Abstract
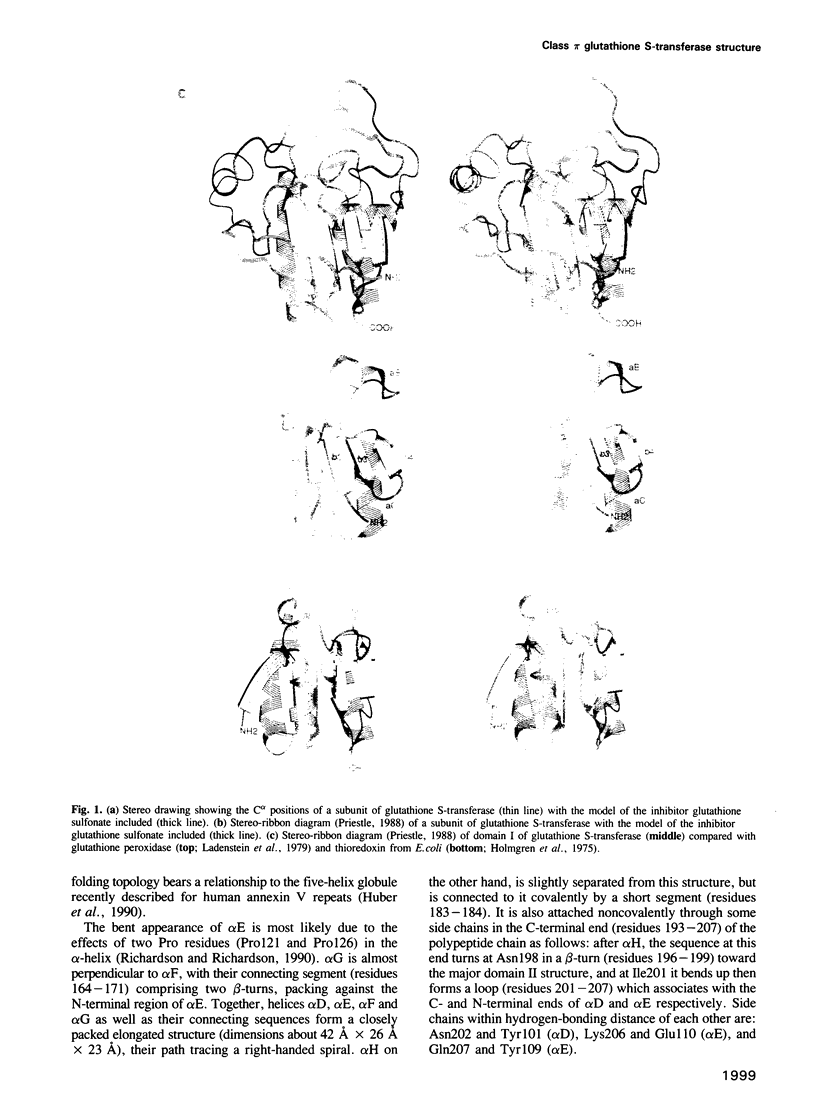
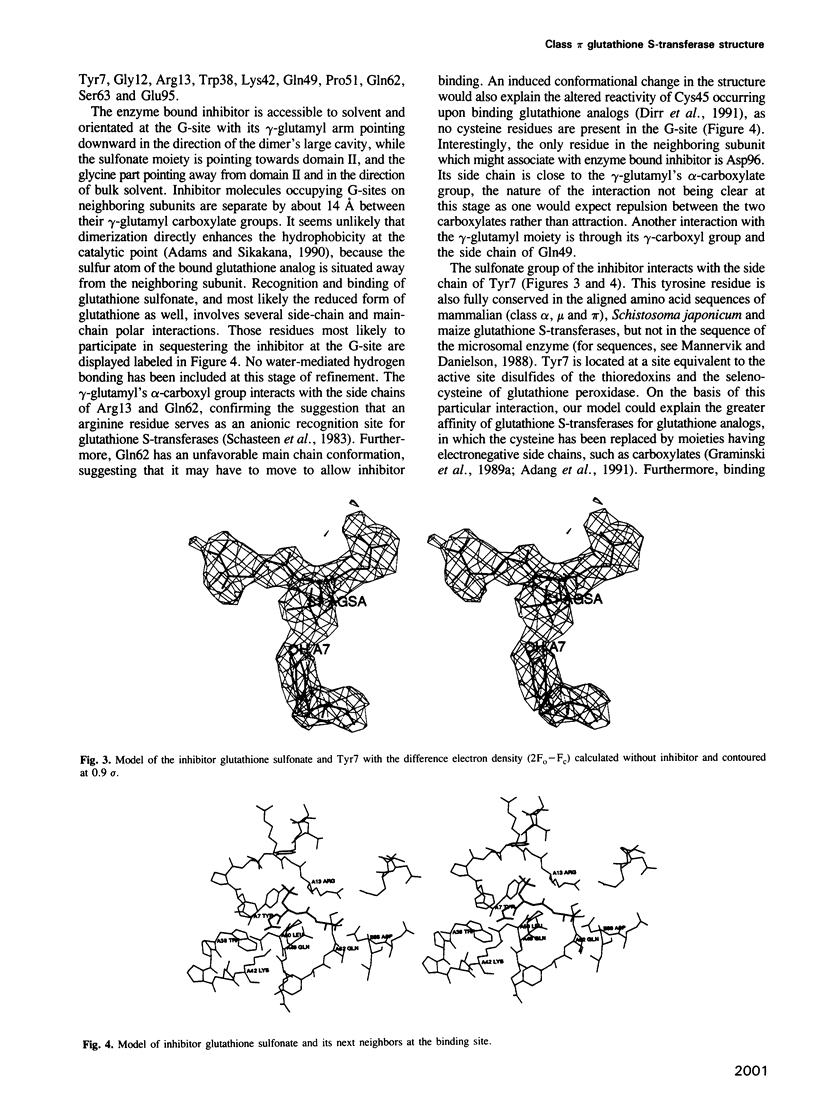
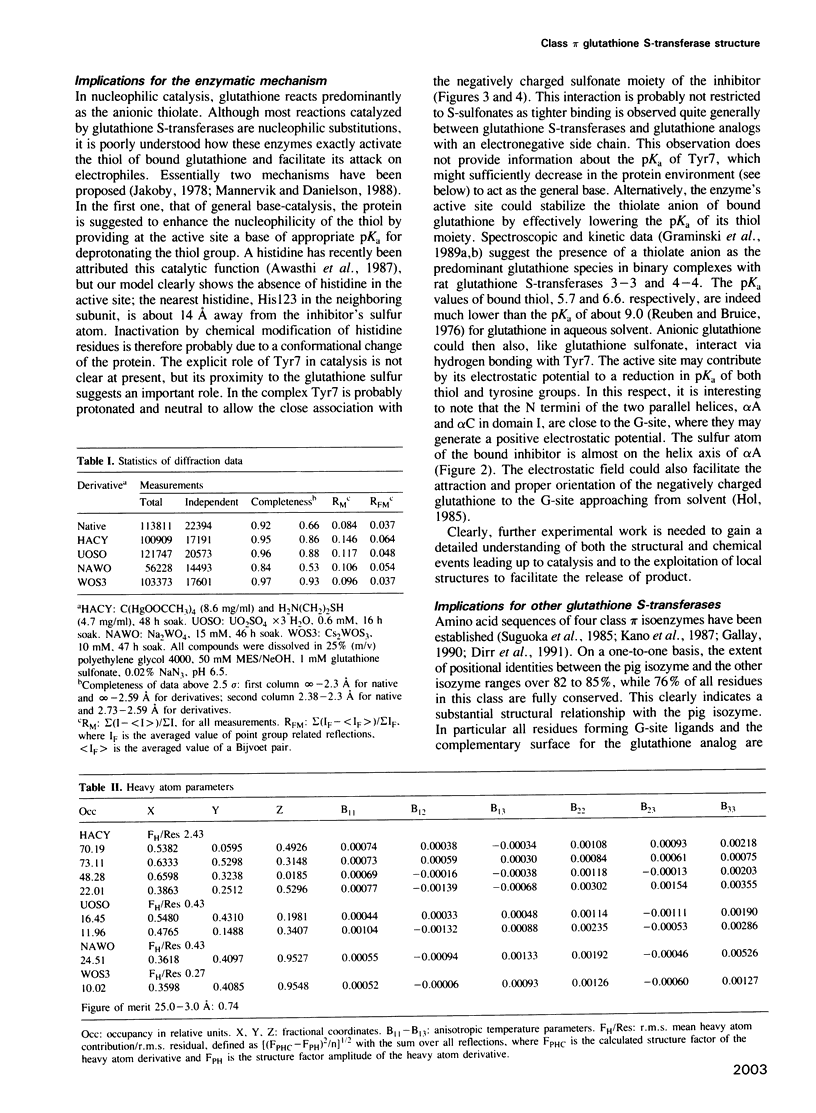
The three-dimensional structure of class pi glutathione S-transferase from pig lung, a homodimeric enzyme, has been solved by multiple isomorphous replacement at 3 A resolution and preliminarily refined at 2.3 A resolution (R = 0.24). Each subunit (207 residues) is folded into two domains of different structure. Domain I (residues 1-74) consists of a central four-stranded beta-sheet flanked on one side by two alpha-helices and on the other side, facing the solvent, by a bent, irregular helix structure. The topological pattern resembles the bacteriophage T4 thioredoxin fold, in spite of their dissimilar sequences. Domain II (residues 81-207) contains five alpha-helices. The dimeric molecule is globular with dimensions of about 55 A x 52 A x 45 A. Between the subunits and along the local diad, is a large cavity which could possibly be involved in the transport of nonsubstrate ligands. The binding site of the competitive inhibitor, glutathione sulfonate, is located on domain I, and is part of a cleft formed between intrasubunit domains. Glutathione sulfonate is bound in an extended conformation through multiple interactions. Only three contact residues, namely Tyr7, Gln62 and Asp96 are conserved within the family of cytosolic glutathione S-transferases. The exact location of the binding site(s) of the electrophilic substrate is not clear. Catalytic models are discussed on the basis of the molecular structure.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams P. A., Sikakana C. N. Factors affecting the inactivation of human placental glutathione S-transferase pi. The kinetic mechanism and pH-dependence of solvational and 1-chloro-2,4-dinitrobenzene-mediated inactivation of the enzyme. Biochem Pharmacol. 1990 Jun 15;39(12):1883–1889. doi: 10.1016/0006-2952(90)90605-k. [DOI] [PubMed] [Google Scholar]
- Adang A. E., Brussee J., Meyer D. J., Coles B., Ketterer B., van der Gen A., Mulder G. J. Substrate specificity of rat liver glutathione S-transferase isoenzymes for a series of glutathione analogues, modified at the gamma-glutamyl moiety. Biochem J. 1988 Oct 15;255(2):721–724. [PMC free article] [PubMed] [Google Scholar]
- Adang A. E., Brussee J., van der Gen A., Mulder G. J. Inhibition of rat liver glutathione S-transferase isoenzymes by peptides stabilized against degradation by gamma-glutamyl transpeptidase. J Biol Chem. 1991 Jan 15;266(2):830–836. [PubMed] [Google Scholar]
- Adang A. E., Brussee J., van der Gen A., Mulder G. J. The glutathione-binding site in glutathione S-transferases. Investigation of the cysteinyl, glycyl and gamma-glutamyl domains. Biochem J. 1990 Jul 1;269(1):47–54. doi: 10.1042/bj2690047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adang A. E., Meyer D. J., Brussee J., Van der Gen A., Ketterer B., Mulder G. J. Interaction of rat glutathione S-transferases 7-7 and 8-8 with gamma-glutamyl- or glycyl-modified glutathione analogues. Biochem J. 1989 Dec 15;264(3):759–764. doi: 10.1042/bj2640759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad H., Wilson D. E., Fritz R. R., Singh S. V., Medh R. D., Nagle G. T., Awasthi Y. C., Kurosky A. Primary and secondary structural analyses of glutathione S-transferase pi from human placenta. Arch Biochem Biophys. 1990 May 1;278(2):398–408. doi: 10.1016/0003-9861(90)90277-6. [DOI] [PubMed] [Google Scholar]
- Awasthi Y. C., Bhatnagar A., Singh S. V. Evidence for the involvement of histidine at the active site of glutathione S-transferase psi from human liver. Biochem Biophys Res Commun. 1987 Mar 30;143(3):965–970. doi: 10.1016/0006-291x(87)90345-7. [DOI] [PubMed] [Google Scholar]
- Bornstein P. Structure of alpha-1-CB8, a large cyanogen bromide produced fragment from the alpha-1 chain of rat collagen. The nature of a hydroxylamine-sensitive bond and composition of tryptic peptides. Biochemistry. 1970 Jun 9;9(12):2408–2421. doi: 10.1021/bi00814a004. [DOI] [PubMed] [Google Scholar]
- Boyer T. D. The glutathione S-transferases: an update. Hepatology. 1989 Mar;9(3):486–496. doi: 10.1002/hep.1840090324. [DOI] [PubMed] [Google Scholar]
- Chen W. J., Graminski G. F., Armstrong R. N. Dissection of the catalytic mechanism of isozyme 4-4 of glutathione S-transferase with alternative substrates. Biochemistry. 1988 Jan 26;27(2):647–654. doi: 10.1021/bi00402a023. [DOI] [PubMed] [Google Scholar]
- Chothia C., Levitt M., Richardson D. Structure of proteins: packing of alpha-helices and pleated sheets. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4130–4134. doi: 10.1073/pnas.74.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles B., Ketterer B. The role of glutathione and glutathione transferases in chemical carcinogenesis. Crit Rev Biochem Mol Biol. 1990;25(1):47–70. doi: 10.3109/10409239009090605. [DOI] [PubMed] [Google Scholar]
- Cowan S. W., Bergfors T., Jones T. A., Tibbelin G., Olin B., Board P. G., Mannervik B. Crystallization of GST2, a human class alpha glutathione transferase. J Mol Biol. 1989 Jul 20;208(2):369–370. doi: 10.1016/0022-2836(89)90398-7. [DOI] [PubMed] [Google Scholar]
- Danielson U. H., Mannervik B. Kinetic independence of the subunits of cytosolic glutathione transferase from the rat. Biochem J. 1985 Oct 15;231(2):263–267. doi: 10.1042/bj2310263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirr H. W., Mann K., Huber R., Ladenstein R., Reinemer P. Class pi glutathione S-transferase from pig lung. Purification, biochemical characterization, primary structure and crystallization. Eur J Biochem. 1991 Mar 28;196(3):693–698. doi: 10.1111/j.1432-1033.1991.tb15867.x. [DOI] [PubMed] [Google Scholar]
- Douglas K. T. Mechanism of action of glutathione-dependent enzymes. Adv Enzymol Relat Areas Mol Biol. 1987;59:103–167. doi: 10.1002/9780470123058.ch3. [DOI] [PubMed] [Google Scholar]
- Epp O., Ladenstein R., Wendel A. The refined structure of the selenoenzyme glutathione peroxidase at 0.2-nm resolution. Eur J Biochem. 1983 Jun 1;133(1):51–69. doi: 10.1111/j.1432-1033.1983.tb07429.x. [DOI] [PubMed] [Google Scholar]
- Graminski G. F., Kubo Y., Armstrong R. N. Spectroscopic and kinetic evidence for the thiolate anion of glutathione at the active site of glutathione S-transferase. Biochemistry. 1989 Apr 18;28(8):3562–3568. doi: 10.1021/bi00434a062. [DOI] [PubMed] [Google Scholar]
- Graminski G. F., Zhang P. H., Sesay M. A., Ammon H. L., Armstrong R. N. Formation of the 1-(S-glutathionyl)-2,4,6-trinitrocyclohexadienate anion at the active site of glutathione S-transferase: evidence for enzymic stabilization of sigma-complex intermediates in nucleophilic aromatic substitution reactions. Biochemistry. 1989 Jul 25;28(15):6252–6258. doi: 10.1021/bi00441a017. [DOI] [PubMed] [Google Scholar]
- Grütter M. G., Priestle J. P., Rahuel J., Grossenbacher H., Bode W., Hofsteenge J., Stone S. R. Crystal structure of the thrombin-hirudin complex: a novel mode of serine protease inhibition. EMBO J. 1990 Aug;9(8):2361–2365. doi: 10.1002/j.1460-2075.1990.tb07410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffken H. W., Knof S. H., Bartlett P. A., Huber R., Moellering H., Schumacher G. Crystal structure determination, refinement and molecular model of creatine amidinohydrolase from Pseudomonas putida. J Mol Biol. 1988 Nov 20;204(2):417–433. doi: 10.1016/0022-2836(88)90586-4. [DOI] [PubMed] [Google Scholar]
- Hoesch R. M., Boyer T. D. Localization of a portion of the active site of two rat liver glutathione S-transferases using a photoaffinity label. J Biol Chem. 1989 Oct 25;264(30):17712–17717. [PubMed] [Google Scholar]
- Hol W. G. The role of the alpha-helix dipole in protein function and structure. Prog Biophys Mol Biol. 1985;45(3):149–195. doi: 10.1016/0079-6107(85)90001-x. [DOI] [PubMed] [Google Scholar]
- Holmgren A., Söderberg B. O., Eklund H., Brändén C. I. Three-dimensional structure of Escherichia coli thioredoxin-S2 to 2.8 A resolution. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2305–2309. doi: 10.1073/pnas.72.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R., Römisch J., Paques E. P. The crystal and molecular structure of human annexin V, an anticoagulant protein that binds to calcium and membranes. EMBO J. 1990 Dec;9(12):3867–3874. doi: 10.1002/j.1460-2075.1990.tb07605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby W. B. The glutathione S-transferases: a group of multifunctional detoxification proteins. Adv Enzymol Relat Areas Mol Biol. 1978;46:383–414. doi: 10.1002/9780470122914.ch6. [DOI] [PubMed] [Google Scholar]
- Kano T., Sakai M., Muramatsu M. Structure and expression of a human class pi glutathione S-transferase messenger RNA. Cancer Res. 1987 Nov 1;47(21):5626–5630. [PubMed] [Google Scholar]
- Karplus P. A., Pai E. F., Schulz G. E. A crystallographic study of the glutathione binding site of glutathione reductase at 0.3-nm resolution. Eur J Biochem. 1989 Jan 2;178(3):693–703. doi: 10.1111/j.1432-1033.1989.tb14500.x. [DOI] [PubMed] [Google Scholar]
- Ladenstein R., Epp O., Bartels K., Jones A., Huber R., Wendel A. Structure analysis and molecular model of the selenoenzyme glutathione peroxidase at 2.8 A resolution. J Mol Biol. 1979 Oct 25;134(2):199–218. doi: 10.1016/0022-2836(79)90032-9. [DOI] [PubMed] [Google Scholar]
- Listowsky I., Abramovitz M., Homma H., Niitsu Y. Intracellular binding and transport of hormones and xenobiotics by glutathione-S-transferases. Drug Metab Rev. 1988;19(3-4):305–318. doi: 10.3109/03602538808994138. [DOI] [PubMed] [Google Scholar]
- Mannervik B., Alin P., Guthenberg C., Jensson H., Tahir M. K., Warholm M., Jörnvall H. Identification of three classes of cytosolic glutathione transferase common to several mammalian species: correlation between structural data and enzymatic properties. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7202–7206. doi: 10.1073/pnas.82.21.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannervik B., Danielson U. H. Glutathione transferases--structure and catalytic activity. CRC Crit Rev Biochem. 1988;23(3):283–337. doi: 10.3109/10409238809088226. [DOI] [PubMed] [Google Scholar]
- Mannervik B., Guthenberg C. Glutathione transferase (human placenta). Methods Enzymol. 1981;77:231–235. doi: 10.1016/s0076-6879(81)77030-7. [DOI] [PubMed] [Google Scholar]
- Mannervik B. The isoenzymes of glutathione transferase. Adv Enzymol Relat Areas Mol Biol. 1985;57:357–417. doi: 10.1002/9780470123034.ch5. [DOI] [PubMed] [Google Scholar]
- Parker M. W., Lo Bello M., Federici G. Crystallization of glutathione S-transferase from human placenta. J Mol Biol. 1990 May 20;213(2):221–222. doi: 10.1016/s0022-2836(05)80183-4. [DOI] [PubMed] [Google Scholar]
- Persson B., Jörnvall H., Alin P., Mannervik B. Structural classes of glutathione transferase: distinctions between isoenzymes and enzymes. Protein Seq Data Anal. 1988 Feb;1(3):183–186. [PubMed] [Google Scholar]
- Pickett C. B., Lu A. Y. Glutathione S-transferases: gene structure, regulation, and biological function. Annu Rev Biochem. 1989;58:743–764. doi: 10.1146/annurev.bi.58.070189.003523. [DOI] [PubMed] [Google Scholar]
- Rosevear P. R., Sellin S., Mannervik B., Kuntz I. D., Mildvan A. S. NMR and computer modeling studies of the conformations of glutathione derivatives at the active site of glyoxalase I. J Biol Chem. 1984 Sep 25;259(18):11436–11447. [PubMed] [Google Scholar]
- Rydel T. J., Ravichandran K. G., Tulinsky A., Bode W., Huber R., Roitsch C., Fenton J. W., 2nd The structure of a complex of recombinant hirudin and human alpha-thrombin. Science. 1990 Jul 20;249(4966):277–280. doi: 10.1126/science.2374926. [DOI] [PubMed] [Google Scholar]
- Schäffer J., Gallay O., Ladenstein R. Glutathione transferase from bovine placenta. Preparation, biochemical characterization, crystallization, and preliminary crystallographic analysis of a neutral class PI enzyme. J Biol Chem. 1988 Nov 25;263(33):17405–17411. [PubMed] [Google Scholar]
- Sesay M. A., Ammon H. L., Armstrong R. N. Crystallization and a preliminary X-ray diffraction study of isozyme 3-3 of glutathione S-transferase from rat liver. J Mol Biol. 1987 Sep 20;197(2):377–378. doi: 10.1016/0022-2836(87)90133-1. [DOI] [PubMed] [Google Scholar]
- Sugimoto M., Kuhlenkamp J., Ookhtens M., Aw T. Y., Reeve J., Jr, Kaplowitz N. Gamma-glutamylcysteine: a substrate for glutathione S-transferases. Biochem Pharmacol. 1985 Oct 15;34(20):3643–3647. doi: 10.1016/0006-2952(85)90224-2. [DOI] [PubMed] [Google Scholar]
- Suguoka Y., Kano T., Okuda A., Sakai M., Kitagawa T., Muramatsu M. Cloning and the nucleotide sequence of rat glutathione S-transferase P cDNA. Nucleic Acids Res. 1985 Sep 11;13(17):6049–6057. doi: 10.1093/nar/13.17.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg B. O., Sjöberg B. M., Sonnerstam U., Brändén C. I. Three-dimensional structure of thioredoxin induced by bacteriophage T4. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5827–5830. doi: 10.1073/pnas.75.12.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- Waxman D. J. Glutathione S-transferases: role in alkylating agent resistance and possible target for modulation chemotherapy--a review. Cancer Res. 1990 Oct 15;50(20):6449–6454. [PubMed] [Google Scholar]