding to assembly intermediates are indicated by arrowheads on the right. Col-0, Ecotype Columbia.
Altered Respiratory Fluxes in ndufv1
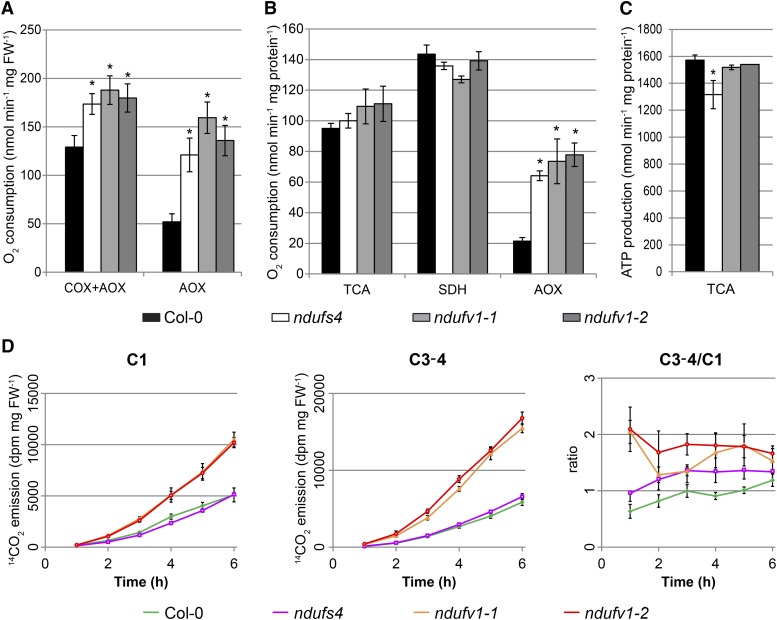
To examine if the absence of complex I had a stronger effect on respiration than its reduction to trace amounts, we measured leaf respiration and moreover assessed the respiratory capacity of isolated mitochondria by quantifying their oxygen consumption. Mutant leaf discs showed higher oxygen consumption than wild-type leaf discs; no difference was detected between ndufv1 and ndufs4 (Fig. 4A). The addition of potassium cyanide (KCN) to inhibit complex IV revealed an increased alternative oxidase (AOX) capacity in all mutants (Fig. 4A), indicating that plants with traces of or no complex I similarly undergo oxidative stress and activate AOX. When using isolated mitochondria, no significant differences were seen between ndufv1, ndufs4, and the wild type in the overall capacity of the TCA cycle or upon feeding of isolated mitochondria with succinate (Fig. 4B). Using an assay designed to estimate the maximum capacity of the AOX, we confirmed that both mild and severe mutants show increased AOX capacity (Fig. 4B). We then compared the ATP production capacities of ndufv1, ndufs4, and wild-type mitochondria. To this end, isolated mitochondria were fed with malate for 10 min; then metabolites were extracted and the ATP levels quantified. The data confirmed our earlier observation (Meyer et al., 2009) that mitochondria from ndufs4 produce less ATP than wild-type mitochondria when respiration is fueled through the TCA cycle (Fig. 4B). Unexpectedly, we did not measure reduced ATP production by ndufv1 mitochondria (Fig. 4C). This indicates that the residual complex I activity present in ndufs4 does not play a major role in ATP production. Moreover, this suggests that ndufv1 mutants activate an alternative route(s) for mitochondrial ATP production to compensate for the complete loss of complex I activity.
Figure 4.
Respiratory parameters of ndufv1. A, Oxygen consumption of leaves. Oxygen consumption of leaf discs was monitored with a Clark-type oxygen electrode maintained in the dark. Respiration was monitored in the absence (cytochrome c oxidase [COX] + AOX) and presence of KCN (AOX). Bars represent the means ± se (n = 9). The asterisk indicates a significant difference from ecotype Columbia (Col-0; Student’s t test, P < 0.05). B, Oxygen consumption of isolated mitochondria fed with malate (TCA) or succinate (succinate dehydrogenase [SDH]) or with succinate in the presence of an inhibitor of complex III to estimate the maximal AOX capacity. Oxygen consumption measurements were done on freshly prepared mitochondria using a Clark-type oxygen electrode. Bars represent the means ± se (n = 3). The asterisk indicates a significant difference from ecotype Columbia (Student’s t test, P < 0.05). C, ATP production of isolated mitochondria. Freshly purified mitochondria were fed with malate for exactly 10 min, and metabolites were extracted. ATP was quantified using a luciferase assay. Bars represent the means ± se (n = 4). The asterisk indicates a significant difference from ecotype Columbia (Student’s t test, P < 0.05). D, 14CO2 evolution experiments to measure fluxes through the respiratory pathways. Glc labeled with 14C at different positions was fed to illuminated leaf discs, and radioactivity in the evolved CO2 was determined. The 14CO2 from Glc labeled at position 1 (C1) or positions 3 and 4 (C3-4) corresponds to the metabolic flux through glycolysis and TCA cycle, respectively. 14CO2 release was measured every hour after the addition of the labeled Glc and summed up to calculate the total 14C evolved. Each point represent the means ± se (n = 6). All measurements for ndufv1-1 and ndufv1-2 are significantly different from those of ecotype Columbia (Student’s t test, P < 0.05). FW, Fresh weight.
We therefore hypothesized that substrate-level phosphorylation at the level of the TCA cycle was increased in ndufv1 compared with ndufs4, which would require a higher flux through the TCA cycle in ndufv1 mutants. To test this hypothesis and gain insight into the in vivo fluxes through the respiratory pathway, we performed a feeding experiment. Leaf discs were incubated with radio-labeled Glc, and the release of radioactivity as CO2 was quantified. Glc labeled at position 1 (C1) was used to estimate fluxes through glycolysis, and Glc labeled at positions 3 and 4 (C3-4) was used to estimate fluxes through the TCA cycle. In the mild mutant ndufs4, fluxes through the TCA cycle and glycolysis differed only slightly from those in the wild type (Fig. 4D). However, the rates of CO2 emission by ndufv1 were substantially higher with both labeled substrates, revealing a dramatic increase in the fluxes through glycolysis and the TCA cycle compared with ndufs4 and the wild type. The TCA cycle-to-glycolysis (C3-4:C1) ratio was also increased in ndufv1 (Fig. 4D). This showed that the fluxes through different pathways of respiration (glycolysis, TCA cycle, and respiratory chain) differed considerably between ndufv1 and ndufs4 and suggested a redirection of carbon metabolic flux from glycolysis to the TCA cycle in ndufv1. As a consequence, some intermediates of the TCA cycle (for example, 2-oxoglutarate and succinate) and some TCA cycle-derived amino acids (for example, Asp and Glu) accumulated to higher levels in the severe mutants (Supplemental Fig. S4; Supplemental Methods S1; Supplemental Table S2). In ndufs4, where the TCA cycle-to-glycolysis (C3-4:C1) ratio was higher than in the wild type but lower than in ndufv1 (Fig. 4D), the accumulation of these metabolites was lower than in ndufv1 (Supplemental Fig. S4).
As the respiratory capacities measured did not show major differences between mild and severe mutants (Fig. 4A), we concluded that the increased fluxes in ndufv1 were due to increased activity and not caused by increased capacities of the various components of the respiratory pathway. This was supported by the BN-PAGE analysis of mitochondrial complexes, which revealed no major changes in the abundances of the main mitochondrial protein complexes (Fig. 3A). We also performed a differential gel analysis to compare the mitochondrial proteomes of ndufv1-1 and ndufs4 and did not detect differences in abundance of the major mitochondrial proteins, including TCA cycle enzymes (Supplemental Fig. S5; Supplemental Methods S1). Thus, the changes in fluxes observed were most likely the consequence of changes in enzyme activities rather than changes in protein abundance.
No Alteration of the NADH-to-NAD+ Ratio
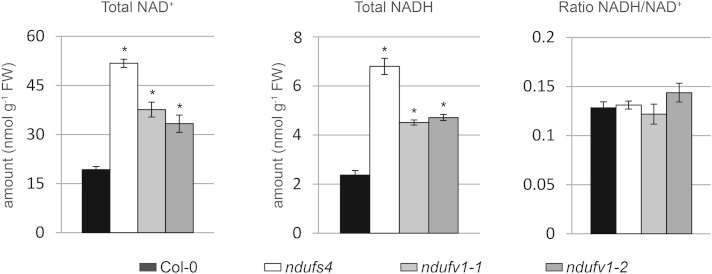
NAD is a key cofactor of several TCA cycle enzymes. The balance between the reduced and oxidized forms of the NAD (also known as the NADH-to-NAD+ ratio) influences the flux through the TCA cycle (Nunes-Nesi et al., 2013). Because complex I plays a central role in the recycling of NAD+ in the mitochondrial matrix, the amounts of NADH and NAD+ and, consequently, the NADH-to-NAD+ ratio might be more strongly affected in ndufv1 than in ndufs4 (in which traces of complex I are still present). We measured the total pools of NAD+ and NADH in leaves. We observed a higher accumulation of these metabolites in both mild and severe mutants compared with the wild type (Fig. 5). Surprisingly, ndufs4 accumulated higher levels of NAD+ and NADH than ndufv1. However, in both mild and severe mutants, the increased pools had no effect on the NADH-to-NAD+ ratio (Fig. 5). Assuming that the measured pools are representative of the pools present in the mitochondrial matrix, we can exclude that the increased flux through the TCA cycle observed in ndufv1 is due to a modified NADH-to-NAD+ ratio.
Figure 5.
NAD+ and NADH levels in complex I mutants. Total amounts of NAD+ and NADH were measured in total leaf extracts. Bars represent the means ± se (n = 4). Asterisks indicate significant differences from ecotype Columbia (Col-0; Student’s t test, P < 0.05). FW, Fresh weight.
ndufv1 and ndufs4 Mutants Exhibit Similar Transcriptomic Response
To get a snapshot of the cellular responses triggered in the severe mutants, we performed a whole-genome expression analysis. Overall, ndufs4 and ndufv1 mutants showed similar transcriptomic responses to the reduction and lack, respectively, in complex I (Table I). These responses included an increased expression of the genes of the respiratory pathways (glycolysis, TCA cycle, and respiratory chain) and specific catabolic pathways, especially amino acid degradation. At the same time, a reduction was seen in the expression of genes encoding components of pathways that are particularly energy demanding (i.e. cell wall biosynthesis and synthesis pathways for secondary metabolites and storage proteins). This indicates that when complex I activity is severely impaired or absent, nuclear gene expression is modified to limit energy usage. We identified no pathway that was differentially expressed in ndufv1 compared with ndufs4 and could potentially contribute to the differences in mutant growth and metabolic phenotype (Supplemental Table S1). Application of the MapMan tool to visualize the response of individual genes within a pathway (Thimm et al., 2004) revealed that the transcriptomic response in ndufv1 was stronger than in ndufs4 (Supplemental Fig. S6). The similar transcriptomic response observed in both severe and mild mutants suggests that this response is triggered by the reduced respiratory chain activity and that the increased fluxes through glycolysis and the TCA cycle do not have a major impact on nuclear gene expression.
Table I. Global changes in the expression of genes grouped by functional categories.
The overall change of the expression of all genes grouped in a functional category (MapMan bin) was statistically evaluated. Only individual genes for which a statistically significant change was recorded in the microarray analysis were considered. The proportion is the ratio of genes considered in the analysis over the total number of genes present in the bin. For each bin, a P value (pval) was calculated to indicate if the genes considered behave in the same way. Only bins for which significant changes for ndufv1-1 and ndufs4 were found (P < 0.05) are displayed.
| MapMan Bin | Direction |
ndufv1-1 |
ndufs4 |
||
|---|---|---|---|---|---|
| Proportion | pval | Proportion | pval | ||
| Oxidative pentose phosphate | Down | 0.53 | 0.011 | 0.47 | 0.001 |
| Cell wall | Down | 0.62 | 0.001 | 0.60 | 0.001 |
| Lipid metabolism | Down | 0.51 | 0.001 | 0.41 | 0.002 |
| Hormone metabolism | Down | 0.51 | 0.001 | 0.44 | 0.001 |
| Polyamine metabolism | Down | 0.67 | 0.001 | 0.44 | 0.022 |
| Development | Down | 0.40 | 0.02 | 0.39 | 0.001 |
| Transport | Down | 0.49 | 0.001 | 0.44 | 0.004 |
| Wax | Down | 0.88 | 0.001 | 0.94 | 0.001 |
| Glutaredoxins | Down | 0.63 | 0.001 | 0.66 | 0.001 |
| Storage proteins | Down | 0.44 | 0.003 | 0.44 | 0.006 |
| Major CHO metabolism | Up | 0.49 | 0.001 | 0.47 | 0.003 |
| Glycolysis | Up | 0.49 | 0.001 | 0.43 | 0.002 |
| Fermentation | Up | 0.64 | 0.004 | 0.64 | 0.001 |
| TCA | Up | 0.71 | 0.001 | 0.52 | 0.031 |
| Mitochondrial electron transport/ATP synthesis | Up | 0.68 | 0.001 | 0.47 | 0.002 |
| Amino acid degradation | Up | 0.51 | 0.001 | 0.43 | 0.001 |
| Cofactor and vitamin metabolism | Up | 0.57 | 0.001 | 0.47 | 0.001 |
| Tetrapyrrole synthesis | Up | 0.41 | 0.013 | 0.44 | 0.018 |
| RNA | Up | 0.45 | 0.002 | 0.43 | 0.018 |
| Cell | Up | 0.48 | 0.001 | 0.49 | 0.001 |
| Metal acquisition | Up | 0.44 | 0.108 | 0.67 | 0.001 |
| Dismutases and catalases | Up | 0.90 | 0.001 | 0.60 | 0.001 |
DISCUSSION
Mitochondrial complex I is the major entry point for electrons into the respiratory chain. Defects in complex I have been associated with reduced growth and metabolic rearrangement. In humans, complex I deficiencies are responsible for several dramatic diseases. In plants, complex I mutants are viable; however, several degrees of growth impairment have been observed. This indicates that complex I deficiencies in plant can differentially affect cellular metabolism and growth and suggests that complex I fulfills an additional role in mitochondrial metabolism. To understand how complex I activity impacts mitochondrial and cellular metabolism, we performed a comparative systems analysis of Arabidopsis lines with traces of or completely lacking complex I activity.
Adjusted Respiration and Metabolic Remodeling in Plants Lacking Complex I
Using classical respiratory measurements that monitor oxygen consumption, we were unable to identify major differences between the different lines. A common feature of mild and severe complex I mutants is the increased capacity of the AOX. This may result from a cellular response to increased oxidative stress. In addition, due to elevated levels of pyruvate (Supplemental Fig. S4), a metabolic activation of AOX is expected to prevent redirection of carbon metabolism (Vanlerberghe et al., 1995). However, our measurements suggest that both mild and severe mutants show a similar AOX response. Therefore, the phenotypic difference between ndufs4 and ndufv1 is not a consequence of altered AOX capacity. By contrast, measurements of the ATP production by isolated mitochondria showed that ndufs4 mutants were slightly impaired in mitochondrial ATP production, whereas, unexpectedly, ndufv1 mitochondria were not (Fig. 5C). Thus, the complete loss of complex I function activates compensatory mechanisms for ATP production that are not induced if traces of complex I are present. Analyses of the accumulation of protein complexes of the OXPHOS system (Fig. 3) and measurements of oxygen consumption by leaf tissue as well as isolated mitochondria (Fig. 4) indicated that compensatory ATP production in ndufv1 was not implemented via the OXPHOS system. Remarkably, ndufv1 mutants displayed substantially higher fluxes through glycolysis and the TCA cycle than ndufs4 plants (Fig. 4D), implying that mutants which fully lack complex I activity compensate for the deficit in ATP production via the respiratory chain by increasing substrate level phosphorylation. As a consequence, efficient systems for the rapid recycling of NAD+ are required to sustain these elevated fluxes. Plants contain alternative NADH dehydrogenases on both sides of the IM that can regenerate NAD+. The activity of these alternative NADH dehydrogenases is likely to be stimulated to compensate for the absence of complex I. An additional contribution to NAD+ recycling could come from the activation of the so-called mitochondrial shuttles. The glycerol-phosphate shuttle recycles NAD+ for use in glycolysis. Similarly, the malate shuttle allows the transfer of electrons from matrix-localized NADH to cytosolic NAD+, resulting in NAD+ recycling in the matrix. Our metabolomic analysis suggests the activation of these shuttles because we detected the presence of glycerol-3-P (glycerol-phosphate shuttle) and a stronger accumulation of 2-oxoglutarate and Asp (malate shuttle) in the ndufv1 mutants (Supplemental Fig. S4). The metabolite profile of ndufs4 suggests that these mechanisms are, however, not activated in this mutant, indicating that the activation of NAD+ recycling systems is a consequence of increased fluxes in ndufv1. In summary, in the absence of complex I activity, an induction of substrate-level phosphorylation and the activation of alternative routes for NAD+ recycling occur.
Regulation of Respiratory Pathways
The increased fluxes through the TCA cycle cannot be explained by an increased TCA cycle capacity, as we did not detect major differences in enzyme abundances between ndufv1 and ndufs4 in our analysis of the mitochondrial proteomes (Supplemental Fig. S5). Our transcriptomic analysis suggests an up-regulation of the expression of genes encoding TCA cycle components. However, changes in gene expression do not always correspond to changes in protein abundance (Fernie and Stitt, 2012). This absence of correlation is especially true for mitochondrial proteins (Lee et al., 2008). Thus, the increased activity of the TCA cycle can only be explained by hyperactivation. Classical enzymatic regulation by substrate/product concentrations is known to regulate the activities of TCA cycle enzymes (Nunes-Nesi et al., 2013). However, to our knowledge, mechanisms involved in the TCA cycle hyperactivation are unknown. Surprisingly, trace amounts of complex I, as present in ndufs4, are sufficient to prevent this metabolic response. This suggests that complex I acts as a negative regulator of respiratory fluxes and plays a role in the control of TCA cycle activity by preventing the hyperactivation of the pathway. Alternatively, the increased TCA cycle activity may be a direct consequence of the higher flux through glycolysis. The molecular mechanism of this regulation remains to be elucidated.
Patients with mitochondrial OXPHOS defects, including patients with complex I deficiencies, almost always suffer from lactic acidosis (Stacpoole, 1997; Robinson, 2006; Distelmaier et al., 2009). Lactic acidosis is characterized by the accumulation of high levels of lactate, which is caused by a higher rate of pyruvate production through glycolysis rather than impaired lactate detoxification (Gore et al., 1996). Our data link increased glycolytic fluxes with the absence of complex I activity, suggesting that complex I activity plays a key role in controlling glycolysis and therefore may be involved in triggering lactic acidosis.
Given the strong phenotypic differences between mild and severe complex I mutants, the increased respiratory fluxes appear to be costly and deleterious for the cell. In rapidly proliferating cells such as cancer cells, a metabolic switch called the Warburg effect is occurring. Under these conditions, glycolysis is uncoupled from the TCA cycle but coupled with lactate fermentation, and glycolytic rates increase to produce high levels of ATP in the cytosol (Upadhyay et al., 2013; Maldonado and Lemasters, 2014). Similarly, when T-cells are proliferating, lower rates of respiration and higher rates of glycolysis are observed (Kamiński et al., 2012). The molecular mechanism responsible for this metabolic switch remains unknown. However, as these metabolic changes are similar to those observed in our study as a consequence of the complete loss of complex I activity, the involvement of complex I in the metabolic rearrangements seen in T-cells appears feasible.
Because of its crucial role in providing ATP for the cell, respiration is tightly controlled. Several regulatory mechanisms controlling respiratory fluxes have been identified. They include the modulation of the activity of key enzymes either by an inhibitory effect of high concentration of the products (Wegener and Krause, 2002; Lenzen, 2014) or by posttranslational modifications such as phosphorylation or redox regulation (Piattoni et al., 2013; Ros and Schulze, 2013; Lushchak et al., 2014; Schmidtmann et al., 2014). Our data suggest an additional regulatory feedback loop involving complex I activity. Thus far, such a regulatory role has not been described for complex I in plants or other organisms. Testing if similar metabolic rearrangements occur in the absence of complex I might identify new avenues for the treatment of complex I deficiencies in humans.
Roles of Accessory Subunits and Subcomplexes
Several complex I mutants have been described in Arabidopsis. In this study, we present an in-depth characterization of severe complex I mutants. Our work confirms that the core subunits, in particular the subunits involved in electron transfer, are crucial for complex I activity. On the other hand, the accessory subunits are important but not essential for the accumulation of complex I. In our previous work, we showed that none of the mutants studied were completely devoid of complex I (Meyer et al., 2011). Here, we show that trace amounts of functional complex I are sufficient to strongly alleviate the severity of the phenotypes of complex I mutants. Therefore, accessory subunits are more important for the assembly and stability of the complex than for its biochemical function. The fact that both severe and mild mutants accumulate the same subcomplexes to similar levels suggests that these subcomplexes are not playing major roles in the phenotypes observed. This observation is in line with the fact that respiratory capacities are similar in mild and severe mutants. We therefore conclude that these subcomplexes do not act as uncouplers of the IM.
Growth Phenotypes of Complex I Mutants
The severity of the growth phenotypes of the various complex I mutants appears to be correlated with the residual levels of complex I. Mutants with no complex I activity exhibit strongly impaired growth. A process critical to these mutants is the establishment of a seedling, which corresponds to the transition between growth fueled by the reserves stored in the endosperm and autotrophic growth. This process requires high amounts of energy, provided through the degradation of stored reserves. Severe complex I mutants are thus likely to be impaired in either the storage or the mobilization of the stored reserves. Mutants impaired in reserve mobilization, such as mutants defective in β-oxidation, also show seedling lethality, although the growth arrest occurs earlier than in complex I mutants (Pinfield-Wells et al., 2005). Therefore, reserve mobilization is unlikely to be impaired in complex I mutants, suggesting that seed maturation is incomplete in these mutants, leading to the production of seeds with reduced reserves and viability. Another observation suggesting lower seed quality is the high variability observed between individuals of the severe mutant lines. Together with our observations on the early growth of severe complex I mutants, this suggests that the fitness of such mutants is strongly compromised. Thus, complex I is essential for plant growth under natural conditions, and complex I mutants can only be maintained under benign growth conditions in the laboratory. This conclusion is in line with studies in mammals describing complex I as essential for growth (Valsecchi et al., 2010). However, we observed that once photosynthesis has been established, growth is no longer dependent on the application of external sugars and the mutants can complete their life cycle. This indicates that ATP production through photosynthesis is sufficient for survival of the mutants but does not compensate the phenotypes caused by the impaired mitochondrial activity.
Among the many complex I mutants identified to date in Arabidopsis, ndufv1 belongs to the group of severe complex I mutants, together with otp43 (de Longevialle et al., 2007), indh (for iron-sulfur protein required for NADH dehydrogenase; Wydro et al., 2013), and mtsf1 (for mitochondrial stability factor1; Haïli et al., 2013). However, ndufv1 is the first-characterized severe complex I mutant directly lacking a subunit of complex I. otp43 and mtsf1 are defective in proteins involved in mitochondrial gene expression, leading to the absence of the mature transcript-encoding subunits Nad1 and Nad4, respectively, and ind1 is unable to assemble mitochondrial Fe-S clusters. While ndufv1, otp43, mtsf4, and ind1 show very similar growth phenotypes, otp43 is unable to produce viable seeds (de Longevialle et al., 2007). Because of the absence of this phenotype in the other severe mutants, this observation suggests either that otp43 harbors additional yet unidentified defects or that Nad1 has an additional function in the mitochondrial membrane. Interestingly, mtsf1 assembles a smaller version of complex I that is able to oxidize NADH in vitro (Haïli et al., 2013). A similar observation was made in the maize ncs2 mutant that is also defective in Nad4 (Karpova and Newton, 1999). In the light of our observation that traces of complex I activity are sufficient to partly revert severe growth defects deriving from the absence of complex I, we conclude that this smaller version of complex I is inactive in vivo. As Nad4 is a core subunit involved in proton pumping, this function is most likely impaired in mtsf1, revealing that the coupling between electron transfer and proton pumping is essential for complex I activity.
CONCLUSION
The characterization of severe complex I mutants indicates that complex I is essential for plant development. Comparing these mutants with plants retaining traces of complex I yielded, to our knowledge, new fundamental insights into the metabolic rearrangements occurring in plants lacking complex I activity, identifying a switch in the metabolic mode of mitochondria and an up-regulation of respiratory fluxes as likely origin for the strong growth defects observed in ndufv1. Partial reversion of these phenotypes when traces of active complex I are present suggests that complex I acts as a negative regulator of respiratory fluxes.
MATERIALS AND METHODS
Plant Material and Growth
Arabidopsis (Arabidopsis thaliana) mutants ndufv1-1 and ndufv1-2 were isolated from T-DNA insertion lines SAIL_319_D07 and GK-062A01, respectively, obtained from the Arabidopsis Biological Resource Centre. Genotyping was performed by PCR using gene- and T-DNA-specific primers (Supplemental Table S3). T-DNA insertion sites were determined by sequencing PCR products with the T-DNA left and right border-specific primers. Mutant lines ndufs4 (Meyer et al., 2009) and ca2 (Perales et al., 2005) have been previously described.
Arabidopsis seeds were surface sterilized in a solution containing 70% (v/v) ethanol and 0.1% (v/v) Tween 20 for 5 min, washed first in 70% (v/v) ethanol and then in 100% (v/v) ethanol and dried and plated onto plates of one-half-strength Murashige and Skoog medium containing 1% (w/v) Suc and 1% (w/v) agar. Plates were stratified at 4°C for 2 d. Seedlings were transferred on soil when leaf 4 was larger than 1 mm. Plants were then grown in a growth chamber under a long-day photoperiod of 16 h of light (150 µE m–2 s–1, 22°C) and 8 h of darkness (18°C).
Transcript Analysis
Total RNA from seedlings (growth stage 1.02, two rosette leaves; Boyes et al., 2001) harvested in the middle of the light period was extracted using the NucleoSpin RNA Plant Kit (Macherey-Nagel), and genomic DNA was removed by two DNase treatments using the RNase Free DNase RQ1 (Promega). Complete removal of both mitochondrial and nuclear DNA was confirmed by PCR on diluted RNA. Microarray Complete Arabidopsis Transcription MicroArray chip hybridizations were conducted by the Complete Arabidopsis Transcription MicroArray team. All experiments were performed with three biological replicates. Gene sets are defined via MapMan (Ath_AGI_ISOFORM_MODEL_TAIR10_Aug2012.txt), whereby the second level of the annotation constitutes a set (for example, all genes in PS.lightreaction is a set, but this set is not split further to PS.lightreaction.photosystem and so on). Sets of less than five genes are excluded from the analysis. Gene set tests are performed using ROAST, as this method accounts for gene-wise correlation (Wu et al., 2010). Microarray data from this article were deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE60533) and at CATdb (http://urgv.evry.inra.fr/CATdb/; project no. RS12-03_Complex-I) according to the Minimum Information About a Microarray Experiment standards.
Isolation of Mitochondria from Arabidopsis Plants
The aerial parts of plants were used as starting material for the isolation of mitochondria. The harvested plants were ground at 4°C in extraction buffer (0.3 m Suc, 5 mm tetrasodium pyrophosphate, 10 mm KH2PO4, pH 7.5, 2 mm EDTA, 1% [w/v] polyvinylpyrrolidone40, 1% [w/v] bovine serum albumin, 5 mm Cys, and 20 mm ascorbic acid) in a mortar (50 mL of buffer for 10 g of plant material). After filtration through two layers of Miracloth, the retained material was ground and filtered another time. The filtrates were pooled and centrifuged for 5 min at 2,000g, and the supernatant was centrifuged for 10 min at 20,000g. The pellet was resuspended in wash buffer (0.3 m Suc, 1 mm EGTA, and 10 mm MOPS/KOH, pH 7.2) and subjected to the same low-speed (2,000g) and high-speed (20,000g) centrifugations. The pellet was then resuspended in a small volume of wash buffer and loaded on top of a Percoll step gradient (from bottom to top: 1 volume, 50% [v/v] Percoll; 5 volumes, 25% [v/v] Percoll; and 1 volume, 18% [v/v] Percoll in wash buffer, equivalent to 20 g of plant loaded per gradient). The gradient was centrifuged at 40,000g for 45 min. The fraction located at the interface between the 50% and 25% layers was collected and washed three times in wash buffer. The mitochondrial suspension was aliquoted and stored at –80°C.
One-Dimensional SDS-PAGE Electrophoresis and Immunodetection
Mitochondrial proteins (20 μg) were solubilized in sample buffer (2% [w/v] SDS, 125 mm Tris-HCl, 10% [w/v] glycerol, 0.04% [v/v] β-mercaptoethanol, and 0.002% [w/v] bromphenol blue, pH 6.8) and heated at 95°C for 5 min. The fractions were loaded on 12% (w/v) polyacrylamide and 0.1% (w/v) SDS gels. For immunodetection, separated proteins were transferred onto polyvinylidene difluoride membranes (Immobilon-P, Millipore) and incubated with the primary antibodies. The primary antibodies anti-CA (Perales et al., 2005), anti-51-kD (Peters et al., 2012), and anti-AtpB (AS05085, Agrisera) were used at a dilution of 1:5,000. Secondary antibodies linked to horseradish peroxidase were used to detect the signal by chemiluminescence using the ECL Prime Detection Kit (GE Healthcare). The signals were recorded using a Luminescent Image Analyzer (G-Box-Chemi XT4, Syngene).
BN-PAGE Electrophoresis and Complex I Activity Assays
BN-PAGE was performed according to the method described by Jänsch et al. (1996). Mitochondrial proteins were solubilized with dodecylmaltoside (1% [w/v] final) in ACA buffer (750 mm amino caproic acid, 0.5 mm EDTA, and 50 mm Tris-HCl, pH 7.0) and incubated 20 min at 4°C. The samples were centrifuged for 10 min at 20,000g, and Serva Blue G (0.2% [v/v] final) was added to the supernatant. The samples were loaded onto a 4.5% to 16% gradient gel. Gels were directly used for in-gel NADH oxidase activity assay according to Zerbetto et al. (1997). The gel was washed three times for 5 min with distilled water and incubated in the reaction medium (0.14 mm NADH, 1.22 mm nitroblue tetrazolium, and 0.1 m Tris-HCl, pH 7.4). When the dark-blue stain was strong enough, the reaction was stopped by transferring the gel to 40% (v/v) methanol-10% (v/v) acetic acid.
Oxygen Consumption Measurements
Oxygen consumption was measured using a Clark-type oxygen electrode (Hansatech Instrument). Respiration on Arabidopsis leaves was measured in the dark at 25°C. Plants were dark adapted for at least 30 min prior to the excision of leaf discs (7 mm in diameter). The leaf discs (between 15 and 25 mg fresh weight) were placed in the chamber containing 1 mL of distilled water. Oxygen consumption was measured for at least 30 min before the addition of 1 mm KCN, and the oxygen consumption was followed for another 30 min. Respiration rates on freshly isolated mitochondria were measured at 25°C using samples equivalent to 100 μg of mitochondrial proteins suspended in 1 mL of reaction medium (0.3 m Suc, 5 mm KH2PO4, 10 mm TES, 10 mm NaCl, 2 mm MgSO4, and 0.1% [w/v] bovine serum albumin, pH 6.8). Different substrates, cofactors, and inhibitors were successively added to the reaction medium to modulate O2 consumption by mitochondria. The respiratory capacity when the electron transport is initiated at the level of the TCA cycle was measured in the presence of Glu (10 mm) and malate (10 mm), CoA (12 μm), thiamine pyrophosphate (0.2 mm), NAD+ (2 mm), and ADP (0.3 mm). The capacity of the succinate dehydrogenase was measured in the presence of succinate (5 mm) and ATP (0.5 mm). The maximal capacity of AOX was measured in the same buffer but at pH 7.2 in the presence of succinate (5 mm) and ATP (0.5 mm) and after successive addition of the complex III inhibitor myxothiazol (2.5 μm) and the AOX activators pyruvate (10 mm) and dithiothreitol (5 mm). Finally, the AOX inhibitor n-propyl gallate (0.5 mm) was added. ATP production was measured using the same cofactors and substrates as for the measurements of the respiratory capacity of mitochondria with optimal TCA cycle, as described previously (Meyer et al., 2009). Mitochondrial integrity was measured following oxygen uptake after the addition of ascorbate (10 mm), cytochrome c (25 μm), and Triton X-100 (0.05% [w/v]). In all experiments, it was consistently better than 90%.
Estimation of Fluxes
Estimation of respiratory flux on the basis of 14CO2 evolution was carried out as described in Nunes-Nesi et al. (2005). Twenty leaf discs (7 mm in diameter) of plants at the same growth stage were incubated in 50 mm MES, pH 6.5, containing 0.3 mm of Glc labeled with 6.2 MBq mmol–1 of 14C at position 1 and positions 3 and 4 (ARC0120A and ARC0211, respectively; American Radiolabeled Chemicals) in closed flasks. Evolved 14CO2 was trapped by 500 μL of 10% (w/v) KOH inserted in the flask, and the trap was replaced by a new one every hour. The entire KOH solution in the trap was mixed with 4 mL of scintillation cocktail (Rotizint Eco Plus, ROTH), and radioactivity was determined by a liquid scintillation counter (LS6500, Beckman Coulter).
Quantification of NAD+ and NADH
Leaves were harvested in the middle of the light period and homogenized in liquid nitrogen. Two aliquots of 25 mg each were used to determine the reduced and the oxidized form, respectively. For quantification of NAD+, the first aliquot was extracted with 250 µL of 0.1 m HClO4. For quantification of NADH, the second aliquot was extracted with 250 µL of 0.1 m KOH. Samples were vortexed and incubated for 10 min on ice and then centrifuged for 10 min at 22,000g at 4°C. The supernatant was boiled for 2 min and neutralized by adding an equal volume of either 0.1 m KOH or HClO4 prepared in 0.2 m Tris, pH 8.4. The quantification was conducted exactly as described by Gibon et al. (2004) using a plate reader (EL808, BioTek Instruments).
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under accession numbers SAIL_319_D07 (ndufv1-1); GK-062A01 (ndufv1-2); SAIL_596_E11 (ndufs4); SALK_010194 (ca2); NDUFV1, At5g08530; NDUFS4, At5g67590; and CA2, At1g47260. The accession numbers for other genes/proteins studied in this work are detailed in Supplemental Figure S3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Growth phenotypes of complex I mutants.
Supplemental Figure S2. Growth phenotypes of complex I mutants in the absence or presence of Suc.
Supplemental Figure S3. Analysis of the early growth stages in severe complex I mutants.
Supplemental Figure S4. Metabolite profiles of complex I mutants.
Supplemental Figure S5. Comparison of the mitochondrial proteomes of ndufv1-1 and ndufs4 using DIGE.
Supplemental Figure S6. MapMan metabolism maps.
Supplemental Table S1. Analysis of the gene categories showing differential response in ndufv1-1 and ndufs4.
Supplemental Table S2. Overview of the metabolite reporting list.
Supplemental Table S3. List of primers used in this study.
Supplemental Methods S1. Methods for metabolite profiling, differential gel electrophoresis, and mass spectrometry.
Supplementary Material
Acknowledgments
We thank Eduardo Zabaleta and Hans-Peter Braun for providing the anti-CA and anti-51-kD antibodies, respectively.
Glossary
- TCA
tricarboxylic acid
- OXPHOS
oxidative phosphorylation
- ETC
electron transfer chain
- IM
inner membrane
- T-DNA
transfer DNA
- BN
blue-native
- CA
carbonic anhydrase
- AOX
alternative oxidase
Footnotes
This work was supported by the Centre National de la Recherche Scientifique, the Max Planck Society, and a Marie Curie International Reintegration Grant (grant no. PIRG256398).
Articles can be viewed without a subscription.
References
- Baradaran R, Berrisford JM, Minhas GS, Sazanov LA (2013) Crystal structure of the entire respiratory complex I. Nature 494: 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt U. (2006) Energy converting NADH:quinone oxidoreductase (complex I). Annu Rev Biochem 75: 69–92 [DOI] [PubMed] [Google Scholar]
- Brangeon J, Sabar M, Gutierres S, Combettes B, Bove J, Gendy C, Chétrit P, Des Francs-Small CC, Pla M, Vedel F, et al. (2000) Defective splicing of the first nad4 intron is associated with lack of several complex I subunits in the Nicotiana sylvestris NMS1 nuclear mutant. Plant J 21: 269–280 [DOI] [PubMed] [Google Scholar]
- Cardol P. (2011) Mitochondrial NADH:ubiquinone oxidoreductase (complex I) in eukaryotes: a highly conserved subunit composition highlighted by mining of protein databases. Biochim Biophys Acta 1807: 1390–1397 [DOI] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Small I (2014) Surrogate mutants for studying mitochondrially encoded functions. Biochimie 100: 234–242 [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Meyer EH, Andrés C, Taylor NL, Lurin C, Millar AH, Small ID (2007) The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 Intron 1 in Arabidopsis thaliana. Plant Cell 19: 3256–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paepe R, Chétrit P, Vitart V, Ambard-Bretteville F, Prat D, Vedel F (1990) Several nuclear genes control both male sterility and mitochondrial protein synthesis in Nicotiana sylvestris protoclones. Mol Gen Genet 222: 206–210 [DOI] [PubMed] [Google Scholar]
- Distelmaier F, Koopman WJ, van den Heuvel LP, Rodenburg RJ, Mayatepek E, Willems PH, Smeitink JA (2009) Mitochondrial complex I deficiency: from organelle dysfunction to clinical disease. Brain 132: 833–842 [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Driscoll S, Cornic G, De Paepe R, Foyer CH, Noctor G (2003) Functional mitochondrial complex I is required by tobacco leaves for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiol 131: 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov RG, Baradaran R, Sazanov LA (2010) The architecture of respiratory complex I. Nature 465: 441–445 [DOI] [PubMed] [Google Scholar]
- Efremov RG, Sazanov LA (2011) Structure of the membrane domain of respiratory complex I. Nature 476: 414–420 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Stitt M (2012) On the discordance of metabolomics with proteomics and transcriptomics: coping with increasing complexity in logic, chemistry, and network interactions scientific correspondence. Plant Physiol 158: 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T. (2014) On the mechanism of respiratory complex I. J Bioenerg Biomembr 46: 255–268 [DOI] [PubMed] [Google Scholar]
- Galkin A, Dröse S, Brandt U (2006) The proton pumping stoichiometry of purified mitochondrial complex I reconstituted into proteoliposomes. Biochim Biophys Acta 1757: 1575–1581 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JH, Palacios N, Cross J, Selbig J, Stitt M (2004) A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore DC, Jahoor F, Hibbert JM, DeMaria EJ (1996) Lactic acidosis during sepsis is related to increased pyruvate production, not deficits in tissue oxygen availability. Ann Surg 224: 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierres S, Combettes B, De Paepe R, Mirande M, Lelandais C, Vedel F, Chétrit P (1999) In the Nicotiana sylvestris CMSII mutant, a recombination-mediated change 5′ to the first exon of the mitochondrial nad1 gene is associated with lack of the NADH:ubiquinone oxidoreductase (complex I) NAD1 subunit. Eur J Biochem 261: 361–370 [DOI] [PubMed] [Google Scholar]
- Gutierres S, Sabar M, Lelandais C, Chetrit P, Diolez P, Degand H, Boutry M, Vedel F, de Kouchkovsky Y, De Paepe R (1997) Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci USA 94: 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haïli N, Arnal N, Quadrado M, Amiar S, Tcherkez G, Dahan J, Briozzo P, Colas des Francs-Small C, Vrielynck N, Mireau H (2013) The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res 41: 6650–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J. (2011) Why does mitochondrial complex I have so many subunits? Biochem J 437: e1–e3 [DOI] [PubMed] [Google Scholar]
- Hirst J. (2013) Mitochondrial complex I. Annu Rev Biochem 82: 551–575 [DOI] [PubMed] [Google Scholar]
- Hunte C, Zickermann V, Brandt U (2010) Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329: 448–451 [DOI] [PubMed] [Google Scholar]
- Jänsch L, Kruft V, Schmitz UK, Braun HP (1996) New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J 9: 357–368 [DOI] [PubMed] [Google Scholar]
- Juszczuk IM, Flexas J, Szal B, Dabrowska Z, Ribas-Carbo M, Rychter AM (2007) Effect of mitochondrial genome rearrangement on respiratory activity, photosynthesis, photorespiration and energy status of MSC16 cucumber (Cucumis sativus) mutant. Physiol Plant 131: 527–541 [DOI] [PubMed] [Google Scholar]
- Kamiński MM, Sauer SW, Kamiński M, Opp S, Ruppert T, Grigaravičius P, Grudnik P, Gröne HJ, Krammer PH, Gülow K (2012) T cell activation is driven by an ADP-dependent glucokinase linking enhanced glycolysis with mitochondrial reactive oxygen species generation. Cell Reports 2: 1300–1315 [DOI] [PubMed] [Google Scholar]
- Karpova OV, Newton KJ (1999) A partially assembled complex I in NAD4-deficient mitochondria of maize. Plant J 17: 511–521 [Google Scholar]
- Kühn K, Carrie C, Giraud E, Wang Y, Meyer EH, Narsai R, des Francs-Small CC, Zhang B, Murcha MW, Whelan J (2011) The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of Arabidopsis thaliana. Plant J 67: 1067–1080 [DOI] [PubMed] [Google Scholar]
- Lee CP, Eubel H, O’Toole N, Millar AH (2008) Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol Cell Proteomics 7: 1297–1316 [DOI] [PubMed] [Google Scholar]
- Lenzen S. (2014) A fresh view of glycolysis and glucokinase regulation: history and current status. J Biol Chem 289: 12189–12194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lushchak OV, Piroddi M, Galli F, Lushchak VI (2014) Aconitase post-translational modification as a key in linkage between Krebs cycle, iron homeostasis, redox signaling, and metabolism of reactive oxygen species. Redox Rep 19: 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado EN, Lemasters JJ (2014) ATP/ADP ratio, the missed connection between mitochondria and the Warburg effect. Mitochondrion (Pt A) 19: 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienfeld JR, Newton KJ (1994) The maize NCS2 abnormal growth mutant has a chimeric nad4-nad7 mitochondrial gene and is associated with reduced complex I function. Genetics 138: 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EH. (2012) Proteomic investigations of complex I composition: how to define a subunit? Front Plant Sci 3: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EH, Solheim C, Tanz SK, Bonnard G, Millar AH (2011) Insights into the composition and assembly of the membrane arm of plant complex I through analysis of subcomplexes in Arabidopsis mutant lines. J Biol Chem 286: 26081–26092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EH, Tomaz T, Carroll AJ, Estavillo G, Delannoy E, Tanz SK, Small ID, Pogson BJ, Millar AH (2009) Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol 151: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton KJ, Coe EH (1986) Mitochondrial DNA changes in abnormal growth (nonchromosomal stripe) mutants of maize. Proc Natl Acad Sci USA 83: 7363–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouws J, Nijtmans LG, Smeitink JA, Vogel RO (2012) Assembly factors as a new class of disease genes for mitochondrial complex I deficiency: cause, pathology and treatment options. Brain 135: 12–22 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Araújo WL, Obata T, Fernie AR (2013) Regulation of the mitochondrial tricarboxylic acid cycle. Curr Opin Plant Biol 16: 335–343 [DOI] [PubMed] [Google Scholar]
- Nunes-Nesi A, Carrari F, Lytovchenko A, Smith AM, Loureiro ME, Ratcliffe RG, Sweetlove LJ, Fernie AR (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137: 611–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagniez-Mammeri H, Rak M, Legrand A, Bénit P, Rustin P, Slama A (2012) Mitochondrial complex I deficiency of nuclear origin. II. Non-structural genes. Mol Genet Metab 105: 173–179 [DOI] [PubMed] [Google Scholar]
- Perales M, Eubel H, Heinemeyer J, Colaneri A, Zabaleta E, Braun HP (2005) Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I + III2 levels and alters mitochondrial physiology in Arabidopsis. J Mol Biol 350: 263–277 [DOI] [PubMed] [Google Scholar]
- Peters K, Niessen M, Peterhänsel C, Späth B, Hölzle A, Binder S, Marchfelder A, Braun HP (2012) Complex I-complex II ratio strongly differs in various organs of Arabidopsis thaliana. Plant Mol Biol 79: 273–284 [DOI] [PubMed] [Google Scholar]
- Piattoni CV, Guerrero SA, Iglesias AA (2013) A differential redox regulation of the pathways metabolizing glyceraldehyde-3-phosphate tunes the production of reducing power in the cytosol of plant cells. Int J Mol Sci 14: 8073–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau B, Mathieu C, Gérard-Hirne C, De Paepe R, Chétrit P (2005) Targeting the NAD7 subunit to mitochondria restores a functional complex I and a wild-type phenotype in the Nicotiana sylvestris CMS II mutant lacking nad7. J Biol Chem 280: 25994–26001 [DOI] [PubMed] [Google Scholar]
- Pinfield-Wells H, Rylott EL, Gilday AD, Graham S, Job K, Larson TR, Graham IA (2005) Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. Plant J 43: 861–872 [DOI] [PubMed] [Google Scholar]
- Pla M, Mathieu C, De Paepe R, Chétrit P, Vedel F (1995) Deletion of the last two exons of the mitochondrial nad7 gene results in lack of the NAD7 polypeptide in a Nicotiana sylvestris CMS mutant. Mol Gen Genet 248: 79–88 [DOI] [PubMed] [Google Scholar]
- Priault P, Fresneau C, Noctor G, De Paepe R, Cornic G, Streb P (2006) The mitochondrial CMSII mutation of Nicotiana sylvestris impairs adjustment of photosynthetic carbon assimilation to higher growth irradiance. J Exp Bot 57: 2075–2085 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Geisler DA, Møller IM (2008) The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 8: 47–60 [DOI] [PubMed] [Google Scholar]
- Robinson BH. (2006) Lactic acidemia and mitochondrial disease. Mol Genet Metab 89: 3–13 [DOI] [PubMed] [Google Scholar]
- Ros S, Schulze A (2013) Balancing glycolytic flux: the role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabar M, De Paepe R, de Kouchkovsky Y (2000) Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol 124: 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazanov LA. (2014) The mechanism of coupling between electron transfer and proton translocation in respiratory complex I. J Bioenerg Biomembr 46: 247–253 [DOI] [PubMed] [Google Scholar]
- Scaglia F, Towbin JA, Craigen WJ, Belmont JW, Smith EO, Neish SR, Ware SM, Hunter JV, Fernbach SD, Vladutiu GD, et al. (2004) Clinical spectrum, morbidity, and mortality in 113 pediatric patients with mitochondrial disease. Pediatrics 114: 925–931 [DOI] [PubMed] [Google Scholar]
- Schmidtmann E, König AC, Orwat A, Leister D, Hartl M, Finkemeier I (2014) Redox regulation of Arabidopsis mitochondrial citrate synthase. Mol Plant 7: 156–169 [DOI] [PubMed] [Google Scholar]
- Shimada S, Shinzawa-Itoh K, Amano S, Akira Y, Miyazawa A, Tsukihara T, Tani K, Gerle C, Yoshikawa S (2014) Three-dimensional structure of bovine heart NADH: ubiquinone oxidoreductase (complex I) by electron microscopy of a single negatively stained two-dimensional crystal. Microscopy (Oxf) 63: 167–174 [DOI] [PubMed] [Google Scholar]
- Stacpoole PW. (1997) Lactic acidosis and other mitochondrial disorders. Metabolism 46: 306–321 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tucker EJ, Compton AG, Calvo SE, Thorburn DR (2011) The molecular basis of human complex I deficiency. IUBMB Life 63: 669–677 [DOI] [PubMed] [Google Scholar]
- Upadhyay M, Samal J, Kandpal M, Singh OV, Vivekanandan P (2013) The Warburg effect: insights from the past decade. Pharmacol Ther 137: 318–330 [DOI] [PubMed] [Google Scholar]
- Valsecchi F, Koopman WJ, Manjeri GR, Rodenburg RJ, Smeitink JA, Willems PH (2010) Complex I disorders: causes, mechanisms, and development of treatment strategies at the cellular level. Dev Disabil Res Rev 16: 175–182 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, Day DA, Wiskich JT, Vanlerberghe AE, McIntosh L (1995) Alternative oxidase activity in tobacco leaf mitochondria (dependence on tricarboxylic acid cycle-mediated redox regulation and pyruvate activation). Plant Physiol 109: 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinothkumar KR, Zhu J, Hirst J (2014) Architecture of mammalian respiratory complex I. Nature 515: 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener G, Krause U (2002) Different modes of activating phosphofructokinase, a key regulatory enzyme of glycolysis, in working vertebrate muscle. Biochem Soc Trans 30: 264–270 [DOI] [PubMed] [Google Scholar]
- Wikström M. (1984) Two protons are pumped from the mitochondrial matrix per electron transferred between NADH and ubiquinone. FEBS Lett 169: 300–304 [DOI] [PubMed] [Google Scholar]
- Wu D, Lim E, Vaillant F, Asselin-Labat ML, Visvader JE, Smyth GK (2010) ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 26: 2176–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wydro MM, Sharma P, Foster JM, Bych K, Meyer EH, Balk J (2013) The evolutionarily conserved iron-sulfur protein INDH is required for complex I assembly and mitochondrial translation in Arabidopsis [corrected]. Plant Cell 25: 4014–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbetto E, Vergani L, Dabbeni-Sala F (1997) Quantification of muscle mitochondrial oxidative phosphorylation enzymes via histochemical staining of blue native polyacrylamide gels. Electrophoresis 18: 2059–2064 [DOI] [PubMed] [Google Scholar]
- Zickermann V, Wirth C, Nasiri H, Siegmund K, Schwalbe H, Hunte C, Brandt U (2015) Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science 347: 44–49 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.