Abstract
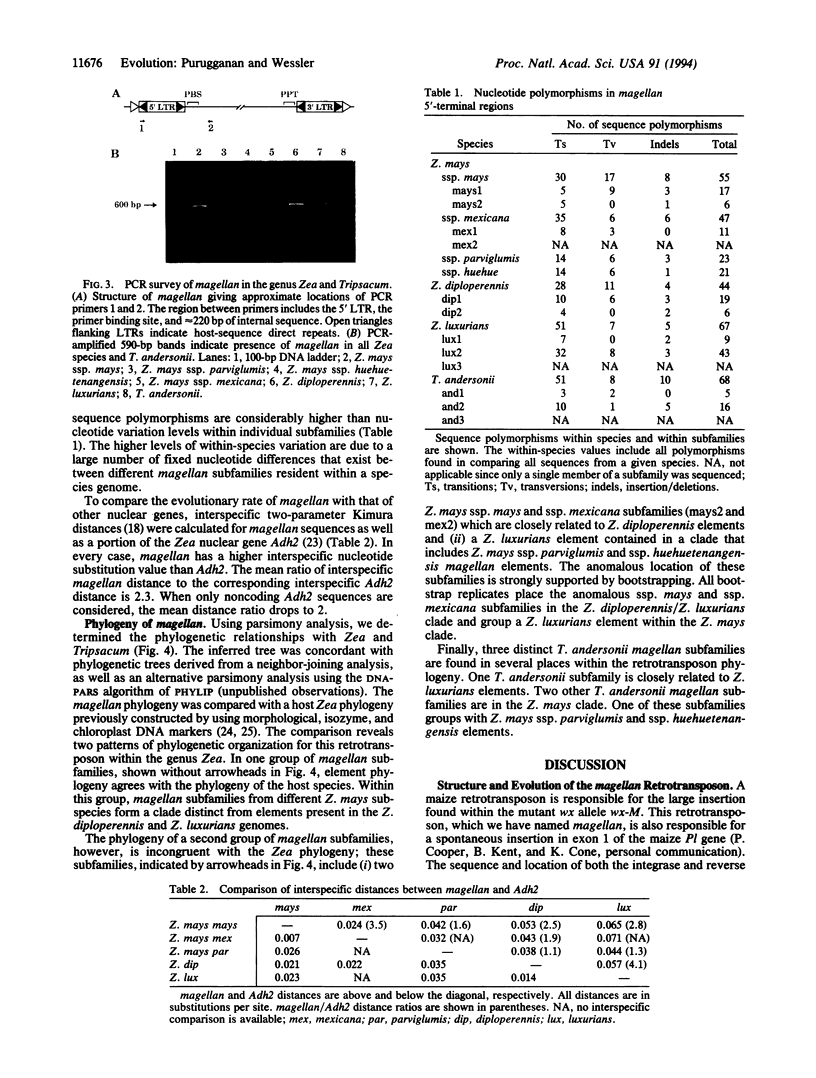
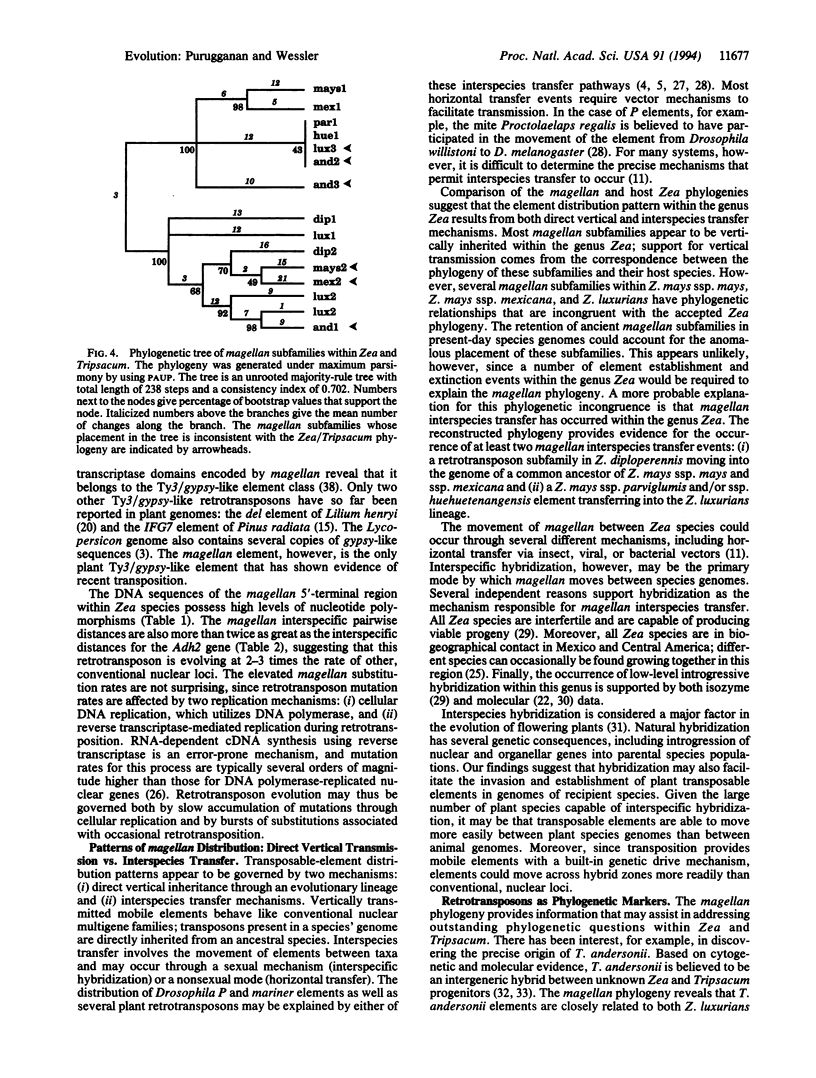
The magellan transposable element is responsible for a spontaneous 5.7-kb insertion in the maize wx-M allele. This element has the sequence and structural characteristics of a Ty3/gypsy-like retrotransposon. The magellan element is present in all Zea species and Tripsacum andersonii; it is absent, however, in the genomes of all other Tripsacum species analyzed. The genetic distances between magellan elements suggest that this retrotransposon is evolving faster than other Zea nuclear loci. The phylogeny of magellan within Zea and T. andersonii also reveals a pattern of interspecies transfers, resulting in the movement of magellan subfamilies between different species genomes. Interspecific hybridization may be a major mechanism by which this retrotransposon invades and establishes itself in new taxa.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boeke J. D., Corces V. G. Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol. 1989;43:403–434. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Langley C. H. The population genetics of Drosophila transposable elements. Annu Rev Genet. 1989;23:251–287. doi: 10.1146/annurev.ge.23.120189.001343. [DOI] [PubMed] [Google Scholar]
- Doebley J., Renfroe W., Blanton A. Restriction site variation in the zea chloroplast genome. Genetics. 1987 Sep;117(1):139–147. doi: 10.1093/genetics/117.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell A. J., Dunbar E., Anderson R., Pearce S. R., Hartley R., Kumar A. Ty1-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res. 1992 Jul 25;20(14):3639–3644. doi: 10.1093/nar/20.14.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut B. S., Clegg M. T. Molecular evolution of the Adh1 locus in the genus Zea. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5095–5099. doi: 10.1073/pnas.90.11.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloubinoff P., Päbo S., Wilson A. C. Evolution of maize inferred from sequence diversity of an Adh2 gene segment from archaeological specimens. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1997–2001. doi: 10.1073/pnas.90.5.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien M. A. Retroelements in higher plants. Trends Genet. 1992 Mar;8(3):103–108. doi: 10.1016/0168-9525(92)90198-d. [DOI] [PubMed] [Google Scholar]
- Holland J., Spindler K., Horodyski F., Grabau E., Nichol S., VandePol S. Rapid evolution of RNA genomes. Science. 1982 Mar 26;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Houck M. A., Clark J. B., Peterson K. R., Kidwell M. G. Possible horizontal transfer of Drosophila genes by the mite Proctolaelaps regalis. Science. 1991 Sep 6;253(5024):1125–1128. doi: 10.1126/science.1653453. [DOI] [PubMed] [Google Scholar]
- Kidwell M. G. Horizontal transfer of P elements and other short inverted repeat transposons. Genetica. 1992;86(1-3):275–286. doi: 10.1007/BF00133726. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- MacRae A. F., Clegg M. T. Evolution of Ac and Dsl elements in select grasses (Poaceae). Genetica. 1992;86(1-3):55–66. doi: 10.1007/BF00133711. [DOI] [PubMed] [Google Scholar]
- Maruyama K., Hartl D. L. Evolution of the transposable element mariner in Drosophila species. Genetics. 1991 Jun;128(2):319–329. doi: 10.1093/genetics/128.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M. The mariner transposable element is widespread in insects. Nature. 1993 Mar 18;362(6417):241–245. doi: 10.1038/362241a0. [DOI] [PubMed] [Google Scholar]
- Shattuck-Eidens D. M., Bell R. N., Neuhausen S. L., Helentjaris T. DNA sequence variation within maize and melon: observations from polymerase chain reaction amplification and direct sequencing. Genetics. 1990 Sep;126(1):207–217. doi: 10.1093/genetics/126.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M., Wessler S., Fedoroff N. Molecular identification and isolation of the Waxy locus in maize. Cell. 1983 Nov;35(1):225–233. doi: 10.1016/0092-8674(83)90225-8. [DOI] [PubMed] [Google Scholar]
- Smyth D. R., Kalitsis P., Joseph J. L., Sentry J. W. Plant retrotransposon from Lilium henryi is related to Ty3 of yeast and the gypsy group of Drosophila. Proc Natl Acad Sci U S A. 1989 Jul;86(13):5015–5019. doi: 10.1073/pnas.86.13.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWiel P. L., Voytas D. F., Wendel J. F. Copia-like retrotransposable element evolution in diploid and polyploid cotton (Gossypium L.). J Mol Evol. 1993 May;36(5):429–447. doi: 10.1007/BF02406720. [DOI] [PubMed] [Google Scholar]
- Varagona M. J., Purugganan M., Wessler S. R. Alternative splicing induced by insertion of retrotransposons into the maize waxy gene. Plant Cell. 1992 Jul;4(7):811–820. doi: 10.1105/tpc.4.7.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Voytas D. F., Cummings M. P., Koniczny A., Ausubel F. M., Rodermel S. R. copia-like retrotransposons are ubiquitous among plants. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7124–7128. doi: 10.1073/pnas.89.15.7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas D. F., Konieczny A., Cummings M. P., Ausubel F. M. The structure, distribution and evolution of the Ta1 retrotransposable element family of Arabidopsis thaliana. Genetics. 1990 Nov;126(3):713–721. doi: 10.1093/genetics/126.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler S. R., Varagona M. J. Molecular basis of mutations at the waxy locus of maize: correlation with the fine structure genetic map. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4177–4181. doi: 10.1073/pnas.82.12.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990 Oct;9(10):3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]



