Abstract
Despite the fact that ferrous myeloperoxidase (MPO) can bind both O2 and NO, its ability to bind CO has been questioned. UV/visible spectroscopy was used to confirm that CO induces small spectral shifts in ferrous MPO, and Fourier transform infrared difference spectroscopy showed definitively that these arose from formation of a heme ferrous–CO compound. Recombination rates after CO photolysis were monitored at 618 and 645 nm as a function of CO concentration and pH. At pH 6.3, kon and koff were 0.14 mM−1·s−1 and 0.23 s−1, respectively, yielding an unusually high KD of 1.6 mM. This affinity of MPO for CO is 10 times weaker than its affinity for O2. The observed rate constant for CO binding increased with increasing pH and was governed by a single protonatable group with a pKa of 7.8. Fourier transform infrared spectroscopy revealed two different conformations of bound CO with frequencies at 1927 and 1942 cm−1. Their recombination rate constants were identical, indicative of two forms of bound CO that are in rapid thermal equilibrium rather than two distinct protein populations with different binding sites. The ratio of bound states was pH-dependent (pKa ≈ 7.4) with the 1927 cm−1 form favored at high pH. Structural factors that account for the ligand-binding properties of MPO are identified by comparisons with published data on a range of other ligand-binding heme proteins, and support is given to the recent suggestion that the proximal His336 in MPO is in a true imidazolate state.
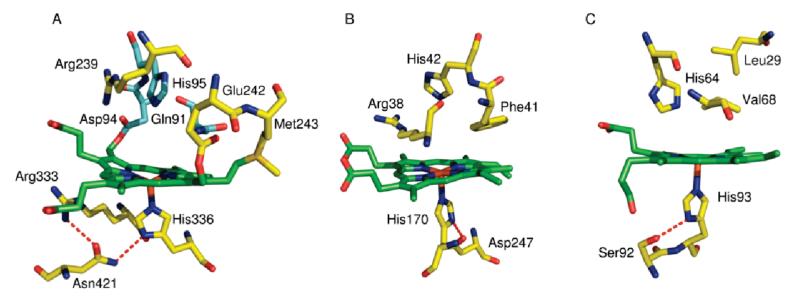
Myeloperoxidase (MPO;1 donor, hydrogen peroxide oxidoreductase, EC 1.11.1.7) is a heme protein found in large concentrations in azurophilic granules of myeloid leukocytes (1). The mature protein is a homodimer linked by a single disulfide bridge (2); each 70 kDa monomeric unit is made up of a light and heavy chain and contains a covalently attached heme (3). MPO plays a central role in host defense and has also been implicated in promoting tissue damage in numerous inflammatory diseases (4–6). MPO has several potential catalytic activities, and despite extensive investigations, its central role in the innate defense system and the exact mode of its antibacterial activity are still a matter of controversy (7, 8). Traditionally, the major role ascribed to MPO is the generation of the potent bleaching agent hypochlorous acid (HOCl) via H2O2-dependent two-electron oxidation of Cl− (9, 10). However, the high catalase activity of MPO (4) has been suggested to provide an additional function of MPO in protecting digestive enzymes from oxidative damage by removing H2O2 from the phagocytic vacuole (11). In MPO and the other mammalian peroxidases (lactoperoxidase (LPO), eosinophil peroxidase (EPO), and thyroid peroxidase) the heme is covalently linked to the protein by two ester bonds between the carboxyl groups of a glutamate (Glu242 in MPO) and an aspartate (Asp94) and the modified methyl groups of the heme (Figure 1A and refs (12–14)). In MPO, a third covalent link is formed between Met243 and the β-carbon of the vinyl group of pyrrole ring A. This positively charged sulfonium linkage causes the heme to assume a bow-shaped structure and is thought to be a major contributor to its unusual optical, redox, and substrate properties (15, 16). For example, the reduction potential at pH 7.0 (Em7) of the FeIII/FeII couple of most peroxidases ranges from −180 to −300 mV (17). In contrast, the Em7 of the FeIII/FeII couple of MPO is +5 mV (18), attributed in large part to the electron-withdrawing sulfonium linkage. This potential is closer to those of the oxygen carriers, hemoglobin (+150 mV (19)) and myoglobin (+46 mV (20)), which need a stable ferrous form under physiological conditions in order to facilitate reversible binding of O2.
Figure 1.
MPO (A), HRP (B), and human myoglobin (C) active sites. In each case the heme is shown in green, and hydrogen bonds are indicated by red dashes. Coordinates are from PDB files 3F9P, 1ATJ, and 4MBN, respectively; drawn using PyMOL.
As is typical for peroxidases, MPO can react with H2O2 to form a reactive intermediate, Compound I. This has a ferryl heme and an associated radical which, as in HRP (21–23), exists at least transiently as a π-cation radical form of the porphyrin ring (24). Further redox reactions and catalytic intermediates also resemble those in other peroxidases (24). Peroxidases also typically bind diatomic ligands such as O2, CO, and NO. Ferrous MPO binds O2 to form an oxyferrous complex, Compound III (25, 26), that is similar to oxymyoglobin. NO also binds to both ferric and ferrous forms of MPO and may act as a modulator of its catalytic activity (27). Despite the fact that MPO can bind both O2 and NO, its ability to bind CO has been less clear, even though CO binds to the structurally related LPO (28–31). Early reports showed that MPO binds CO weakly and induces a small blue shift in the UV/visible spectra (32), and CO-induced difference spectra of intact neutrophils also exhibited bands attributable to ferrous MPO–CO (33). However, more recently it has been proposed that CO binding to ferrous MPO is prevented altogether, either by a conformational change that narrows the heme pocket or by binding of a sixth ligand that prevents access to the heme (28). Static and transient UV/visible and Fourier transform infrared (FTIR) spectroscopies were used here to show that ferrous MPO does indeed bind CO, albeit with a very low affinity that is even lower than its affinity for O2. A possible structural basis for this unusual property of MPO and implications regarding its physiological functions are discussed by comparisons with other types of CO- and O2-binding heme proteins.
EXPERIMENTAL PROCEDURES
Chemicals were purchased from Sigma-Aldrich, and carbon monoxide was from BOC Ltd., U.K.
Isolation and Purification of MPO from Human Neutrophils
MPO was extracted from human leukocytes as described previously (34). Crude MPO was exchanged into equilibration buffer (500 mL, 25 mM sodium acetate, 0.2 M NaCl, pH 4.7), and initial purification was carried out by ion-exchange chromatography using a CM-Sepharose column (50 × 2 cm). The column was washed with 2 L of 25 mM sodium acetate and 0.5 M NaCl at pH 4.7, and MPO was eluted as a single dark green band with 25 mM sodium acetate and 1 M NaCl at pH 4.7. The eluant was concentrated and exchanged into 50 mM sodium phosphate at pH 7.0. The crude MPO was further purified by hydrophobic interaction chromatography using a phenyl-Sepharose 6 Fast Flow column (GE healthcare) (50 × 2 cm). MPO was eluted using an ammonium sulfate gradient (0.7–1.7 M), and fractions with a purity index (A430/A280) >0.8 were collected, concentrated, and exchanged into 50 mM sodium phosphate buffer at pH 7.3. Ferric MPO concentration was determined from its absorbance at 430 nm using ε = 91000 M−1·cm−1 per heme (34).
CO Photolysis/Recombination
UV/visible spectra were recorded with a single beam spectrometer constructed in-house and equipped with a stepped dispersive monochromator and photomultiplier detector. Ferrous MPO was prepared by addition of 1 mM sodium dithionite to ferric MPO in 50 mM sodium phosphate buffer at the required pH in a 1 cm quartz cuvette under a N2 atmosphere. The ferrous–CO adduct was formed by saturating the reduced sample with CO gas. Kinetics measurements were performed with 1.6 μM MPO in 50 mM sodium phosphate at appropriate pH values. Photolysis was achieved with a train of five flashes at 25 Hz from a xenon flashlamp (20 J/flash; 6 μs duration at half-maximum). The flash and photomultiplier were screened with BG39 (cutoff >580 nm) and RG610 (cutoff <610 nm) filters, respectively. Photolysis and subsequent recombination kinetics at room temperature were monitored over 8 s at 645 nm minus 618 nm, and 10 individual transients were averaged to improve the signal/noise ratio. Averaged data were fitted to a single exponential decay using a nonlinear least-squares method. For CO concentration dependency, the sample was equilibrated with CO that was premixed with N2 to give 20%, 40%, 50%, 80%, or 100% CO. Concentrations were calculated assuming that the solubility of CO in aqueous solution at 25 °C is 1 mM·atm−1. The pH dependency of the observed rate constant of CO reassociation, kobs, was determined in samples saturated with CO at pH values between 5.75 and 10.5.
FTIR Spectroscopy
Mid-IR spectra were recorded in transmission mode with a Bruker IFS/66S FTIR spectrophotometer fitted with a liquid nitrogen-cooled MCT-A detector at 4 cm−1 resolution, giving an accuracy of cited frequencies of ±1 cm−1. Five millimolar MPO was washed in 50 mM sodium phosphate at the required pH by repeated concentration and redilution cycles using ultrafiltration (Vivaspin, 10 kDa cutoff). Three microliters was then mixed with 1 μL of CO-saturated 0.5 M sodium dithionite in 1 M sodium phosphate at the same pH. The sample was placed on a CaF2 window and exposed to a stream of H2O-saturated CO for 1 min, before placing a second window on top and sealing with silicon vacuum grease. Optimal samples had an absorption peak at 1650 cm−1 (amide I + water) around 1.0 and a 1550 cm−1 amide II peak around 0.5. Actinic light was provided by a 250 W quartz–iodine lamp, filtered with glass, water, and a BG39 cutoff filter, and delivered to the sample via a light pipe. “Light minus dark” spectra were recorded at 277 K as follows: 100 interferograms were averaged to provide an initial dark baseline, after which the light was switched on and a sample spectrum recorded after 1 s. Finally, the light was switched off, and the recording was repeated after 3 s dark relaxation in order to provide an indication of relaxation rate and sample baseline drift. Light/dark cycles were repeated up to 2000 times and signal averaged to produce the data shown.
Time-resolved spectra were recorded in rapid-scan mode at 277 K. A prephotolysis dark baseline was recorded prior to each flash. Photodissociation of the ferrous MPO–CO compound was achieved with a train of five flashes from a xenon flashlamp as described above that was synchronized with initiation of recording of single interferograms at 4 cm−1 resolution over the next 28 s. This cycle was repeated 1000 times, and data were averaged before Fourier transformation to generate a 3D data block of absorbance versus frequency versus time. Difference spectra at specific times after CO photodissociation and kinetics at specific frequencies were extracted from the 3D data blocks with Bruker OPUS 6.5 software and fitted with monoexponential decay functions.
RESULTS
UV/Visible Analysis of CO Binding to Ferrous MPO
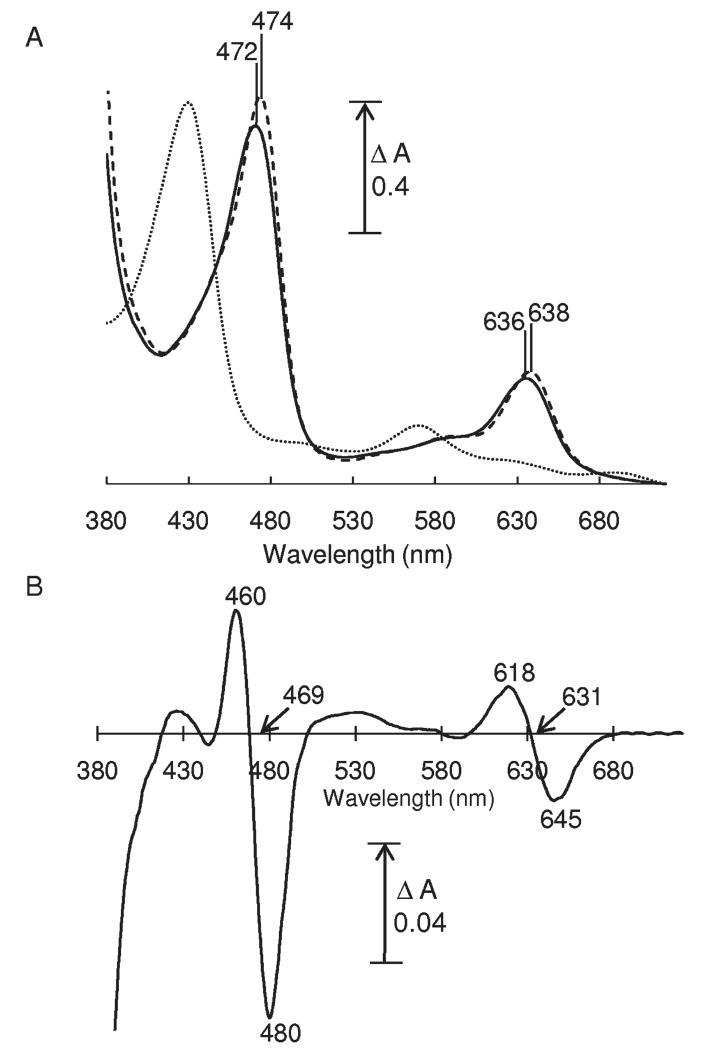
UV/visible absorption spectroscopy confirmed that CO addition to ferrous MPO induced small blue shifts (Figure 2A) of the Soret band from 474 to 472 nm and of the α-band from 638 to 636 nm. These shifts are clearer in the ferrous–CO minus ferrous difference spectrum (Figure 2B), which displayed a peak and trough in the Soret region at 460 and 480 nm (isosbestic at 469 nm) and in the visible region at 618 and 645 nm (isosbestic at 631 nm). Similar blue shifts of Soret and visible bands occur when CO binds to other ferrous heme proteins (35), though the magnitudes of the changes in MPO are much smaller.
Figure 2.
UV/visible spectra of MPO. (A) Absolute spectra of 12.7 μM MPO in 50 mM sodium phosphate buffer at pH 6.8: ferric (dotted line), ferrous (dashed line), and ferrous–CO with saturating CO (solid line). (B) Ferrous–CO minus ferrous difference spectrum.
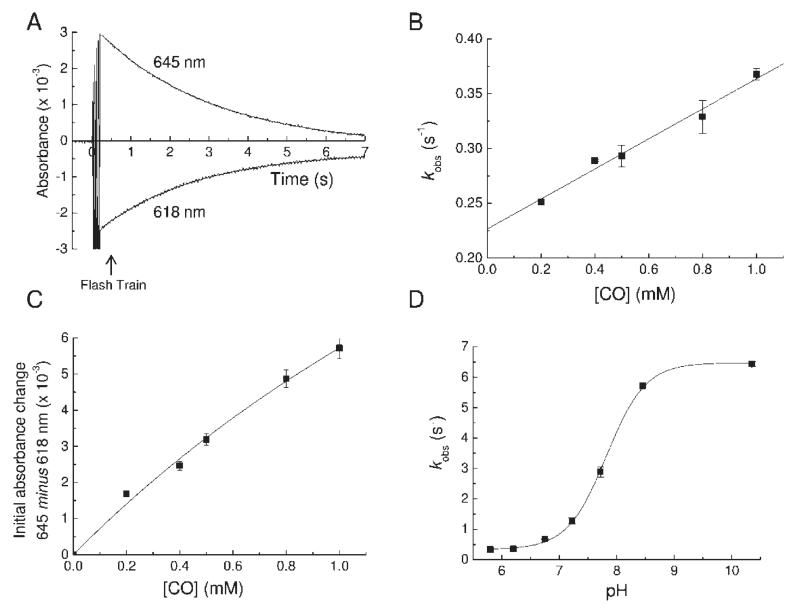
Flash photolysis was used to dissociate bound CO from ferrous MPO, and the kinetics of recombination at room temperature were monitored as loss of ferrous MPO at 645 nm or regeneration of ferrous MPO–CO at 618 nm (Figure 3A). Transients were fitted to single exponentials to obtain pseudo-first-order rate constants of recombination (kobs). A plot of kobs versus CO concentration at pH 6.3 (Figure 3B) was linear with a positive y-intercept, indicating that CO binding is reversible and can be modeled by a simple one-step binding mechanism. The slope gives a second-order binding rate constant of 0.14 ± 0.01 mM−1·s−1 and the y-intercept a dissociation rate constant of 0.23 ± 0.01 s−1, yielding a dissociation constant, KD (koff/kon), of 1.6 ± 0.2 mM at pH 6.3. The extent of formation of the CO compound, obtained from the initial absorbance change at 645 nm minus 618 nm with multiple flashes, was CO concentration dependent. A plot of the initial absorbance change versus CO concentration at pH 6.3 (Figure 3C) could be fitted to a hyperbolic function to yield an approximate KD of 3.3 ± 1.4 mM, consistent with the more accurate, kinetically determined value. The KD of 1.6 mM means that the extent of formation of the ferrous–CO compound is only ~30% at 1 atm (1 mM) of CO. By extrapolation, an extinction coefficient of 8.9 mM−1·cm−1 was calculated for full formation of the ferrous–CO minus ferrous difference spectrum at 618 nm minus 645 nm, a value similar to that of the equivalent shift in other peroxidases (35). The binding rate increased with increasing pH values, as has also been observed for CO binding to LPO (31). A plot of kobs versus pH at saturating CO (Figure 3D) could be fitted with a Henderson–Hasselbalch equation for a single acid–base group with a pKa of 7.8.
Figure 3.
Photolysis and recombination of ferrous MPO–CO. For all experiments the MPO concentration was 1.6 μM. (A) Typical transient absorption changes at 618 and 645 nm following flash photolysis of the ferrous MPO–CO complex at pH 6.3. The transient decay kinetics follows a single exponential with a pseudo-first-order rate constant (kobs). (B) Plot of kobs as a function of CO concentration at pH 6.3. The slope of a linear fit to the data gave the true second-order rate constant for CO binding (kon), and its y-axis intercept gave the dissociation rate constant (koff). (C) Plot of the initial absorbance change at 645 nm minus 618 nm after a train of flashes as a function of CO concentration at pH 6.3. (D) pH dependency of kobs. The points are overlaid with a Henderson–Hasselbalch curve for an acid/base group with a pK of 7.8.
FTIR Analysis of CO Binding to Ferrous MPO
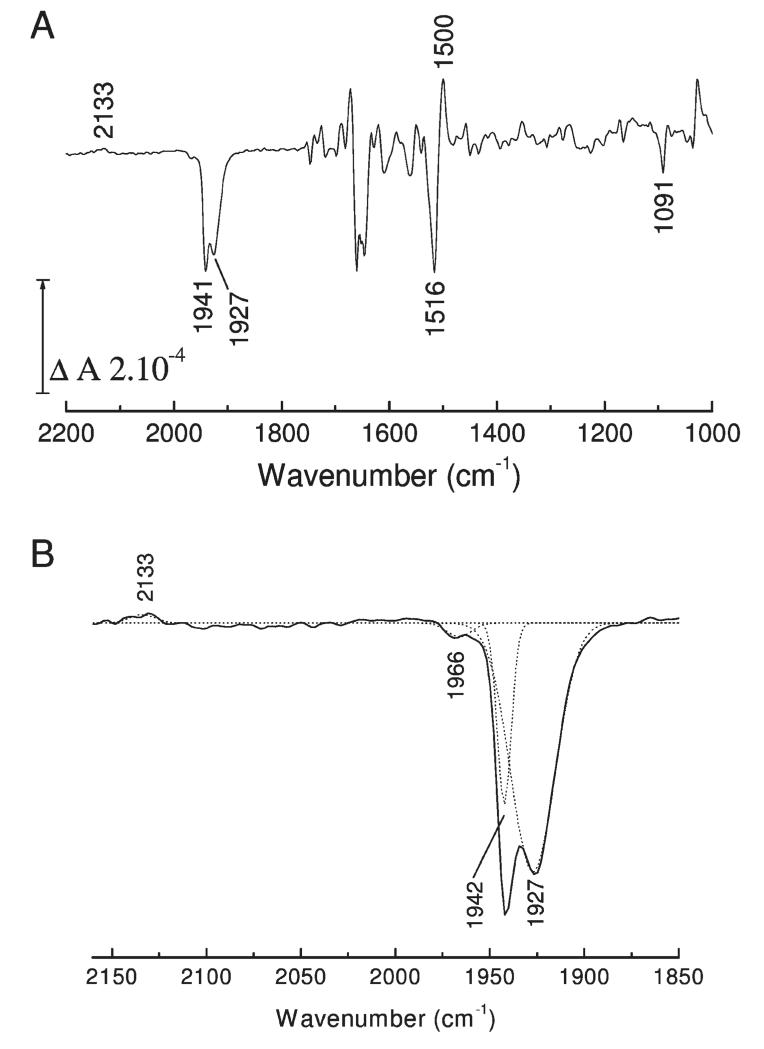
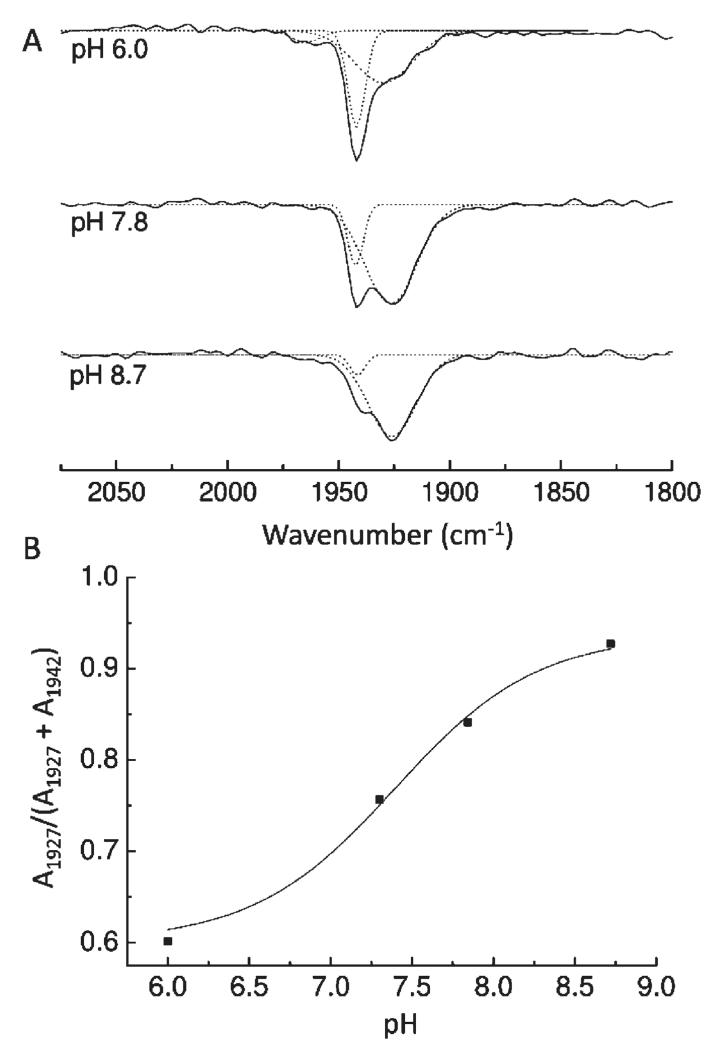
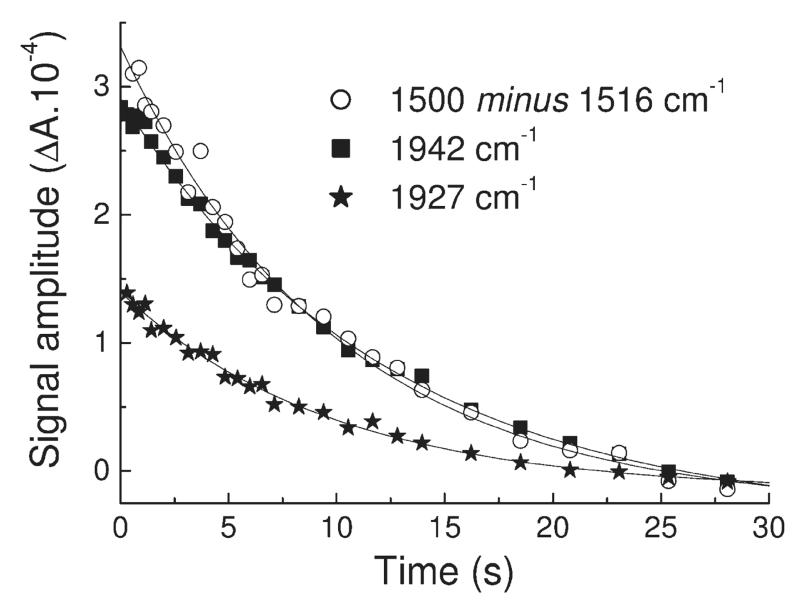
Photolability of the ferrous heme–CO compound allowed ferrous minus ferrous–CO FTIR difference spectra to be generated at high signal/noise ratio by averaging cycles of light minus dark difference spectra. Typical data for MPO are presented in Figure 4. Changes in heme and protein result in multiple overlapping bands below 1800 cm−1. Loss of heme-bound CO appears as a narrow (fwhm 7 cm−1) trough at 1942 cm−1 together with a much broader one (fwhm 22 cm−1) at 1927 cm−1. A third very minor component at 1966 cm−1 is present at acidic pH. A weak positive signal at 2133 cm−1 is also present. A similar band has also been observed on CO photolysis from myoglobin (36, 37), NO synthase (38), and HRP (39) and may represent a protein-bound form of CO that occurs transiently during photolysis and recombination (40, 41) or a weak affinity surface site that reacts to the increased solution CO concentration after photolysis (38). The principal troughs indicate two major binding modes of CO in the MPO active site, as is the case for CO bound to LPO (30, 42). The narrowness of the higher frequency band suggests a conformationally restricted site whereas the broadness and lower frequency of the 1927 cm−1 band suggest a more flexible orientation of CO in stronger H-bonding interaction and/or a more positive pocket polarity (42, 43). Multiple conformations of bound CO are commonly seen in other heme proteins, for example, in myoglobin (44), HRP (45), CcP (46), and NO synthase (38). The relative amounts of these two forms in MPO is pH-dependent (Figure 5A); the 1927 cm−1 form is favored at higher pH and is controlled by an acid–base group with a pKa of roughly 7.4 when CO is bound (Figure 5B). The kinetics of recombination of the two bands of bound CO, determined by rapid-scan FTIR measurements (Figure 6), were the same (as is also the case for the two CO conformers in HRP (39)), suggesting strongly that they arise from a thermally distributed mixture of states in the whole MPO population rather than from heterogeneous types of ligand-binding pockets. The features below 1800 cm−1 arise from vibrational changes of the polypeptide backbone, specific amino acids, and heme. These relaxed with the same kinetics as the bands of bound CO, demonstrating that any major protein conformational changes must occur synchronously with rebinding of CO (in Figure 6, this is illustrated with a plot of the kinetics at 1500 minus 1516 cm−1, a change that is likely to arise from both heme and amide II band shifts). In relation to FTIR spectra of CO photolysis in other heme proteins, one general observation is that these changes are large in relation to the size of the troughs from the bound CO. This suggests that heme/protein perturbations caused by CO ligation are more extensive in MPO. Assignments can be made based on extensive studies of other heme proteins and related models (47–51) and from a prior IR study of the heme ester linkages in MPO (12). For example, features in the 1750–1720 cm−1 region, possibly together with at least part of those at 1160 cm−1, arise from the covalent ester linkages between heme and protein (12). The large changes in the 1700–1600 and 1570–1500 cm−1 ranges are generally amide I and II shifts, respectively, due to multiple small changes of the polypeptide backbone. In HRP, the 1700–1600 cm−1 region also contains bands from the distal arginine (39), and it is possible that this region of MPO also contains changes from distal pocket Arg239 and/or Gln91. Numerous small porphyrin/vinyl modes are also expected in predictable ranges throughout the 1700–1000 cm−1 region. Their details are beyond the scope of the present study, but identification of likely bands and their assignments to specific normal modes can be inferred from extensive published data on model hemes. Bands in the 1100 cm−1 region in other histidine-ligated heme proteins have been assigned to the ring C5N1 bond of the proximal histidine (49). In both myoglobin and HRP, its frequency is above 1100 cm−1. This is consistent with the imidazole state of their proximal histidines in both cases, despite the fact that both Raman and NMR studies have shown that sharing of the Nτ proton through H-bonding with Asp247 in HRP (Figure 1) can impart a strong imidazolate “character” (52, 53). In MPO, however, the 1091 cm−1 frequency is more consistent with a true imidazolate form of the proximal histidine (His336) (54, 55). In the most recent published structure of MPO (56) Asn421 is orientated with its carbonyl group H-bonded to Arg333, leaving its amine group facing the Nτ hydrogen atom of proximal His336 (Figure 1A). If the Nτ hydrogen atom of proximal His336 were protonated, this would create an unfavorable interaction between them. As a result, it was proposed that His336 is in its imidazolate state, thus enabling it to act as an H-bond acceptor with the amine group of Asn421. Of LPO structures in the PDB database (57–61), 2NQX, 2O86, 2IKC, and 3I6N have the proximal Asn modeled in this same orientation whereas the majority shows the carbonyl group orientated toward the proximal histidine, which would require the neutral imidazole form. Hence, it is not clear at present whether an imidazolate proximal histidine is a general structural feature of the mammalian peroxidases. These points are relevant to the discussion below of the factors controlling CO binding affinity.
Figure 4.
FTIR difference spectrum of CO photolysis. (A) Ferrous minus ferrous–CO spectra of MPO were recorded at pH 7.3 as described in Experimental Procedures. CO and protein modes discussed in the text are labeled. (B) Enlargement of the 2160–1850 cm−1 spectral region. CO bands were simulated with four Gaussian functions centered at 2133, 1966 (2%), 1942 (18%), and 1927 (80%) cm−1 with fwhm of 14 ± 4, 13 ± 2, 7, and 22 cm−1, respectively.
Figure 5.
pH dependency of the IR bands of ferrous MPO–CO. (A) CO photolysis difference spectra were recorded at pH 6.0, 7.8, and 8.7 as for Figure 4. CO bands were simulated with two or three Gaussian functions as necessary. (B) pH effect on the proportion of the 1927 cm−1 form, expressed as a fraction of the total integrated areas of the two main 1927 and 1942 cm−1 bands. Data are fitted with a Henderson–Hasselbalch equation for a single acid–base transition and gave a pKa of 7.4 ± 0.2.
Figure 6.
Time-resolved rapid-scan FTIR transients during CO recombination with ferrous MPO at pH 7.8 and 277 K. Plots of absorbance versus time were extracted from the 3D data set of spectra after CO photolysis at 1942 cm−1 (squares) and 1927 cm−1 (stars) for the two main CO bands and at 1500 cm−1 minus 1516 cm−1 (open circles) for a typical example of band relaxation within the protein/heme region. Monoexponential fits of kobs ≈ 0.10 s−1 are overlaid.
DISCUSSION
The UV/visible and FTIR spectra provide definitive identification of the CO compound of ferrous MPO. Uncertainty over this issue most probably has arisen because only partial saturation can be achieved with CO at atmospheric pressure due to an unusually low affinity (high KD). The rates of CO binding, together with the ratio of bound conformers, are governed by a protonatable group with a pK around 7.8 in the ferrous form and roughly the same in the ferrous–CO form. Such behavior is seen in other peroxidases such as HRP (62) and CcP (46) where the pK has been attributed to the distal histidine whose Nτ can provide an H-bond to the CO when it is in its protonated imidazolium form. The higher pH form of MPO reacts faster with CO (Figure 3), as is also the case in HRP (62), CcP (46), and LPO (31). By analogy, the 1942 cm−1 form involves H-bonding to distal His95 (whose pH is 7.8 in ferrous MPO) whereas at high pH the CO interacts with nearby residues Arg239 and Gln91 and/or water molecules to give the broader (less conformationally restricted) 1927 cm−1 form. Hence the binding of CO to MPO in two pH-dependent conformations resembles CO binding to other peroxidases. However, in HRP and CcP the lower pH form of bound CO has a lower frequency, consistent with stronger H-bonding to the bound CO. In MPO (and also in LPO (29, 42)), in contrast, the lower pH form has a higher frequency despite the fact that the distal His is similarly providing extra H-bonding. The structural basis for this unexpected direction of frequency shift in MPO and LPO is not clear. Protonation of the distal His95 to form its charged, imidazolium state might reasonably be expected to induce an increased polarity of the distal pocket, an effect that is also expected to increase the CO stretch frequency. Hence, additional structural changes must occur within the distal pocket that more than counteract any frequency-lowering H-bonding and/or polarity changes. Some support for more extensive structural changes in the distal heme pocket of MPO comes from the larger CO-induced IR changes in heme and protein that occur in MPO compared to other peroxidases (Figure 4, 1800–1000 cm−1 region); the nature of such changes will require further IR or crystallographic studies.
What is particularly distinct in MPO in comparison to other peroxidases including LPO is the weakness of binding of CO. This arises both from a lower bimolecular association rate constant and a faster dissociation rate constant (Table 1). Weak binding of diatomic ligands to ferrous heme appears to be a property of the mammalian peroxidases more generally. For example, the KDs of LPO and EPO for CO and O2 are 500–300-fold higher than those of myoglobin, as is the KD of MPO for O2. However, the KD of MPO for CO is around 50000-fold higher and almost 100-fold higher than that of EPO and LPO. As a result, and in contrast to most other heme proteins including LPO, MPO has a higher KD for CO than for O2. This discrimination between ligands is often defined as the O2/CO partition coefficient, M, and is equivalent to the dissociation constant ratio (KO2/KCO). The M value for sterically unhindered model heme compounds is ~4000 (63) and ~25 for myoglobin (64) but is only 0.1 for MPO (Table 1).
Table 1.
CO, O2, and NO Binding to Ferrous Heme Complexesa
| CO (FeII) | O2 (FeII) | NO (FeII) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||
| k on | k off | K D | k on | k off | K D | M | k on | k off | K D | E m7 | ref | |
| chelated protoheme | 11000 | 25 × 10−3 | 2.3 × 10−3 | 62000 | 4200 | 68 | 29500 | 180 | 2.9 × 10−4 | 1.6 × 10−6 | −80 | 83 – 85 |
| MPO | 0.14 | 226 × 10−3 | 1650 | 11 | 1.92 | 173 | 0.10 | 100 | 4.6 | 46 | 5 | 25, 2828 |
| EPO | 0.65 | 12 × 10−3 | 19 | 37 | nd | nd | 1300 | 24 | 19 | nd | 28 | |
| LPO | 0.59 | 1.3 × 10−3 | 2.2 | 38 | 11.2 | 295 | 134 | 140 | 0.25 | 1.8 | −191 | 28, 86, 87 |
| 1.4 | 29 × 10−3 | 21 | ||||||||||
| HRP | 3 | 0.1 × 10−3 | 33 × 10−3 | 53 | 0.1 | 1.9 | 58 | nd | nd | nd | −278 | 62, 88, 89 |
| CcP | 2 | 9.2 × 10−3 | 4.6 | 45 | 0.22 | nd | nd | −194 | 90 – 92 | |||
| Mb | 990 | 36 × 10−3 | 37 × 10−3 | 16000 | 15.6 | 0.98 | 26 | 17000 | 1.2 × 10−4 | 7 × 10−6 | 46 | 92 – 95 |
| trHbO | 14 | 4 × 10−3 | 0.29 | 110 | 1.4 × 10−3 | 13 × 10−3 | 0.05 | 180 | nd | nd | nd | 73 |
| alHbO | 190 | 18 × 10−3 | 95 × 10−3 | 1500 | 4 × 10−3 | 2.7 × 10−3 | 0.03 | 5600 | nd | nd | nd | 74 |
| CcO | 70 | 23 × 10−3 | 0.30 | 100000 | 45000 | 450 | 1500 | 100000 | 0.13 | 0.013 | 350 | 95 – 97 |
| chloroperoxidase | 72 | 2.2 | 31 | 550 | 12 | 22 | 0.71 | nd | nd | nd | −140 | 92, 98, 99 |
| cytochrome P450cam | 37 | 0.14 | 3.8 | 770 | 1.1 | 1.4 | 0.37 | −170 | 98 – 100 | |||
Data cited are the association (kon, ×103 M−1 s−1) and dissociation (koff, s−1) rate constants, the corresponding dissociation constant (KD, μM), the partition coefficient (M) for discrimination of CO versus O2 (from ratio of KD values), and the FeIII/FeII midpoint potential measured at pH 7.0 (Em7, mV); nd, not determined.
The physical factors that control ligand binding to hemes have been studied in detail (65–67). Steric factors can provide discrimination (68) since optimal binding requires a linear configuration for Fe–CO but a bent one for Fe–O2. This arises because the two extra electrons of dioxygen are antibonding in the linear conformation but are a nonbonding lone pair when Fe–O2 is bent (69). However, calculations show that the bonding orbitals of CO can distort to some extent to retain optimal binding even when the Fe–CO atomic coordinates are bent (66). Also, studies of site-directed mutants of myoglobin (70, 71) have shown that steric effects in this protein are small, and the lower M value (~25) compared to free heme (~4000) arises predominantly from stronger electrostatic and H-bonding interactions between the more polar Fe–O2 species and distal pocket residues (72). Examples of heme proteins in which oxygen stabilization occurs to such an extent that their M values are below 1 include the truncated hemoglobin, trHbO, from Mycobacterium tuberculosis (73) and a hemoglobin, alHbO, from Ascaris lumbricoides (74). In both cases, the heme-bound O2 is stabilized particularly strongly by hydrogen bonds from a distal tyrosine and a second residue. However, the same types of distal pocket factors cannot provide a full explanation of ligand-binding properties of the mammalian peroxidases because in trHbO and alHbO (Table 1) both ligands are strongly bound (KDs ≪ μM), whereas in MPO both CO and oxygen are particularly weakly bound (KDs > 100 μM).
The structures of the distal heme pockets of ferric MPO (56) and LPO (57) do not reveal major steric constraints. However, both resonance Raman spectroscopy (75) and X-ray crystallographic data (76) of the cyanide-ligated form of ferric MPO show an FeCN angle of ≈155°, compared to 170° for HRP, which allows the cyanide to form H-bonds with two of the highly conserved active site water molecules and the distal histidine. Such H-bonding to a similarly bent Fe–O2 could provide preferential stabilization of oxygen in comparison to CO. However, there are no obvious further constraints in the distal pockets that could account for the acute overall weakening of binding of both ligands. In addition, the structures of the distal heme pockets of ferric MPO (56) and LPO (57) are very similar, and yet the M value of LPO (~135) is more than 1000-fold greater than that of MPO (Table 1).
It is possible that distal pocket structural factors occur in the ferrous forms of MPO and LPO, whose structures have not yet been determined, which could result in severe weakening of ferrous–ligand binding. Table 2 shows the association rate constants (kon) for binding of NO and cyanide to ferric MPO and several other representative ferric heme proteins. The second-order rate constant for NO binding to ferric MPO is 10-fold greater than to ferrous MPO. It has been proposed that this is due to a change in the distal heme pocket upon reduction of MPO (28) although the nature of the change remains to be determined. Most importantly, rate constants for the binding of both NO and cyanide to ferric MPO are comparable to those of other ferric heme proteins. By contrast with the uniformly lower rate constants for binding of ligands to ferrous MPO, this suggests that reduction of the heme in MPO does introduce constraints to ligand binding, resulting in lowered ligand affinities. Again, however, it is difficult to understand how distal factors alone could cause the extreme M value of MPO.
Table 2.
NO and Cyanide Binding to Ferric Heme Proteinsa
| NO (FeIII) |
CN (FeIII) |
||||||
|---|---|---|---|---|---|---|---|
| k on | k off | K D | k on | k off | K D | ref | |
| MPO | 1070 | 11 | 10 | 1300 | 0.56 | 0.43 | 28, 101 |
| LPO | 3000 | nd | nd | 1100 | 20 | 18 | 28, 102 |
| EPO | 1350 | 65 | 48 | nd | nd | nd | 28 |
| HRP | 190 | 75 | 400 | 98 | 0.28 | 2.9 | 103, 104 |
| CcP | nd | nd | nd | 110 | 0.9 | 8.2 | 105 |
| Mb | 190 | 14 | 74 | 0.17 | 3.8 × 10−4 | 2.2 | 106, 107 |
Data cited are the association (kon, ×103 M−1 s−1) and dissociation (koff, s−1) rate constants, together with the corresponding dissociation constant (KD, μM); nd, not determined.
Additional factors that can affect the metal–ligand bond are the heme substituents, heme ring distortions, and the nature of the proximal heme ligand. Ring substituent effects occur through their electron-withdrawing/donating effects of the heme potential, which in turn affects the degree of electron sharing with the ligand. Accordingly, binding of oxygen is far more affected than is binding of CO. Due to its sulfonium linkage, the heme potential of MPO is higher than other peroxidases but similar to myoglobin (18), and so substituent effects, although unusual in MPO, are unlikely to be a major cause of its low M value. Crystallographic data also reveal heme ring plane distortions in MPO that are caused by the covalent heme linkages, though their effects on ligand binding are difficult to predict.
Particularly pertinent to MPO are considerations of the known strong trans effect of proximal ligands (69). The metal bond consists of σ-bonding together with any back-bonding from metal dπ orbitals to ligand π* orbitals, which strengthens the Fe–ligand bond. Histidine or imidazole has a positive trans effect when bound opposite CO and O2; i.e., the two bonds trans to each other are both strengthened and CO dissociation is slowed, while NO appears to have a negative trans effect (77). The extent of this effect is dependent on the proximal histidine/heme geometry. Indeed, in the classical R/T transition of hemoglobin, strain on the proximal histidine in the T state causes the iron to move out of the heme plane and resulting in an 800-fold decrease in oxygen affinity (78, 79). In leghemoglobin, it has been suggested that histidine in the “eclipsed” orientation (i.e., over the heme nitrogens) exacerbates this effect and that rotation by 45° will minimize it (80). In MPO, however, the proximal histidine is already in an orientation with respect to the heme nitrogens that is optimal for ligand binding, and so this cannot be a weakening factor. However, stronger donor ligands such as thiolate or imidazolate also lower the Fe–CO bond strength, an effect arising from competition between the donor σ-orbitals of CO and the proximal ligand for the available ferrous iron dz2 acceptor orbital (66). This can be seen, for example, in the low affinities for CO of thiolate-ligated P450 and chloroperoxidase (Table 1). Recent crystallographic data on MPO (56) indicate that the proximal His336 must be in its imidazolate form because of H-bond donation from the NH of Asn421 (whose carbonyl is an H-bond acceptor from Arg333). The band at 1091 cm−1 in Figure 4 arises primarily from the C1N5 bond of the proximal histidine. The intensity of this normal mode band increases when the histidine is metal-ligated (81). Its frequency is significantly lower than that of the proximal histidine of either myoglobin or HRP. Studies of protoporphyrin IX imidazole compounds have shown that its frequency is lower in the imidazolate form (49), and DFT calculations of model zinc imidazole compounds predict the same shift (54). Hence, its lower frequency in MPO could be consistent with a true imidazolate state of the proximal histidine as has been suggested from crystallographic data. It is also evident that the iron is 0.2 Å out of the heme plane in the ferric MPO structure (56). An imidazolate form of the proximal histidine could well provide a major factor for this displacement and for the low binding constants for CO and O2. It might be noted that the proximal histidine of HRP can also have strong imidazolate character through H-bonding with Asp247 (Figure 1B). However, the presence of the proton means that, in contrast to MPO, ligand binding can still be stabilized by a ligand-induced shift to greater imidazole character.
This work highlights the low affinities for binding of diatomic ligands by ferrous MPO, coupled to an extremely low M value that particularly disfavors CO ligation. A variety of causative factors can be identified. Those on the distal side include H-bonding and electrostatic factors that favor oxygen over CO but may also include as yet uncharacterized steric constraints in the ferrous protein that disfavor both ligands. On the proximal side, ligation by imidazolate histidine is likely to provide an additional major factor that disfavors binding of both CO and O2. These factors combine to give the unusual combination of low ligand affinities and ligand-discriminating M value displayed by MPO. They allow ferric MPO to react rapidly with superoxide to form an oxyferrous compound III that is relatively stable in comparison to other peroxidases. Compound III has been shown to accumulate during the respiratory superoxide burst within neutrophil vacuoles (82). This might be an intermediate in a protective catalatic pathway but will also tend to slowly rerelease oxygen and/or superoxide and hence prolong the respiratory burst response. In addition, any ferrous MPO formed when it dissociates from Compound III will be particularly insensitive to poisoning by any CO that might arise during degradation of bacterial cells. More direct in vivo spectroscopic studies are needed to establish whether these could add to the physiological catalatic and chloride-oxidizing roles of MPO that have already been described.
ACKNOWLEDGMENT
We are very grateful to Dr. Christa Jakopitsch for preliminary experiments and useful discussions, Mr. Santiago Garcia for specialist electronic support, and Mr. Thomas Warelow for technical assistance.
Footnotes
This work was funded by an MRC grant (G0700050). A.M. is supported by a BBSRC research grant (BB/H000097/1).
Abbreviations: (ATR)-FTIR spectroscopy, (attenuated total reflection)-Fourier transform infrared spectroscopy; EPO, eosinophil peroxidase; fwhm, full width at half-maximum; LPO, lactoperoxidase; MPO, myeloperoxidase.
REFERENCES
- 1.Schultz J, Kaminker K. Myeloperoxidase of the leukocyte of normal human blood. I. Content and localization. Arch. Biochem. Biophys. 1962;96:465–467. doi: 10.1016/0003-9861(62)90321-1. [DOI] [PubMed] [Google Scholar]
- 2.Fiedler TJ, Davey CA, Fenna RE. X-ray crystal structure and characterization of halide-binding sites of human myeloperoxidase at 1.8 Å resolution. J. Biol. Chem. 2000;275:11964–11971. doi: 10.1074/jbc.275.16.11964. [DOI] [PubMed] [Google Scholar]
- 3.Taylor KL, Strobel F, Yue KT, Ram P, Pohl J, Woods AS, Kinkade JM. Isolation and identification of a protoheme-IX derivative released during autolytic cleavage of human myeloperoxidase. Arch. Biochem. Biophys. 1995;316:635–642. doi: 10.1006/abbi.1995.1083. [DOI] [PubMed] [Google Scholar]
- 4.Kettle AJ, Winterbourn CC. A kinetic analysis of the catalase activity of myeloperoxidase. Biochemistry. 2001;40:10204–10212. doi: 10.1021/bi010940b. [DOI] [PubMed] [Google Scholar]
- 5.Edwards SW, Hallett MB. Seeing the wood for the trees: the forgotten role of neutrophils in rheumatoid arthritis. Immunol. Today. 1997;18:320–324. doi: 10.1016/s0167-5699(97)01087-6. [DOI] [PubMed] [Google Scholar]
- 6.Buss IH, Darlow BA, Winterbourn CC. Elevated protein carbonyls and lipid peroxidation products correlating with myeloperoxidase in tracheal aspirates from premature infants. Pediatr. Res. 2000;47:640–645. doi: 10.1203/00006450-200005000-00014. [DOI] [PubMed] [Google Scholar]
- 7.Nauseef WM. How human neutrophils kill and degrade microbes: an integrated view. Immunol. Rev. 2007;219:88–102. doi: 10.1111/j.1600-065X.2007.00550.x. [DOI] [PubMed] [Google Scholar]
- 8.Ghibaudi E, Laurenti E. Unraveling the catalytic mechanism of lactoperoxidase and myeloperoxidase. Eur. J. Biochem. 2003;270:4403–4412. doi: 10.1046/j.1432-1033.2003.03849.x. [DOI] [PubMed] [Google Scholar]
- 9.Harrison JE, Schultz J. Studies on chlorinating activity of myeloperoxidase. J. Biol. Chem. 1976;251:1371–1374. [PubMed] [Google Scholar]
- 10.Klebanoff SJ. Myeloperoxidase: friend and foe. J. Leukocyte Biol. 2005;77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- 11.Cross AR, Segal AW. The NADPH oxidase of professional phagocytes—prototype of the NOX electron transport chain systems. Biochim. Biophys. Acta. 2004;1657:1–22. doi: 10.1016/j.bbabio.2004.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kooter IM, Pierik AJ, Merkx M, Averill BA, Moguilevsky N, Bollen A, Wever R. Difference Fourier transform infrared evidence for ester bounds linking the heme group in myeloperoxidase, lactoperoxidase, and eosinophil peroxidase. J. Am. Chem. Soc. 1997;119:11542–11543. [Google Scholar]
- 13.Suriano G, Watanabe S, Ghibaudi EM, Bollen A, Ferrari RP, Moguilevsky N. Glu375Gln and Asp225Val mutants: about the nature of the covalent linkages between heme group and apo-protein in bovine lactoperoxidase. Bioorg. Med. Chem. Lett. 2001;11:2827–2831. doi: 10.1016/s0960-894x(01)00533-9. [DOI] [PubMed] [Google Scholar]
- 14.Oxvig C, Thomsen AR, Overgaard MT, Sorensen ES, Hojrup P, Bjerrum MJ, Gleich GJ, Sottrup-Jensen L. Biochemical evidence for heme linkage through esters with Asp-93 and Glu-241 in human eosinophil peroxidase. The ester with Asp-93 is only partially formed in vivo. J. Biol. Chem. 1999;274:16953–16958. doi: 10.1074/jbc.274.24.16953. [DOI] [PubMed] [Google Scholar]
- 15.Newton N, Morell DB, Clarke L, Clezy PS. Haem prosthetic groups of some animal peroxidases. 2. Myeloperoxidase. Biochim. Biophys. Acta. 1965;96:476–486. doi: 10.1016/0005-2787(65)90564-2. [DOI] [PubMed] [Google Scholar]
- 16.Wever R, Plat H. Spectral properties of myeloperoxidase and its ligand complexes. Biochim. Biophys. Acta. 1981;661:235–239. doi: 10.1016/0005-2744(81)90009-7. [DOI] [PubMed] [Google Scholar]
- 17.Dunford HB. Heme Peroxidases. Wiley-VCH; New York: 1999. [Google Scholar]
- 18.Battistuzzi G, Bellei M, Zederbauer M, Furtmuller PG, Sola M, Obinger C. Redox thermodynamics of the Fe(III)/Fe(II) couple of human myeloperoxidase in its high-spin and low-spin forms. Biochemistry. 2006;45:12750–12755. doi: 10.1021/bi061647k. [DOI] [PubMed] [Google Scholar]
- 19.Antonini E, Brunori M, Rossifanelli A, Wyman J, Taylor JF, Caputo A. Studies on oxidation-reduction potentials of heme proteins. I. Human hemoglobin. J. Biol. Chem. 1964;239:907–912. [PubMed] [Google Scholar]
- 20.Varadarajan R, Zewert TE, Gray HB, Boxer SG. Effects of buried ionizable amino acids on the reduction potention of recombinant myoglobin. Science. 1989;243:69–72. doi: 10.1126/science.2563171. [DOI] [PubMed] [Google Scholar]
- 21.Schulz CE, Devaney PW, Winkler H, Debrunner PG, Doan N, Chiang R, Rutter R, Hager LP. Horseradish peroxidase compound I: evidence for spin coupling between the heme iron and a “free” radical. FEBS Lett. 1979;103:102–105. doi: 10.1016/0014-5793(79)81259-4. [DOI] [PubMed] [Google Scholar]
- 22.Roberts JE, Hoffman BM, Rutter R, Hager LP. Electron-nuclear double resonance of horseradish peroxidase compound I. J. Biol. Chem. 1981;256:2118–2121. [PubMed] [Google Scholar]
- 23.Rutter R, Valentine M, Hendrich MP, Hager LP, Debrunner PG. Chemical nature of the porphyrin π cation radical in horseradish peroxidase compound I. Biochemistry. 1983;22:4769–4774. doi: 10.1021/bi00289a024. [DOI] [PubMed] [Google Scholar]
- 24.Furtmüller PG, Zederbauer M, Jantschko W, Helm J, Bogner M, Jakopitsch C, Obinger C. Active site structure and catalytic mechanisms of human peroxidases. Arch. Biochem. Biophys. 2006;445:199–213. doi: 10.1016/j.abb.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 25.Jantschko W, Furtmuller PG, Zederbauer M, Jakopitsch C, Obinger C. Kinetics of oxygen binding to ferrous myeloperoxidase. Arch. Biochem. Biophys. 2004;426:91–97. doi: 10.1016/j.abb.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 26.Abu-Soud HM, Raushel FM, Hazen SL. A novel multistep mechanism for oxygen binding to ferrous hemoproteins: rapid kinetic analysis of ferrous-dioxy myeloperoxidase (compound III) formation. Biochemistry. 2004;43:11589–11595. doi: 10.1021/bi049541h. [DOI] [PubMed] [Google Scholar]
- 27.Abu-Soud HM, Hazen SL. Nitric oxide modulates the catalytic activity of myeloperoxidase. J. Biol. Chem. 2000;275:5425–5430. doi: 10.1074/jbc.275.8.5425. [DOI] [PubMed] [Google Scholar]
- 28.Abu-Soud HM, Hazen SL. Interrogation of heme pocket environment of mammalian peroxidases with diatomic ligands. Biochemistry. 2001;40:10747–10755. doi: 10.1021/bi010478v. [DOI] [PubMed] [Google Scholar]
- 29.Smith ML, Paul J, Ohlsson PI, Paul KG. Correlations between reduction potential, CO stretch frequency, and CO half-bandwidth in hemoproteins. Biochemistry. 2010;23:6776–6785. [Google Scholar]
- 30.Hu S, Treat RW, Kincaid JR. Distinct heme active-site structure in lactoperoxidase revealed by resonance Raman spectroscopy. Biochemistry. 1993;32:10125–10130. doi: 10.1021/bi00089a031. [DOI] [PubMed] [Google Scholar]
- 31.Ciaccio C, De SG, Marini S, Sinibaldi F, Santucci R, Arcovito A, Bellelli A, Ghibaudi E, Ferrari RP, Coletta M. Proton linkage for CO binding and redox properties of bovine lactoperoxidase. Biophys. J. 2004;86:448–454. doi: 10.1016/S0006-3495(04)74121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sono M, Dawson JH, Ikeda-Saito M. Characterization of the spleen green hemeprotein with magnetic and natural circular dichroism spectroscopy: positive evidence for a myeloperoxidase-type active site. Biochim. Biophys. Acta. 1986;873:62–72. doi: 10.1016/0167-4838(86)90190-1. [DOI] [PubMed] [Google Scholar]
- 33.Edwards SW, Lloyd D. CO-reacting hemoproteins of neutrophils—Evidence for cytochrome b245 and myeloperoxidase as potential oxidases during the respiratory burst. Biosci. Rep. 1987;7:193–199. doi: 10.1007/BF01124789. [DOI] [PubMed] [Google Scholar]
- 34.Odajima T, Yamazaki I. Myeloperoxidase of leukocyte of normal blood 0.1. reaction of myeloperoxidase with hydrogen peroxide. Biochim. Biophys. Acta. 1970;206:71–77. doi: 10.1016/0005-2744(70)90083-5. [DOI] [PubMed] [Google Scholar]
- 35.Horie S. On the carbone monoxide-shift of absorption spectrum of cytochrome a3. J. Biochem. 1964;56:113–121. doi: 10.1093/oxfordjournals.jbchem.a127967. [DOI] [PubMed] [Google Scholar]
- 36.Ansari A, Berendzen J, Braunstein D, Cowen BR, Frauenfelder H, Hong MK, Iben IET, Johnson JB, Ormos P, Sauke TB, Scholl R, Schulte A, Steinbach PJ, Vittitow J, Young RD. Rebinding and relaxation in the heme pocket. Biophys. Chem. 1987;26:337–355. doi: 10.1016/0301-4622(87)80034-0. [DOI] [PubMed] [Google Scholar]
- 37.Mourant JR, Braunstein DP, Chu K, Frauenfelder H, Nienhaus GU, Ormos P, Young RD. Ligand binding to heme proteins. 2. Transitions in the heme pocket of myoglobin. Biophys. J. 1993;65:1496–1507. doi: 10.1016/S0006-3495(93)81218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ingledew WJ, Smith SME, Gao YT, Jones RJ, Salerno JC, Rich PR. Ligand, cofactor, and residue vibrations in the catalytic site of endothelial nitric oxide synthase. Biochemistry. 2005;44:4238–4246. doi: 10.1021/bi047891y. [DOI] [PubMed] [Google Scholar]
- 39.Maréchal A, Ingledew WJ, Rich PR. Time-resolved FTIR study of CO recombination with horseradish peroxidase. Biochem. Soc. Trans. 2008;36:1165–1168. doi: 10.1042/BST0361165. [DOI] [PubMed] [Google Scholar]
- 40.Rohlfs RJ, Olson JS, Gibson QH. A comparison of the geminate recombination kinetics of several monomeric heme-proteins. J. Biol. Chem. 1988;263:1803–1813. [PubMed] [Google Scholar]
- 41.Braunstein DP, Chu K, Egeberg KD, Frauenfelder H, Mourant JR, Nienhaus GU, Ormos P, Sligar SG, Springer BA, Young RD. Ligand binding to heme proteins. 3. FTIR studies of His-E7 and Val-E11 mutants of carbonmonoxy-myoglobin. Biophys. J. 1993;65:2447–2454. doi: 10.1016/S0006-3495(93)81310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paul KG, Smith ML, Ohlsson PI. Infrared spectra of carbonyl lactoperoxidase. Inorg. Chim. Acta. 1983;79:169. [Google Scholar]
- 43.Vogel KM, Kozlowski PM, Zgierski MZ, Spiro TG. Role of the axial ligand in heme-CO backbonding; DFT analysis of vibrational data. Inorg. Chim. Acta. 2000;297:11–17. [Google Scholar]
- 44.Mayer E. FTIR spectroscopic study of the dynamics of conformational substates in hydrated carbonyl-myoglobin films via temperature-dependence of the CO stretching band parameters. Biophys. J. 1994;67:862–873. doi: 10.1016/S0006-3495(94)80547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingledew WJ, Rich PR. A study of the horseradish peroxidase catalytic site by FTIR spectroscopy. Biochem. Soc. Trans. 2005;33:886–889. doi: 10.1042/BST0330886. [DOI] [PubMed] [Google Scholar]
- 46.Iizuka T, Makino R, Ishimura Y, Yonetani T. Reversible acidic-alkaline transistion of the carbon monoxide complex of cytochrome c peroxidase. J. Biol. Chem. 1985;260:1407–1412. [PubMed] [Google Scholar]
- 47.Abe M, Kitagawa T, Kyogoku Y. Resonance Raman spectra of octaethylporphyrinato-Ni(II) and meso-deuterated and 15N substituted derivatives. II. A normal coordinate analysis. J. Chem. Phys. 1978;69:4526–4534. [Google Scholar]
- 48.Moss D, Nabedryk E, Breton J, Mäntele W. Redox-linked conformational changes in proteins detected by a combination of IR spectroscopy and protein electrochemistry. Evaluation of the technique with cytochrome c. Eur. J. Biochem. 1990;187:565–572. doi: 10.1111/j.1432-1033.1990.tb15338.x. [DOI] [PubMed] [Google Scholar]
- 49.Berthomieu C, Boussac A, Mäntele W, Breton J, Nabedryk E. Molecular changes following oxidoreduction of cytochrome b559 characterized by Fourier transform infrared difference spectroscopy and electron paramagnetic resonance: photooxidation in photosystem II and electrochemistry of isolated cytochrome b559 and iron protoporphyrin IX-bisimidazole model compounds. Biochemistry. 1992;31:11460–11471. doi: 10.1021/bi00161a026. [DOI] [PubMed] [Google Scholar]
- 50.de Meis L, Blanpain J-P, Goffeau A. P1⃡ATP exchange in the absence of proton gradient by the H+-ATPase from yeast plasma membranes. FEBS Lett. 1987;212:323–327. doi: 10.1016/0014-5793(87)81369-8. [DOI] [PubMed] [Google Scholar]
- 51.Rich PR, Breton J. Attenuated total reflection Fourier transform infrared studies of redox changes in bovine cytochrome c oxidase: resolution of the redox Fourier transform infrared difference spectrum of heme a3. Biochemistry. 2002;41:967–973. doi: 10.1021/bi0109717. [DOI] [PubMed] [Google Scholar]
- 52.Teraoka J, Kitagawa T. Structural implication of the heme-linked ionization of horseradish peroxidase probed by the Fe-histidine streching raman line. J. Biol. Chem. 1981;256:3969–3977. [PubMed] [Google Scholar]
- 53.Fujii H. 13C NMR signal detection of iron-bound cyanide ions in ferric cyanide complexes of heme proteins. J. Am. Chem. Soc. 2002;124:5936–5937. doi: 10.1021/ja025737y. [DOI] [PubMed] [Google Scholar]
- 54.Hasegawa K, Ono T-A, Noguchi T. Ab initio density functional theory calculations and vibrational analysis of zinc-bound 4-methylimidazole as a model of a histidine ligand in metalloenzymes. J. Phys. Chem. A. 2002;106:3377–3390. [Google Scholar]
- 55.Hasegawa K, Ono T-A, Noguchi T. Vibrational spectra and ab initio DFT calculations of 4-methylimidazole and its different protonation forms: infrared and Raman markers of the protonation state of a histidine side chain. J. Phys. Chem. B. 2000;104:4253–4265. [Google Scholar]
- 56.Carpena X, Vidossich P, Schroettner K, Callisto BM, Banerjee S, Stampler J, Soudi M, Furtmueller PG, Rovira C, Fita I, Obinger C. Essential role of proximal histidine-asparagine interaction in mammalian peroxidases. J. Biol. Chem. 2009;284:25929–25937. doi: 10.1074/jbc.M109.002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh AK, Singh N, Sharma S, Singh SB, Kaur P, Bhushan A, Srinivasan A, Singh TP. Crystal structure of lactoperoxidase at 2.4 Å resolution. J. Mol. Biol. 2008;376:1060–1075. doi: 10.1016/j.jmb.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 58.Singh AK, Singh N, Sharma S, Shin K, Takase M, Kaur P, Srinivasan A, Singh TP. Inhibition of lactoperoxidase by its own catalytic product: crystal structure of the hypothiocyanate-inhibited bovine lactoperoxidase at 2.3-Å resolution. Biophys. J. 2009;96:646–654. doi: 10.1016/j.bpj.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheikh IA, Singh AK, Singh N, Sinha M, Singh SB, Bhushan A, Kaur P, Srinivasan A, Sharma S, Singh TP. Structural evidence of substrate specificity in mammalian peroxidases: structure of the thiocyanate complex with lactoperoxidase and its interactions at 2.4 Å resolution. J. Biol. Chem. 2009;284:14849–14856. doi: 10.1074/jbc.M807644200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh AK, Singh N, Sinha M, Bhushan A, Kaur P, Srinivasan A, Sharma S, Singh TP. Binding modes of aromatic ligands to mammalian heme peroxidases with associated functional implications: crystal structures of lactoperoxidase complexes with acetylsalicylic acid, salicylhydroxamic acid, and benzylhydroxamic acid. J. Biol. Chem. 2009;284:20311–20318. doi: 10.1074/jbc.M109.010280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh AK, Kumar RP, Pandey N, Singh N, Sinha M, Bhushan A, Kaur P, Sharma S, Singh TP. Mode of binding of the tuberculosis prodrug isoniazid to heme peroxidases: binding studies and crystal structure of bovine lactoperoxidase with isoniazid at 2.7 Å resolution. J. Biol. Chem. 2010;285:1569–1576. doi: 10.1074/jbc.M109.060327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coletta M, Ascoli F, Brunori M, Traylor TG. pH dependence of carbon monoxide binding to ferrous horseradish peroxidase. J. Biol. Chem. 1986;261:9811–9814. [PubMed] [Google Scholar]
- 63.Lavalette D, Tetreau C, Mispelter J, Momenteau M, Lhoste JM. Linear free-energy relationships in binding of oxygen and carbon-monoxide with heme model compounds and heme-proteins. Eur. J. Biochem. 1984;145:555–565. doi: 10.1111/j.1432-1033.1984.tb08592.x. [DOI] [PubMed] [Google Scholar]
- 64.Carver TE, Brantley RE, Singleton EW, Arduini RM, Quillin ML, Phillips GN, Olson JS. A novel site-directed mutant of myoglobin with an unusually high O2 affinity and low autooxidation rate. J. Biol. Chem. 1992;267:14443–14450. [PubMed] [Google Scholar]
- 65.Sage JT. Myoglobin and CO: structure, energetics, and disorder. J. Biol. Inorg. Chem. 1997;2:537–543. [Google Scholar]
- 66.Spiro TG, Zgierski MZ, Kozlowski PM. Stereoelectronic factors in CO, NO and O2 binding to heme from vibrational spectroscopy and DFT analysis. Coord. Chem. Rev. 2001;219:923–936. [Google Scholar]
- 67.Spiro TG, Kozlowski PM. Is the CO adduct of myoglobin bent, and does it matter? Acc. Chem. Res. 2001;34:137–144. doi: 10.1021/ar000108j. [DOI] [PubMed] [Google Scholar]
- 68.Collman JP, Brauman JI, Halbert TR, Suslick KS. Nature of O2 and CO binding to metalloporphyrins and heme proteins. Proc. Natl. Acad. Sci. U.S.A. 1976;73:3333–3337. doi: 10.1073/pnas.73.10.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Traylor TG, Traylor PS. Considerations for the design of useful synthetic oxygen carriers. Annu. Rev. Biophys. Bioeng. 1982;11:105–127. doi: 10.1146/annurev.bb.11.060182.000541. [DOI] [PubMed] [Google Scholar]
- 70.Kim K, Fettinger J, Sessler JL, Cyr M, Hugdahl J, Collman JP, Ibers JA. Structural characterization of a sterically encumbered iron(II) porphyrin CO complex. J. Am. Chem. Soc. 1989;111:403–405. [Google Scholar]
- 71.Kim K, Ibers JA. Structure of a carbon monoxide adduct of a capped porphyrin—Fe(C2-Cap)(CO)(1-methylimidazole) J. Am. Chem. Soc. 1991;113:6077–6081. [Google Scholar]
- 72.Springer BA, Sligar SG, Olson JS, Phillips GN., Jr. Mechanisms of ligand recognition in myoglobin. Chem. Rev. 1994;94:699–714. [Google Scholar]
- 73.Ouellet H, Juszczak L, Dantsker D, Samuni U, Ouellet YH, Savard PY, Wittenberg JB, Wittenberg BA, Friedman JM, Guertin M. Reactions of Mycobacterium tuberculosis truncated hemoglobin O with ligands reveal a novel ligand-inclusive hydrogen bond network. Biochemistry. 2003;42:5764–5774. doi: 10.1021/bi0270337. [DOI] [PubMed] [Google Scholar]
- 74.Goldberg DE. Oxygen-avid hemoglobin of Ascaris. Chem. Rev. 1999;99:3371–3378. doi: 10.1021/cr970152l. [DOI] [PubMed] [Google Scholar]
- 75.Lopez-Garriga JJ, Oertling WA, Kean RT, Hoogland H, Wever R, Babcock GT. Metal-ligand vibrations of cyanoferric myeloperoxidase and cyanoferric horseradish peroxidase: evidence for a constrained heme pocket in myeloperoxidase. Biochemistry. 1990;29:9387–9395. doi: 10.1021/bi00492a012. [DOI] [PubMed] [Google Scholar]
- 76.Blair-Johnson M, Fiedler T, Fenna R. Human myeloperoxidase: structure of a cyanide complex and its interaction with bromide and thiocyanate substrates at 1.9 Å resolution. Biochemistry. 2001;40:13990–13997. doi: 10.1021/bi0111808. [DOI] [PubMed] [Google Scholar]
- 77.Traylor TG, Sharma VS. Why NO? Biochemistry. 1992;31:2847–2849. doi: 10.1021/bi00126a001. [DOI] [PubMed] [Google Scholar]
- 78.Mathews AJ, Olson JS. Assignment of rate constants for O2 and CO binding to alpha and beta subunits within R- and T-state human hemoglobin. Methods Enzymol. 1994;232:363–386. doi: 10.1016/0076-6879(94)32055-1. [DOI] [PubMed] [Google Scholar]
- 79.Perutz MF, Wilkinson AJ, Paoli M, Dodson GG. The stereochemical mechanism of the cooperative effects in hemoglobin revisited. Annu. Rev. Biophys. Biomol. Struct. 1998;27:1–34. doi: 10.1146/annurev.biophys.27.1.1. [DOI] [PubMed] [Google Scholar]
- 80.Kundu S, Snyder B, Das K, Chowdhury P, Park J, Petrich JW, Hargove MS. The leghemoglobin peroximal heme pocket directs oxygen dissociation and stabilizes bound heme. Proteins: Struct., Funct., Energ. 2002;46:268–277. doi: 10.1002/prot.10048. [DOI] [PubMed] [Google Scholar]
- 81.Dupeyat F, Vidaud C, Lorphelin A, Berthomieu C. Long distance charge redistribution upon Cu,Zn-superoxide dismutase reduction. J. Biol. Chem. 2004;279:48091–48101. doi: 10.1074/jbc.M402728200. [DOI] [PubMed] [Google Scholar]
- 82.Winterbourn CC, Garcia RC, Segal AW. Production of the superoxide adduct of myeloperoxidase (compound III) by stimulated human neutrophils and its reactivity with hydrogen peroxide and chloride. Biochem. J. 1985;228:583–592. doi: 10.1042/bj2280583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tani F, Matsu-ura M, Ariyama K, Setoyama T, Shimada T, Kobayashi S, Hayashi T, Matsuo T, Hisaeda Y, Naruta Y. Iron twin-coronet porphyrins as models of myoglobin and hemoglobin: amphibious electrostatic effects of overhanging hydroxyl groups for successful CO/O2 discrimination. Chemistry. 2003;9:862–870. doi: 10.1002/chem.200390096. [DOI] [PubMed] [Google Scholar]
- 84.Rose EJ, Hoffman BM. Nitric oxide ferrohemes—Kinetics of formation and photo-dissociation quantum yields. J. Am. Chem. Soc. 1983;105:2866–2873. [Google Scholar]
- 85.Jones SE, Srivatsa GS, Sawyer DT, Traylor TG, Mincey TC. Redox chemistry of iron tetraphenylporphyrin, imidazolate-chelated protoheme, and thiolate-chelated protoheme and of their Iron(II) superoxide adducts in dimethylsulfoxide. Inorg. Chem. 1983;22:3903–3910. [Google Scholar]
- 86.Galijasevic S, Saed GM, Diamond MP, Abu-Soud HM. High dissociation rate constant of ferrous-dioxy complex linked to the catalase-like activity in lactoperoxidase. J. Biol. Chem. 2004;279:39465–39470. doi: 10.1074/jbc.M406003200. [DOI] [PubMed] [Google Scholar]
- 87.Ohlsson PI, Paul KG. The reduction potential of lactoperoxidase. Acta Chem. Scand. B. 1983;37:917–921. doi: 10.3891/acta.chem.scand.37b-0917. [DOI] [PubMed] [Google Scholar]
- 88.Rodriguez-Lopez JN, Smith AT, Thorneley RN. Effect of distal cavity mutations on the binding and activation of oxygen by ferrous horseradish peroxidase. J. Biol. Chem. 1997;272:389–395. doi: 10.1074/jbc.272.1.389. [DOI] [PubMed] [Google Scholar]
- 89.Yamada H, Makino R, Yamazaki I. Effects of 2,4-substituents of deuteroheme upon redox potentials of horseradish peroxidases. Arch. Biochem. Biophys. 1975;169:344–353. doi: 10.1016/0003-9861(75)90350-1. [DOI] [PubMed] [Google Scholar]
- 90.Babcock GT, Wikström M. Oxygen activation and the conservation of energy in cell respiration. Nature. 1992;356:301–309. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]
- 91.Mims MP, Porras AG, Olson JS, Noble RW, Peterson JA. Ligand binding to heme proteins. An evaluation of distal effects. J. Biol. Chem. 1983;258:14219–14232. [PubMed] [Google Scholar]
- 92.Millis CD, Cai D, Stankovich MT, Tien M. Oxidation-reduction potentials and ionization states of extracellular peroxidases from the lignin-degrading fungus Phanerochaete chrysosporium. Biochemistry. 1989;28:8484–8489. doi: 10.1021/bi00447a032. [DOI] [PubMed] [Google Scholar]
- 93.Ishikawa H, Uchida T, Takahashi S, Ishimori K, Morishima I. Ligand migration in human myoglobin: steric effects of isoleucine 107(G8) on O2 and CO binding. Biophys. J. 2001;80:1507–1517. doi: 10.1016/S0006-3495(01)76123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Balasubramanian S, Lambright DG, Simmons JH, Gill SJ, Boxer SG. Determination of the carbon monoxide binding constants of myoglobin mutants: comparison of kinetic and equilibrium methods. Biochemistry. 1994;33:8355–8360. doi: 10.1021/bi00193a024. [DOI] [PubMed] [Google Scholar]
- 95.Cooper CE. Nitric oxide and iron proteins. Biochim. Biophys. Acta. 1999;1411:290–309. doi: 10.1016/s0005-2728(99)00021-3. [DOI] [PubMed] [Google Scholar]
- 96.Verkhovsky MI, Morgan JE, Wikström M. Oxygen binding and activation: early steps in the reaction of oxygen with cytochrome c oxidase. Biochemistry. 1994;33:3079–3086. doi: 10.1021/bi00176a042. [DOI] [PubMed] [Google Scholar]
- 97.Gorbikova EA, Vuorilehto K, Wikström M, Verkhovsky MI. Redox titration of all electron carriers of cytochrome c oxidase by Fourier transform infrared spectroscopy. Biochemistry. 2006;45:5641–5649. doi: 10.1021/bi060257v. [DOI] [PubMed] [Google Scholar]
- 98.Campbell BN, Jr., Araiso T, Reinisch L, Yue KT, Hager LP. A kinetic study of the binding of carbon monoxide to ferrous chloroperoxidase. Biochemistry. 1982;21:4343–4349. doi: 10.1021/bi00261a025. [DOI] [PubMed] [Google Scholar]
- 99.Yeh HC, Hsu PY, Tsai AL, Wang LH. Spectroscopic characterization of the oxyferrous complex of prostacyclin synthase in solution and in trapped sol-gel matrix. FEBS J. 2008;275:2305–2314. doi: 10.1111/j.1742-4658.2008.06385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Honeychurch MJ, Hill HAO, Wong LL. The thermodynamics and kinetics of electron transfer in the cytochrome P450(cam) enzyme system. FEBS Lett. 1999;451:351–353. doi: 10.1016/s0014-5793(99)00610-9. [DOI] [PubMed] [Google Scholar]
- 101.Bolscher BG, Wever R. A kinetic study of the reaction between human myeloperoxidase, hydroperoxides and cyanide. Inhibition by chloride and thiocyanate. Biochim. Biophys. Acta. 1984;788:1–10. doi: 10.1016/0167-4838(84)90290-5. [DOI] [PubMed] [Google Scholar]
- 102.Dolman D, Dunford HB, Chowdhury DM, Morrison M. The kinetics of cyanide binding by lactoperoxidase. Biochemistry. 1968;7:3991–3996. doi: 10.1021/bi00851a028. [DOI] [PubMed] [Google Scholar]
- 103.Glover RE, Koshkin V, Dunford HB, Mason RP. The reaction rates of NO with horseradish peroxidase compounds I and II. Nitric Oxide. 1999;3:439–444. doi: 10.1006/niox.1999.0256. [DOI] [PubMed] [Google Scholar]
- 104.Ellis WD, Dunford HB. The kinetics of cyanide and fluoride binding by ferric horseradish peroxidase. Biochemistry. 1968;7:2054–2062. doi: 10.1021/bi00846a006. [DOI] [PubMed] [Google Scholar]
- 105.Erman JE. Kinetic and equilibrium studies of cyanide binding by cytochrome c peroxidase. Biochemistry. 1974;13:39–44. doi: 10.1021/bi00698a007. [DOI] [PubMed] [Google Scholar]
- 106.Radi R. Reactions of nitric oxide with metalloproteins. Chem. Res. Toxicol. 1996;9:828–835. doi: 10.1021/tx950176s. [DOI] [PubMed] [Google Scholar]
- 107.Motie M, Kassner RJ, Meyer TE, Cusanovich MA. Kinetics of cyanide binding to Chromatium vinosum ferricytochrome c′. Biochemistry. 1990;29:1932–1936. doi: 10.1021/bi00459a038. [DOI] [PubMed] [Google Scholar]