Abstract
Since Ramon y Cajal’s examination of the cellular makeup of the cerebral cortex, it has been appreciated that this tissue exhibits some of the greatest degrees of cellular heterogeneity in the entire nervous system. This intricate structure emerges during a well-choreographed developmental process. Here we review current classifications of the cellular constituents of the cerebral cortex and examine how these building blocks are forged during development. We also look at how basic developmental features underlying cortex formation in vivo have been applied to protocols aimed at generating cortical tissue in vitro.
Introduction
The neocortex is the most complex part of the brain with enormous diversity in cell type, morphology, connectivity and function and the region that has undergone the largest expansion in volume during mammalian evolution1, 2. This structure mediates high-level cognitive processing, including sensory perception and skilled motor planning, as well as attention, language, emotion and potentially even consciousness itself3. The neocortex develops from the prosencephalon (forebrain), which is derived from the anterior portion of the neural tube. The prosencephalon gives rise to the telencephalon (cerebrum) and diencephalon. The telencephalon develops into the neocortex (referred to as “cerebral cortex” from here on), the allocortex and the striatum, while the diencephalon develops into the thalamus and surrounding nuclei. The neocortex forms a bilaterally symmetric structure consisting of two hemispheres. In mammals, neocortical architecture is typically structured into six radial layers of cells, as opposed to the allocortex, which has fewer layers. Neocortical layers are functionally divided into supragranular layers (I-III) that process intracortical information, the granular layer (IV) that receives input from other brain regions, notably the thalamus, and the infragranular layers (V-VI) that are the primary output layers that send information to other brain regions. The neocortex is also divided tangentially into functionally defined regions called areas4, 5.
Cells of the mammalian cerebral cortex have classically been categorized using a wide range of functional, structural and molecular characteristics (Figure 1)6-9. However, it is important to consider that these classification schemes are somewhat artificially imposed on an underlying biological and functional reality that often does not strictly fit within categorical subdivisions. In addition, it is clear that the field has just scratched the surface in defining the true cellular diversity of the cortex, and how this cellular diversity is manifested in different species. While still a work in progress, knowledge of the cellular composition of this tissue provides a fascinating window into the developmental processes that produce the cellular architecture underlying the very essence of human behavior. Moreover, such knowledge may inform future restorative strategies to repair cortical damage in vivo and to model cortical disease in the dish.
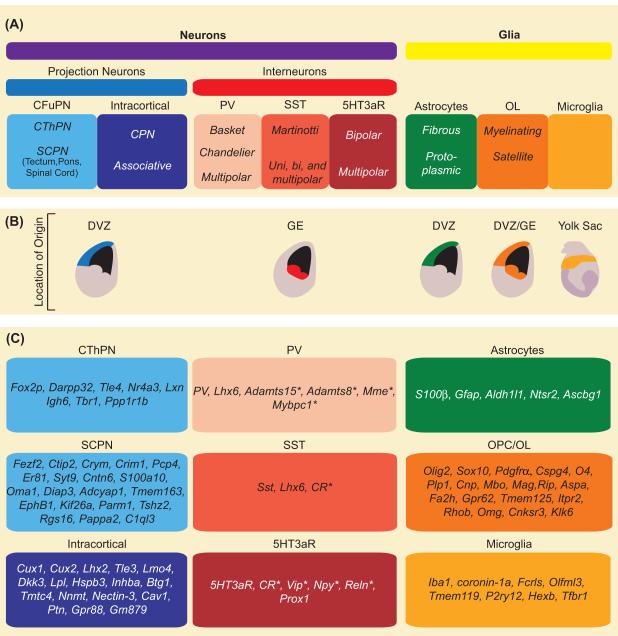
Figure 1. Cells of the Cerebral Cortex.
(A) Classification of established cell types in the cerebral cortex. These cells are broadly divided into neurons and glia. Neurons can be classified into glutamatergic projection neurons and GABAergic interneurons, each of which is highly heterogeneous and can be subdivided into many different subtypes, according to morphological, molecular and functional features. Glia can be divided into astrocytes, oligodendrocytes and microglia, which can also be categorized into several subtypes. (B) Developmental origin of the various cortical cell types. (C) Selected cell type markers of projection neurons21, 24, 25, 136, interneurons40, 41, 137, 138 and glia139-142. * indicates markers labeling a subset of the respective cell type. Abbreviations: CFuPN, corticofugal projection neuron; CThPN, corticothalamic projection neuron; SCPN, subcerebral projection neuron; CPN, callosal projection neuron; DVZ, dorsal ventricular zone; GE, ganglionic eminences.
Here we will take a holistic approach and review the fundamental cellular building blocks of the cerebral cortex, describe their developmental origin and consider the progress made in recapitulating this cellular diversity in vitro by applying developmental principles to pluripotent stem cells.
Neurons of the mammalian cerebral cortex: cellular diversity at its best
Neurons of the cerebral cortex have been classified according to a large number of parameters including the anatomical location of the cell body, destination of axonal projections, somatodendritic morphology, electrophysiological characteristics, molecular signatures and developmental origin10, 11. At the highest level, cortical neurons are divided into glutamatergic, excitatory projection neurons (PNs), which make up approximately 80% of all cortical neurons and form long range connections, and inhibitory interneurons (INs), which comprise the remaining 20%12, form local connections within the cortical parenchyma and provide the inhibitory drive to the cortical network through gamma-aminobutyric acid (GABA)-mediated neurotransmission (Figure 1A)13.
Projection neurons compose the entirety of the cortical output circuit
Historically, PNs have been categorized by the targets of their axonal projections and can be broadly divided into intracortical and corticofugal neurons (Figure 1A).
Intracortical projection neurons can be broken down into associative and commissural projection neurons. Associative PNs connect different cortical areas within the same hemisphere, or different layers within the same area (or even within the same cortical column)14. Commissural projection neurons connect the two cortical hemispheres by projecting axons through the dorsally located corpus callosum (CC), the major fiber commissure of the brain, or through the ventrally located anterior commissure (AC). Fiber commissures are bundles of axons that connect the two cerebral hemispheres. The CC is a relatively recent evolutionary invention, it is present only in placental mammals, and the majority of commissural neurons in rodents and primates send projections through the CC15. The cell bodies of intracortical PNs reside in all six layers, although they are present predominantly in the upper cortical layers (layers II/III)16 and can be recognized by the expression of Satb2 and Cux1, among other subtype-specific genes (Figure 1C)14.
Another category of PNs are corticofugal projection neurons (CFuPNs), which project to brain structures outside of the cortex, and which can be further classified into corticothalamic projection neurons (CThPNs) and subcerebral projection neurons (SCPNs). CThPNs connect the cortex to different thalamic nuclei, have their cell bodies primarily in layer VI, the innermost cortical layer, and express high levels of Tbr1 and Tle4, among other genes14, 17, 18. In contrast, SCPNs include, among others, the corticotectal neurons that project to the superior colliculus, corticopontine neurons that project to the pons in the hindbrain and corticospinal motor neurons that project to the spinal cord8, 19. SCPN cell bodies are primarily located in layer V and these cells express high levels of Ctip2 and Fezf2, among other marker genes (Figure 1C)20-22.
Another class of cortical PNs are corticostriatal projection neurons (CStrPNs), which have projections to the striatum as well as contralateral cortex. Their cell bodies are found primarily in layers II-VI, although a large number is found in layer Va. These neurons are often referred to as intratelencephalic corticostriatal projection neurons (CStrPNi), since, like commissural neurons, they send projections to the contralateral cortex, although they also have collaterals innervating the ipsi- and contralateral striatum23.
Molecular profiling of purified populations of projection neurons has led to the identification of molecular signatures that define some of the classical subtypes8, 14, 21, 24. More recently, some of the classic projection neuron classes have been profiled, for the first time, over a critical window of early fate specification in the developing embryo, using high-throughput methods. Specifically, purified populations of CPNs, CthPNs and SCPNs were isolated from developing cortex and compared by RNA sequencing at several early time points immediately after fate specification25. The data generated a new database of early expressed transcripts with exquisite, early profiles of gene expression within distinct populations. In addition, the work provides evidence that beyond differential expression of coding genes, non-coding transcripts (e.g. lncRNAs) and a whole spectrum of transcriptional dynamics (e.g. alternative promoter, CDS and isoform usage etc.) are differentially used in the cortex during projection neuron lineage selection. The data and analysis is now publicly available through the newly created “Developing Cortical Neuron Transcriptome Resource” (DeCoN) (http://decon.fas.harvard.edu/) dataset. This tool facilitates integrated, multidimensional data mining and provides a powerful new resource to generate insights into the transcriptional regulation underlying projection neuron diversity in the developing cortex. A selection of markers from this and prior datasets is included in (Figure 1C).
It is now clear that only combinatorial gene expression, and not single molecules, can distinguish one population of projection neurons from another and that molecular signatures are dynamic (i.e. the same population expresses distinct combinations and levels of marker genes at distinct stages of development). Intriguingly, molecular analysis suggests the existence of many more classes of projection neurons than currently recognized. For example, while CPNs across multiple layers express certain genes (e.g. Satb2, Lpl and Hspb3) that distinguish them from corticofugal neurons (CThPNs and SCPNs) of the deep layers, other markers have been identified that are only expressed in defined subsets of CPNs24. For example, Cux2 and Inhba mostly label CPNs of the upper layers and, additionally, within the layer II/III CPN population there is remarkable sublaminar specificity of gene expression patterns24. EphA3 and Nnmt are expressed in CPNs within a thin superficial sublamina of layer II/III; Nectin-3 and Chn2 label a middle sublamina; and Ptn and Cav1 are expressed in the deepest portion of layer II/III24. Finally, there is regional molecular heterogeneity within defined populations, such that neurons in distinct cortical areas have unique gene signatures. For example, Diap3 is expressed specifically in layer V SCPNs of the sensorimotor cortex, while Crim1 is highly expressed in the rostral motor cortex21. Thus it appears that overlapping and combinatorial patterns of gene expression define distinct neuronal subsets. With the advent of high throughput single cell methods26, we are bound to learn substantially more about the true molecular heterogeneity of these neuronal populations.
All projection neurons of the cortex develop from progenitors located in the germinal zone of the dorsal telencephalon, within the anterior neural tube. Beginning at around embryonic day 9.5 (E9.5) in mice, neuroepithelial cells (NE) form a pseudostratified layer of early progenitors, that compose the entire thickness of the neural tube separating the inner ventricular lumen from the outer pial surface. Later, NE cells differentiate into more committed progenitors called radial glia cells (RGCs), whose cell bodies lie within the ventricular zone (VZ) (lining the ventricle) and which have processes extending radially to the pial surface (Figure 2)27. As in other structures of the body, early progenitors are progressively patterned during development by diffusible molecules that are secreted in gradients along the antero-posterior (AP) and dorsoventral (DV) axis of the telencephalic vesicles. Molecules such as bone morphogenetic proteins (BMPs), sonic hedgehog (SHH), fibroblast growth factor (FGF) 8, retinoic acid (RA) and members of the Wingless-Int (WNT) family28, 29 propagate from signaling centers located at the boundaries of the tissue and pattern the progenitors to express specific codes of transcription factors, which are themselves expressed in distinct DV and AP concentration gradients14. These transcription factors include LIM homeobox 2 (LHX2), empty spiracles homologue 2 (EMX2), paired box 6 (PAX6) and chicken ovalbumin upstream promoter (COUP-TFI)30. Collectively, these signals pattern the progenitors of the dorsal telencephalon towards a cortical fate. Cortical progenitors must then expand and begin to give rise to the heterogeneous panel of projection neurons present in the mature cortex and to some of its macroglia constituents (i.e. astrocytes and subsets of oligodendrocytes).
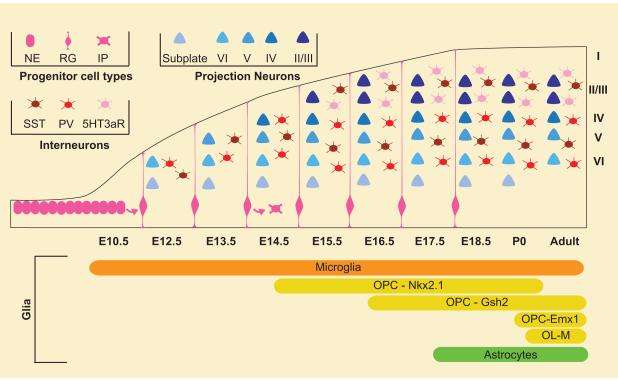
Figure 2. Schematic Timeline of Arrival of All Major Cell Types in the Cerebral Cortex.
The cortex is built through an intricately choreographed process that involves the integration of many different cell types. The cortex begins as a layer of neuroepithelial cells. Microglia from the yolk sac arrive early in development and invade the developing cortical plate. Projection neurons are generated in an inside out fashion beginning at E12.5. Interneurons migrate to the cortex from the ventral forebrain. Oligodendrocyte precursors arrive in the cortex in three waves from the MGE, LGE/CGE and from within the cortex itself. Later in corticogenesis, progenitors initiate production of astrocytes. Abbreviations: IP, intermediate progenitor; NE, neuroepithelial cell; OL, oligodendrocyte; OPC, oligodendrocyte precursor cell; PV, parvalbumin; RG, radial glia; SST, somatostatin; 5HT3aR, serotonin receptor 3a.
At mid-stages of corticogenesis, radial glia gives rise to a third type of progenitor cell called intermediate progenitors (IPs), which facilitate generation of large numbers of cells in a relatively short window of time (E10.5 to E18.5). IPs expand around E14.5 and form a second germinal zone, the subventricular zone (SVZ)31-33. The vast majority of cortical projection neurons are born either directly from radial glial cells or indirectly via intermediate progenitors in the dorsal pallium itself, the region of the telencephalon that gives rise to the neocortex (Figure 1B)27. Most projection neuron classes are born between E12.5 and E16.5, during a series of temporally restricted, but overlapping waves of neurogenesis33. Following their birth in the VZ and SVZ, these neurons migrate radially toward the pial surface along radial glial processes and seed a region called the cortical plate34, 35. CThPNs and deep layer commissural neurons arrive first, forming layer VI9, 34, 36. Layer V SCPNs and CPNs are among the neurons born next, followed by CPNs of the upper layers (II/III). These neurons migrate radially to the cortical plate, continue through the previously established deep layers, and seed the more superficial layers (“inside-out” mode of migration; Figure 2)36. The molecular basis of the cell fate decisions that progenitors undergo to generate the different classes of projection neurons is only beginning to be understood, and is an area of extensive investigation, which has been thoroughly reviewed elsewhere14.
Cortical interneurons modulate the function of projection neurons within the local microcircuit
While projection neurons communicate information between distant regions of the cortex and the CNS, interneurons typically modulate and coordinate delivery of this information by tuning the activity of projection neurons within the local microcircuitry37. Much work has been done attempting to classify the enormous heterogeneity of interneurons, however a universally accepted classification scheme has yet to emerge38. Here, we will briefly review a commonly used framework for classifying interneuron diversity.
While projection neurons have typically been categorized primarily based on hodology (connectivity), classification of interneurons has relied on a combination of molecular markers, electrophysiological properties and dendritic arbor morphologies, and attempts have been made to standardize these classifications39. Molecular markers can be used to divide GABAergic cortical interneurons into three comprehensive, non-overlapping groups, demarcated by expression of either parvalbumin (PV+), somatostatin (SST+) or the ionotropic serotonin receptor 5HT3a (5HT3aR) (Figure 1A)40. These interneuron subtypes are found in all cortical layers, although their relative proportions vary by layer. SST+ and PV+ interneurons are found primarily in the deep layers, while 5HT3aR+ interneurons are more abundant in the upper layers40-42.
Within these three broad classes, there is tremendous diversity of cell types. PV+ interneurons make up about 40% of cortical interneurons40, 41, are fast spiking and can be morphologically divided into basket and chandelier cells39, 43. SST+ cells represent about 30% of cortical interneurons40, 41, and are primarily Martinotti cells, although several groups have reported one to two additional subtypes of SST+ cells44, 45. 5HT3aR+ interneurons comprise the remaining 30% of cortical interneurons and due to the recent identification of this population, the classification of the relevant subtypes is still ongoing40, 41, 46. While the vast majority GABAergic neurons form local connections47, GABA expressing neurons with long-range projections have been identified and have a wide diversity of targets including distant regions of ipsi-48-52 and contralateral51, 53-56 cortex and striatum57, 58. It has been speculated that long-range GABAergic projections synchronize activity across remote networks59.
As with projection neurons, there are numerous molecular markers that are expressed in overlapping, often functionally-distinct, subsets of these broad groups of interneurons (Figure 1C)60. For instance, vasoactive intestinal protein (VIP+) interneurons comprise a subset of 5HT3aR+ interneurons40 that are thought to play an important role in disinhibition of cortical circuits61. SST+ Martinotti cells can be divided into calretinin (CR)+ vs CR− cells62. Compared to CR− Martinotti cells, CR+ interneurons are located in more superficial layers, have larger and broader dendritic trees and fire wider action potentials62. Many other molecular markers including neuropeptide Y, nNOS, reelin and cholesystokinin can be similarly used to label distinct interneuron subsets37, 60. For a more in depth description of cortical interneuron classification we refer the readers to excellent prior review articles38, 39.
As a more definitive framework for interneuron types emerges, our understanding of their developmental origins improves as well63-65. While projection neurons are born along the dorsolateral wall of the early telencephalon, interneurons are derived from a transient outpouching of the ventral telencephalon, named the ganglionic eminence, which is divided into medial, lateral and caudal regions (Figure 1B)66. PV+ and SST+ interneurons originate primarily from Nkx2.1+ progenitors in the medial ganglionic eminence (MGE)67-69. In contrast, 5HT3aR+ interneurons arise from progenitors expressing COUP-TFII in the caudal ganglionic eminence (CGE)41. From their place of birth in the ventral telencephalon, cortical interneurons migrate tangentially to the dorsal telencephalon where they then radially invade the developing cortical layers70. Like projection neurons, early born interneurons tend to seed the deep cortical layers, while later born neurons tend to populate the upper layers (Figure 2)71-74. Interestingly, laminar positioning of interneurons is altered by changes in projection neuron identity and location, suggesting that projection neurons play an important role in guiding interneurons to their appropriate radial destinations42.
Glial cells: much more than just “glue”
The critical role of glial cells in supporting and complementing neuronal function has been increasingly recognized. Indeed, the proportion of glial cells has increased tremendously in the nervous system during evolution, paralleling increased brain size75. As in other regions of the CNS, cortical glial cells are divided into astrocytes, oligodendrocytes and microglia (Figure 1A)76.
Astrocytes contribute to a wide range of cortical homeostatic functions including establishment of the blood brain barrier, regulation of ionic and water content and mediation of cell-cell calcium signaling. Most notably, these cells also play important roles in information processing by influencing neuronal activity directly through secretion of neuromodulatory molecules and regulation of synaptic transmission and plasticity76, 77. Astrocytes have been broadly broken down into two categories based on morphology and location (Figure 1A). Fibrous astrocytes are located in the white matter and are characterized by a uniform shape reminiscent of a star (thus the name “astrocytes”). They express high levels of glial fibrilary acidic protein (GFAP). Protoplasmic astrocytes are found in the grey matter, contact blood capillaries and ensheath synapses with more irregular processes76, 78.
Our understanding of astrocyte development has been limited by an inability to specifically define stages of astrocyte differentiation. Astrocytes can be distinguished by their characteristic morphologies only upon maturation, and studies typically rely solely on a limited number of molecular markers to track astrocyte development79. However, fate mapping experiments have shown that cortical astrocytes are born from radial glia and their progenitors, primarily after the completion of neurogenesis (Figure 2)27, 76. Classification of astrocyte diversity is just beginning, but there is general consensus that many distinct classes likely exist, which remain to be characterized.
Oligodendrocytes (OLs) can be distinguished from astrocytes morphologically and on the basis of cytological and molecular features80. While the cellular heterogeneity of oligodendrocytes has also not been fully investigated81, two types of functionally distinct OLs have been identified: myelinating and satellite oligodendrocytes (Figure 1A). Myelinating oligodendrocytes ensheath cortical axons, giving rise to myelin, an “insulating” lining made of tightly juxtaposed sheets of oligodendrocyte membranes. Myelin is well known to increase the efficiency and speed of action potential conduction and is a distinctive structural feature of the vertebrate nervous system80. Interestingly, while it is generally accepted that myelin has played a central role in allowing the evolution of increasingly complex brains, it is intriguing that the cerebral cortex is not uniformly myelinated (upper layers are less myelinated than deep layers)82. Recent work has demonstrated that oligodendrocyte progenitor cells (OPCs) are distributed in all cortical layers, while mature, myelinating OLs are present in higher numbers within the deep layers82. This results in uneven myelination of the layers, an observation that has led to the discovery that projection neurons in different layers have distinct profiles of distribution of myelin along their axons, with layer II/III projection neurons presenting the most heterogeneous profiles of myelination82.
Another type of oligodendrocytes, known as satellite oligodendrocytes, appears to be functionally distinct from the myelinating population. These are located in the grey matter, closely juxtaposed to neuronal cell bodies, and are more common in deep cortical layers83. They do not produce myelin, but rather are believed to regulate the levels of metabolites present in the local environment surrounding projection neuron cell bodies83, 84. Much of their functional roles remains a mystery83.
Whether oligodendrocytes are derived from dorsal or ventral telencephalic progenitors has been the subject of much debate85. Kessaris and colleagues used Cre-lox mediated fate mapping to demonstrate that cortical oligodendrocytes are generated in three consecutive waves, and belong to at least three different lineages86. The first wave originates in the MGE (Nkx2.1 lineage), followed by a second wave born in the LGE (Gsh2 lineage) and finally the third wave originates dorsally within the cortical wall (Emx1 lineage) (Figure 2)86. Interestingly, MGE-derived oligodendrocytes are completely lost at around postnatal day 10 (P10) in mice and the OLs myelinating the adult cortex belong exclusively to the Gsh2 and the Emx1 lineages86. The difference, if any, between these three lineages of cortical oligodendrocytes is unknown. They may, however, provide a mechanism for ensuring sufficient oligodendrocyte production, since ablation of any one of the three OL sources does not result in observable deficits in myelination, suggesting that the other progenitor populations can compensate86.
In addition to astrocytes and oligodendrocytes, which constitute the macroglia, microglia also have important roles in cortical function. Microglia are the resident tissue macrophages of the brain and have adapted accordingly. Microglia performs an astonishing range of functions in the cortex, including initiating defensive responses to injury or infection87-89, and regulating homeostatic mechanisms of neuronal migration, survival and death90-92 as well as important roles in synaptic pruning93-95. While traditionally thought of as a homogenous population, microglia have begun to be classified by differential responses to stimuli and diverse molecular profiles96. For instance, lineage tracing from the Hoxb8 locus labels 40% of brain microglia, suggesting that it may define a developmentally distinct microglial subtype96.
Unlike the neurons and macroglia discussed above, microglia are not born within the nervous system97, 98. Instead, they are derived from hematopoietic progenitors located in the embryonic yolk sac (Figure 1B)98, 99. These progenitors line the outer surface of the primitive neuroepithelium and invade the cortical wall at very early stages of development (E10.5)100. These cells mature in the VZ/SVZ101 before migrating into the deep layers first (at E18.5) and into the upper layers later (Figure 2)91, 100.
Glial cells perform a wide array of functions required for both cortical information processing and homeostatic maintenance of the cortical tissue77, 102. Glia have been understudied compared to neurons, and we are only just beginning to understand the extent of the glial cellular diversity required to perform all of their duties. If glia are more specialized than previously appreciated, it is likely that understanding of glial diversity will highlight new and fundamental organizing principles of cortical structure and function.
Building cortex in the dish
Recently, the ability to manipulate embryonic and induced pluripotent stem cells has led to a large body of work aimed at creating cells of the central nervous system in vitro. These directed differentiation strategies allow for the generation of large numbers of disease-relevant cell types from a variety of cellular sources, including both healthy and patient-specific somatic cells, and thus provide a powerful strategy for modeling human CNS development and disease in the dish. Given the large number of cells that can be potentially generated, this approach can be applied to high-throughput drug screening and may in theory provide a source of specific cell types for autologous transplantation. Can the cellular diversity of the mammalian cerebral cortex be recreated in the dish?
Directed differentiation strategies for cell types of the cerebral cortex have taken many forms and have been met with varied degrees of success. Here we will discuss some of the progress made in generating different classes of cortical glia and neurons from pluripotent cells, highlighting how developmental strategies and signals that shape these cellular identities during corticogenesis in the embryo have informed protocols to recapitulate the process in the dish.
Pluripotent stem cells have the potential to generate a plethora of cell fates, although it appears that when cultured as single cells without exogenous patterning signals they choose to default to a neural fate103-105. In intact embryos, specification to a neural fate is permitted through inhibition of morphogens, including bone morphogenic proteins (BMPs)106. Similar strategies have been successfully reproduced in vitro to pattern pluripotent stem cells to a neural fate by inhibition of BMP and TGF-β signaling, a treatment known as Dual SMAD inhibition (Figure 3)107.
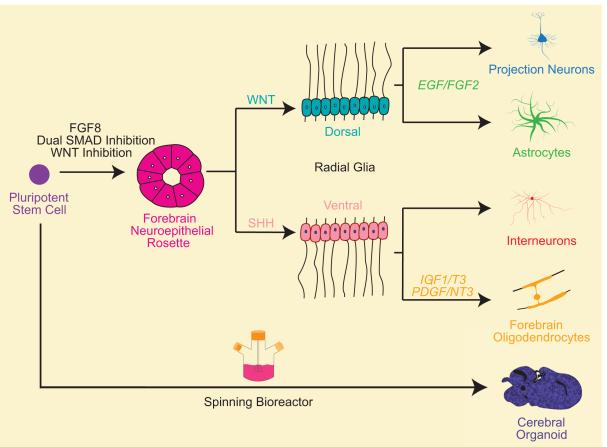
Figure 3. In vitro Generation of Cortical Cells.
Strategies to differentiate cortical cell types in vitro follow similar developmentally inspired trajectories. Pluripotent cells, either embryonic or induced, form forebrain neuroepithelial cells even in the absence of factors. This default fate decision can be reinforced with FGF8 or dual SMAD/WNT inhibition. For directed differentiation of interneurons and oligodendrocytes, progenitors are ventrally patterned, while for astrocytes and projection neurons, progenitors are dorsally patterned. Once this regional specification is established, developmental programs unfold in typical order such that neurons are generated first, followed by astrocytes and oligodendrocytes. Different culture conditions and durations allow for enrichment of the desired population, although all strategies give rise to mixtures of neurons, glia and progenitors. For cerebral organoids, pluripotent stem cells are grown in spinning bioreactors to allow natural developmental programs to unfold.
Later in development, a process of regional patterning by morphogens leads to the parcellation of neural progenitors into distinct domains fated to form different parts of the central nervous system108-110. The AP axis of the neural tube is patterned by a gradient of retinoic acid, which specifies a posterior (hindbrain/spinal cord) fate, while FGF8, released by the commissural plate, reinforces the anterior (forebrain) fate108, 109, 111, 112. Within the rostral neural tube, the DV axis is patterned ventrally by sonic hedgehog (SHH) released from the ventral telencephalon113, 114, and dorsal identity is mediated by FGFs and WNT signaling (Figure 3)115. This knowledge has informed strategies to pattern progenitors in the dish to either an anterior dorsal fate (primed to generate cortical projection neurons and astrocytes) or an anterior ventral fate (primed to form cortical interneurons and oligodendrocytes) (reviewed in Hansen et al. 2011116).
It is striking that neural progenitors that are patterned by these methods to a dorsal, “cortical”, fate in the dish subsequently execute a program of neuronal differentiation such that different classes of projection neurons are produced following a similar temporal order as observed in the intact developing cortex. Generation of neurons expressing deep layer markers occurs first, followed by generation of neurons with molecular features of upper layers (albeit not all these cellular identities are generated in each protocol)117-123. As in development, newly generated neurons appear able to migrate along the processes of the radial glia and build layer-like structures populated by different neurons120, 121, 123. These data demonstrate the impressive self-specifying capacity of pluripotent stem cell-derived neural progenitors, which are intrinsically primed to recapitulate critical aspects of cortical neurogenesis in the dish.
It is also notable that cortical projection neurons generated through such directed differentiation protocols survive upon transplantation in the early postnatal mouse cortex, send projections to multiple distal targets and form functional synapses119.
This shows that at least some basic aspects of early cortical development and generation of projection neuron diversity can be achieved. Questions of course remain about whether these neurons truly reflect specific endogenous classes or whether they could be functionally equivalent to their endogenous counterparts. In addition, the generation of one specific class of neurons by biasing progenitors to one or a limited set of neuronal identities remains an unmet challenge in the field. It is likely that this task will require a deeper understanding of how progenitor lineage-bifurcation decisions that shape individual projection neuron identities are orchestrated in the first place, in the embryo.
Cortical astrocytes have also been generated from dorsally patterned neural progenitors that are prevented from differentiating during an early neurogenic window of time causing them to “skip” neuron generation and acquire a gliogenic identity (Figure 3)124. These methods are effective to produce cultures in which more than 90% of the differentiated cells express the astrocytic markers GFAP and S100β125. Functionally, these cells can perform many astrocyte-specific functions in vitro, including glutamate uptake, propagation of calcium waves and facilitation of synapse formation125. Furthermore, intraventricular transplantation of these cells resulted in engraftment, association with blood vessels and maintenance of an astrocytic identity in the corpus callosum of mouse125. Interestingly, regional identity could be imposed as cells with features of astrocytes of the cortex or astrocytes of the spinal cord could be generated using the same morphogens that normally specify these regional identities in vivo125.
Unlike projection neurons or astrocytes, cortical interneurons arise from the ventral telencephalon, within the ganglionic eminences66. Thus, to create interneurons in vitro, progenitors have been first patterned to a ventral fate by SHH signaling and to an anterior fate by different methods, before allowing neurogenesis126-129. This approach can generate cultures enriched for GABA+ cells (Figure 3)126. These cells include a highly heterogeneous assortment of neuronal subtypes, characterized by the expression of distinct markers including SST and, to a lesser extent PV126-129. Interneurons generated by these methods developed appropriate electrophysiological traits and GABAergic synapses emerged when co-cultured with human or mouse fetal cortical neurons126, 128. Interneurons with features resembling those of CGE origin (i.e. Calretinin+ and COUP-TFII+) could also be derived from ventralized progenitors exposed to activin130. In the future it will be exciting to explore ways to expand the repertoire of interneuron subtypes that are produced and to fully define their functional properties in vivo.
Generation of oligodendrocytes in vitro has been highly sought after, given their great clinical impact in prominent diseases of the white matter (e.g. multiple sclerosis). Embryonically, cortical oligodendrocytes first arise in the ganglionic eminences and a second wave is born later from progenitors of the dorsal pallium. So far, protocols have specified progenitors to a ventral fate and, using a strategy similar to that used to produce astrocytes, neural progenitors are kept from differentiation during the neurogenic period by addition of mitogenic growth factors (Figure 3)131. Once mitogens are removed oligodendroglia differentiation is facilitated by culturing cells for extended periods of time in the presence of thyroid hormone or platelet derived growth factor (PDGF), which promote the differentiation of immature oligodendrocyte precursors into mature oligodendrocytes132, 133. These methods produce mostly immature oligodendrocytes, but a smaller subset of cells express more mature markers, exhibit a multibranched morphology and are capable of myelinating axons in vitro131. It is important to note that despite being distinct from spinal cord oligodendrocytes, these cells share features of oligodendrocytes found in both the dorsal and ventral forebrain.
All together these experiments indicate that in their simplest incarnation, components of the cellular diversity found in the cerebral cortex can be generated in the dish starting from pluripotent stem cells of both murine and human origin. Although much work remains to be done, this paves the way for the exciting opportunity to model cortical development and disease in vitro.
Concluding remarks and perspective
The cerebral cortex is one of the most extraordinarily complex structures of the nervous system. Presumably, its cellular diversity evolved to obtain great specialization to execute the unmatched functional capacities of the cortex. Defining and classifying the basic cellular components of the cortex is fundamental to understanding how this tissue is made, how it functions, and how it succumbs to disease. While the process of cell classification remains a daunting task, the past decade has seen a surge of molecular studies that integrate combinatorial marker expression with analysis of more classical traits (i.e. connectivity, electrophysiological properties etc.) to better resolve individual cell identities. While still incomplete, it is fair to say that classification of cortical neuron subtypes is more advanced than that of glia, and it is likely that the next few years will see a surge in studies revealing the true diversity of both oligodendrocytes and astrocytes.
A better understanding of normal embryonic development of the cortex is clearly facilitating a large range of studies aimed at “programming” pluripotent stem cells to generate this cellular diversity in the dish. Developmental signals have informed these directed differentiation protocols and knowledge of endogenous cellular diversity has helped understand the identity of the cell produced in vitro, even if several questions remain. The field has so far failed to produce individual classes of cortical neurons (and glia), as single pure populations. In addition, in most cases it is unclear to what extent cells obtained by these methods resemble their endogenous counterparts134. Finally, as it is the case for other types of cells made in the dish, pluripotent stem cell-derived neurons and glia appear immature.
Development of the cerebral cortex is a choreographed interplay of multiple cell types born in specific temporal order, from often-distant germinal zones, and that ultimately migrate to very precise relative locations to build the 3D tissue architecture of the adult. Will the field ever be able to mimic such grace and complexity in the dish? A mere few years ago the answer would have been “likely not”. But last year Lancaster and colleagues showed the world a tiny piece of cerebral tissue completely produced from human pluripotent stem cells and looking strikingly similar to early fetal human brain (Figure 3)121. Forebrain, midbrain and hindbrain structures were all present and the forebrain showed regionalization121. Interneurons were born in regions expressing ventral forebrain markers and migrated to regions expressing dorsal markers121. Within the cortex, appropriate lamination patterns were observed121. Progenitors lined a central cavity, in a region reminiscent of the ventricular zone, and newly born neurons migrated radially along glial processes121. Building on seminal prior demonstrations of the extensive self-organizing capabilities of neural progenitors when culture in the absence of specific extrinsic cues120, 123, 135, this work offered a glimpse of what is possible. Much work remains to be done to improve these cerebral organoids so that they include cell types born late during cortical development (i.e. upper layer neurons, astrocytes and oligodendrocytes), as well as cells that originate outside of the nervous system (i.e. microglia). It is our opinion however, that these in vitro approaches will grow into valuable, realistic models to study human cortical development and complex disease.
Acknowledgements
We would like to thank Simona Lodato, Juliana Brown and Emanuela Zuccaro for their very helpful feedback. Work in the Arlotta laboratory is funded by the NIH, the Harvard Stem Cell Institute, Target ALS and the New York Stem Cell Foundation. P.A. is a New York Stem Cell Foundation-Robertson Investigator. J.H. supported by award number T32GM007753 from the National Institute of General Medical Science.
References
- 1.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. doi: 10.1242/dev.090571. 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- 3.Fuster JM. Cortex and mind : unifying cognition. Oxford University Press; Oxford ; New York: 2003. [Google Scholar]
- 4.Grove EA, Fukuchi-Shimogori T. Generating the cerebral cortical area map. Annu Rev Neurosci. 2003;26:355–380. doi: 10.1146/annurev.neuro.26.041002.131137. 10.1146/annurev.neuro.26.041002.131137. [DOI] [PubMed] [Google Scholar]
- 5.Alfano C, Studer M. Neocortical arealization: evolution, mechanisms, and open questions. Dev Neurobiol. 2013;73:411–447. doi: 10.1002/dneu.22067. 10.1002/dneu.22067. [DOI] [PubMed] [Google Scholar]
- 6.Peters A, Jones EG. Cerebral cortex. Plenum Press; New York: 1984. [Google Scholar]
- 7.Ramón y Cajal S. Histology of the nervous system of man and vertebrates. Oxford University Press; New York: 1995. [Google Scholar]
- 8.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 9.Bayer SA, Altman J. Neocortical development. Raven Press; New York, N.Y.: 1991. [Google Scholar]
- 10.Toyama K, Matsunami K, Ono T, Tokashiki S. An intracellular study of neuronal organization in the visual cortex. Exp Brain Res. 1974;21:45–66. doi: 10.1007/BF00234257. [DOI] [PubMed] [Google Scholar]
- 11.Migliore M, Shepherd GM. Opinion: an integrated approach to classifying neuronal phenotypes. Nat Rev Neurosci. 2005;6:810–818. doi: 10.1038/nrn1769. 10.1038/nrn1769. [DOI] [PubMed] [Google Scholar]
- 12.Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, Kaneko T. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol. 2003;467:60–79. doi: 10.1002/cne.10905. 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- 13.Parnavelas JG. The origin and migration of cortical neurones: new vistas. Trends Neurosci. 2000;23:126–131. doi: 10.1016/s0166-2236(00)01553-8. [DOI] [PubMed] [Google Scholar]
- 14.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aboitiz F, Montiel J. One hundred million years of interhemispheric communication: the history of the corpus callosum. Braz J Med Biol Res. 2003;36:409–420. doi: 10.1590/s0100-879x2003000400002. [DOI] [PubMed] [Google Scholar]
- 16.Fame RM, MacDonald JL, Macklis JD. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 2011;34:41–50. doi: 10.1016/j.tins.2010.10.002. 10.1016/j.tins.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, Beyer RP, Bammler TK, Rubenstein JL, Hevner RF. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci U S A. 2010;107:13129–13134. doi: 10.1073/pnas.1002285107. 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, Chen B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molnar Z, Cheung AF. Towards the classification of subpopulations of layer V pyramidal projection neurons. Neurosci Res. 2006;55:105–115. doi: 10.1016/j.neures.2006.02.008. 10.1016/j.neures.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci U S A. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 22.Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 23.Sohur US, Padmanabhan HK, Kotchetkov IS, Menezes JR, Macklis JD. Anatomic and molecular development of corticostriatal projection neurons in mice. Cereb Cortex. 2014;24:293–303. doi: 10.1093/cercor/bhs342. 10.1093/cercor/bhs342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molyneaux BJ, Arlotta P, Fame RM, MacDonald JL, MacQuarrie KL, Macklis JD. Novel subtype-specific genes identify distinct subpopulations of callosal projection neurons. J Neurosci. 2009;29:12343–12354. doi: 10.1523/JNEUROSCI.6108-08.2009. 10.1523/JNEUROSCI.6108-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molyneaux BJ, Goff LA, Brettler AC, Chen HH, Brown JR, Hrvatin S, Rinn JL, Arlotta P. DeCoN: Genome-wide Analysis of In Vivo Transcriptional Dynamics during Pyramidal Neuron Fate Selection in Neocortex. Neuron. 2015;85:275–288. doi: 10.1016/j.neuron.2014.12.024. 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–630. doi: 10.1038/nrg3542. 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 27.Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- 28.Shimogori T, Banuchi V, Ng HY, Strauss JB, Grove EA. Embryonic signaling centers expressing BMP, WNT and FGF proteins interact to pattern the cerebral cortex. Development. 2004;131:5639–5647. doi: 10.1242/dev.01428. 10.1242/dev.01428. [DOI] [PubMed] [Google Scholar]
- 29.Campbell K. Dorsal-ventral patterning in the mammalian telencephalon. Curr Opin Neurobiol. 2003;13:50–56. doi: 10.1016/s0959-4388(03)00009-6. [DOI] [PubMed] [Google Scholar]
- 30.O’Leary DD, Sahara S. Genetic regulation of arealization of the neocortex. Curr Opin Neurobiol. 2008;18:90–100. doi: 10.1016/j.conb.2008.05.011. 10.1016/j.conb.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Embryonic vertebrate central nervous system: revised terminology. The Boulder Committee. Anat Rec. 1970;166:257–261. doi: 10.1002/ar.1091660214. 10.1002/ar.1091660214. [DOI] [PubMed] [Google Scholar]
- 32.Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2008;30:24–32. doi: 10.1159/000109848. 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- 33.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 34.Angevine JB, Jr., Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 35.Rakic P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- 36.Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- 37.Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeFelipe J, Lopez-Cruz PL, Benavides-Piccione R, Bielza C, Larranaga P, Anderson S, Burkhalter A, Cauli B, Fairen A, Feldmeyer D, et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat Rev Neurosci. 2013;14:202–216. doi: 10.1038/nrn3444. 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petilla Interneuron Nomenclature G. Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsaki G, Cauli B, Defelipe J, et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 2008;9:557–568. doi: 10.1038/nrn2402. 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudy B, Fishell G, Lee S, Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011;71:45–61. doi: 10.1002/dneu.20853. 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S, Hjerling-Leffler J, Zagha E, Fishell G, Rudy B. The largest group of superficial neocortical GABAergic interneurons expresses ionotropic serotonin receptors. J Neurosci. 2010;30:16796–16808. doi: 10.1523/JNEUROSCI.1869-10.2010. 10.1523/JNEUROSCI.1869-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lodato S, Rouaux C, Quast KB, Jantrachotechatchawan C, Studer M, Hensch TK, Arlotta P. Excitatory projection neuron subtypes control the distribution of local inhibitory interneurons in the cerebral cortex. Neuron. 2011;69:763–779. doi: 10.1016/j.neuron.2011.01.015. 10.1016/j.neuron.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawaguchi Y, Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex. 1997;7:476–486. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- 44.Ma Y, Hu H, Berrebi AS, Mathers PH, Agmon A. Distinct subtypes of somatostatin-containing neocortical interneurons revealed in transgenic mice. J Neurosci. 2006;26:5069–5082. doi: 10.1523/JNEUROSCI.0661-06.2006. 10.1523/JNEUROSCI.0661-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGarry LM, Packer AM, Fino E, Nikolenko V, Sippy T, Yuste R. Quantitative classification of somatostatin-positive neocortical interneurons identifies three interneuron subtypes. Front Neural Circuits. 2010;4:12. doi: 10.3389/fncir.2010.00012. 10.3389/fncir.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelsom C, Lu W. Development and specification of GABAergic cortical interneurons. Cell Biosci. 2013;3:19. doi: 10.1186/2045-3701-3-19. 10.1186/2045-3701-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamamaki N, Tomioka R. Long-Range GABAergic Connections Distributed throughout the Neocortex and their Possible Function. Front Neurosci. 2010;4:202. doi: 10.3389/fnins.2010.00202. 10.3389/fnins.2010.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albus K, Wahle P. The topography of tangential inhibitory connections in the postnatally developing and mature striate cortex of the cat. Eur J Neurosci. 1994;6:779–792. doi: 10.1111/j.1460-9568.1994.tb00989.x. [DOI] [PubMed] [Google Scholar]
- 49.Fabri M, Manzoni T. Glutamate decarboxylase immunoreactivity in corticocortical projecting neurons of rat somatic sensory cortex. Neuroscience. 1996;72:435–448. doi: 10.1016/0306-4522(95)00568-4. [DOI] [PubMed] [Google Scholar]
- 50.McDonald CT, Burkhalter A. Organization of long-range inhibitory connections with rat visual cortex. J Neurosci. 1993;13:768–781. doi: 10.1523/JNEUROSCI.13-02-00768.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Higo S, Akashi K, Sakimura K, Tamamaki N. Subtypes of GABAergic neurons project axons in the neocortex. Front Neuroanat. 2009;3:25. doi: 10.3389/neuro.05.025.2009. 10.3389/neuro.05.025.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomioka R, Rockland KS. Long-distance corticocortical GABAergic neurons in the adult monkey white and gray matter. J Comp Neurol. 2007;505:526–538. doi: 10.1002/cne.21504. 10.1002/cne.21504. [DOI] [PubMed] [Google Scholar]
- 53.Gonchar YA, Johnson PB, Weinberg RJ. GABA-immunopositive neurons in rat neocortex with contralateral projections to S-I. Brain Res. 1995;697:27–34. doi: 10.1016/0006-8993(95)00746-d. [DOI] [PubMed] [Google Scholar]
- 54.Fabri M, Manzoni T. Glutamic acid decarboxylase immunoreactivity in callosal projecting neurons of cat and rat somatic sensory areas. Neuroscience. 2004;123:557–566. doi: 10.1016/j.neuroscience.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 55.Tomioka R, Okamoto K, Furuta T, Fujiyama F, Iwasato T, Yanagawa Y, Obata K, Kaneko T, Tamamaki N. Demonstration of long-range GABAergic connections distributed throughout the mouse neocortex. Eur J Neurosci. 2005;21:1587–1600. doi: 10.1111/j.1460-9568.2005.03989.x. 10.1111/j.1460-9568.2005.03989.x. [DOI] [PubMed] [Google Scholar]
- 56.Peters A, Payne BR, Josephson K. Transcallosal non-pyramidal cell projections from visual cortex in the cat. J Comp Neurol. 1990;302:124–142. doi: 10.1002/cne.903020110. 10.1002/cne.903020110. [DOI] [PubMed] [Google Scholar]
- 57.Lee AT, Vogt D, Rubenstein JL, Sohal VS. A class of GABAergic neurons in the prefrontal cortex sends long-range projections to the nucleus accumbens and elicits acute avoidance behavior. J Neurosci. 2014;34:11519–11525. doi: 10.1523/JNEUROSCI.1157-14.2014. 10.1523/JNEUROSCI.1157-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jinno S, Kosaka T. Parvalbumin is expressed in glutamatergic and GABAergic corticostriatal pathway in mice. J Comp Neurol. 2004;477:188–201. doi: 10.1002/cne.20246. 10.1002/cne.20246. [DOI] [PubMed] [Google Scholar]
- 59.Uhlhaas PJ, Singer W. Neuronal dynamics and neuropsychiatric disorders: toward a translational paradigm for dysfunctional large-scale networks. Neuron. 2012;75:963–980. doi: 10.1016/j.neuron.2012.09.004. 10.1016/j.neuron.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 60.Gonchar Y, Wang Q, Burkhalter A. Multiple distinct subtypes of GABAergic neurons in mouse visual cortex identified by triple immunostaining. Front Neuroanat. 2007;1:3. doi: 10.3389/neuro.05.003.2007. 10.3389/neuro.05.003.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pi HJ, Hangya B, Kvitsiani D, Sanders JI, Huang ZJ, Kepecs A. Cortical interneurons that specialize in disinhibitory control. Nature. 2013;503:521–524. doi: 10.1038/nature12676. 10.1038/nature12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu X, Roby KD, Callaway EM. Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. J Comp Neurol. 2006;499:144–160. doi: 10.1002/cne.21101. 10.1002/cne.21101. [DOI] [PubMed] [Google Scholar]
- 63.Marin O, Muller U. Lineage origins of GABAergic versus glutamatergic neurons in the neocortex. Curr Opin Neurobiol. 2014;26:132–141. doi: 10.1016/j.conb.2014.01.015. 10.1016/j.conb.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- 65.Gelman DM, Marin O. Generation of interneuron diversity in the mouse cerebral cortex. Eur J Neurosci. 2010;31:2136–2141. doi: 10.1111/j.1460-9568.2010.07267.x. 10.1111/j.1460-9568.2010.07267.x. [DOI] [PubMed] [Google Scholar]
- 66.Corbin JG, Butt SJ. Developmental mechanisms for the generation of telencephalic interneurons. Dev Neurobiol. 2011;71:710–732. doi: 10.1002/dneu.20890. 10.1002/dneu.20890. [DOI] [PubMed] [Google Scholar]
- 67.Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 69.Fogarty M, Grist M, Gelman D, Marin O, Pachnis V, Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J Neurosci. 2007;27:10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- 71.Miller MW. Cogeneration of retrogradely labeled corticocortical projection and GABA-immunoreactive local circuit neurons in cerebral cortex. Brain Res. 1985;355:187–192. doi: 10.1016/0165-3806(85)90040-9. [DOI] [PubMed] [Google Scholar]
- 72.Lopez-Bendito G, Sturgess K, Erdelyi F, Szabo G, Molnar Z, Paulsen O. Preferential origin and layer destination of GAD65-GFP cortical interneurons. Cereb Cortex. 2004;14:1122–1133. doi: 10.1093/cercor/bhh072. 10.1093/cercor/bhh072. [DOI] [PubMed] [Google Scholar]
- 73.Fairen A, Cobas A, Fonseca M. Times of generation of glutamic acid decarboxylase immunoreactive neurons in mouse somatosensory cortex. J Comp Neurol. 1986;251:67–83. doi: 10.1002/cne.902510105. 10.1002/cne.902510105. [DOI] [PubMed] [Google Scholar]
- 74.Valcanis H, Tan SS. Layer specification of transplanted interneurons in developing mouse neocortex. J Neurosci. 2003;23:5113–5122. doi: 10.1523/JNEUROSCI.23-12-05113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sherwood CC, Stimpson CD, Raghanti MA, Wildman DE, Uddin M, Grossman LI, Goodman M, Redmond JC, Bonar CJ, Erwin JM, et al. Evolution of increased glia-neuron ratios in the human frontal cortex. Proc Natl Acad Sci U S A. 2006;103:13606–13611. doi: 10.1073/pnas.0605843103. 10.1073/pnas.0605843103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 77.Clarke LE, Barres BA. Emerging roles of astrocytes in neural circuit development. Nat Rev Neurosci. 2013;14:311–321. doi: 10.1038/nrn3484. 10.1038/nrn3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller RH, Raff MC. Fibrous and protoplasmic astrocytes are biochemically and developmentally distinct. J Neurosci. 1984;4:585–592. doi: 10.1523/JNEUROSCI.04-02-00585.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Molofsky AV, Krencik R, Ullian EM, Tsai HH, Deneen B, Richardson WD, Barres BA, Rowitch DH. Astrocytes and disease: a neurodevelopmental perspective. Genes Dev. 2012;26:891–907. doi: 10.1101/gad.188326.112. 10.1101/gad.188326.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- 81.Tomassy GS, Fossati V. How big is the myelinating orchestra? Cellular diversity within the oligodendrocyte lineage: facts and hypotheses. Front Cell Neurosci. 2014;8:201. doi: 10.3389/fncel.2014.00201. 10.3389/fncel.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tomassy GS, Berger DR, Chen HH, Kasthuri N, Hayworth KJ, Vercelli A, Seung HS, Lichtman JW, Arlotta P. Distinct profiles of myelin distribution along single axons of pyramidal neurons in the neocortex. Science. 2014;344:319–324. doi: 10.1126/science.1249766. 10.1126/science.1249766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takasaki C, Yamasaki M, Uchigashima M, Konno K, Yanagawa Y, Watanabe M. Cytochemical and cytological properties of perineuronal oligodendrocytes in the mouse cortex. Eur J Neurosci. 2010;32:1326–1336. doi: 10.1111/j.1460-9568.2010.07377.x. 10.1111/j.1460-9568.2010.07377.x. [DOI] [PubMed] [Google Scholar]
- 84.van Landeghem FK, Weiss T, von Deimling A. Expression of PACAP and glutamate transporter proteins in satellite oligodendrocytes of the human CNS. Regul Pept. 2007;142:52–59. doi: 10.1016/j.regpep.2007.01.008. 10.1016/j.regpep.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 85.Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Suzuki Y, Claflin J, Wang X, Lengi A, Kikuchi T. Microglia and macrophages as innate producers of interferon-gamma in the brain following infection with Toxoplasma gondii. Int J Parasitol. 2005;35:83–90. doi: 10.1016/j.ijpara.2004.10.020. 10.1016/j.ijpara.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 88.Smith C, Gentleman SM, Leclercq PD, Murray LS, Griffin WS, Graham DI, Nicoll JA. The neuroinflammatory response in humans after traumatic brain injury. Neuropathol Appl Neurobiol. 2013;39:654–666. doi: 10.1111/nan.12008. 10.1111/nan.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morrison HW, Filosa JA. A quantitative spatiotemporal analysis of microglia morphology during ischemic stroke and reperfusion. J Neuroinflammation. 2013;10:4. doi: 10.1186/1742-2094-10-4. 10.1186/1742-2094-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M, Yamashita T. Layer V cortical neurons require microglial support for survival during postnatal development. Nat Neurosci. 2013;16:543–551. doi: 10.1038/nn.3358. 10.1038/nn.3358. [DOI] [PubMed] [Google Scholar]
- 91.Squarzoni P, Oller G, Hoeffel G, Pont-Lezica L, Rostaing P, Low D, Bessis A, Ginhoux F, Garel S. Microglia modulate wiring of the embryonic forebrain. Cell Rep. 2014;8:1271–1279. doi: 10.1016/j.celrep.2014.07.042. 10.1016/j.celrep.2014.07.042. [DOI] [PubMed] [Google Scholar]
- 92.Arno B, Grassivaro F, Rossi C, Bergamaschi A, Castiglioni V, Furlan R, Greter M, Favaro R, Comi G, Becher B, et al. Neural progenitor cells orchestrate microglia migration and positioning into the developing cortex. Nat Commun. 2014;5:5611. doi: 10.1038/ncomms6611. 10.1038/ncomms6611. [DOI] [PubMed] [Google Scholar]
- 93.Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 95.Roumier A, Bechade C, Poncer JC, Smalla KH, Tomasello E, Vivier E, Gundelfinger ED, Triller A, Bessis A. Impaired synaptic function in the microglial KARAP/DAP12-deficient mouse. J Neurosci. 2004;24:11421–11428. doi: 10.1523/JNEUROSCI.2251-04.2004. 10.1523/JNEUROSCI.2251-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen SK, Tvrdik P, Peden E, Cho S, Wu S, Spangrude G, Capecchi MR. Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell. 2010;141:775–785. doi: 10.1016/j.cell.2010.03.055. 10.1016/j.cell.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nayak D, Roth TL, McGavern DB. Microglia development and function. Annu Rev Immunol. 2014;32:367–402. doi: 10.1146/annurev-immunol-032713-120240. 10.1146/annurev-immunol-032713-120240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 100.Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, Wieghofer P, Heinrich A, Riemke P, Holscher C, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci. 2013;16:273–280. doi: 10.1038/nn.3318. 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 101.Swinnen N, Smolders S, Avila A, Notelaers K, Paesen R, Ameloot M, Brone B, Legendre P, Rigo JM. Complex invasion pattern of the cerebral cortex bymicroglial cells during development of the mouse embryo. Glia. 2013;61:150–163. doi: 10.1002/glia.22421. 10.1002/glia.22421. [DOI] [PubMed] [Google Scholar]
- 102.Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 103.Tropepe V, Hitoshi S, Sirard C, Mak TW, Rossant J, van der Kooy D. Direct neural fate specification from embryonic stem cells: a primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron. 2001;30:65–78. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 104.Hemmati-Brivanlou A, Melton DA. Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell. 1994;77:273–281. doi: 10.1016/0092-8674(94)90319-0. [DOI] [PubMed] [Google Scholar]
- 105.Grunz H, Tacke L. Neural differentiation of Xenopus laevis ectoderm takes place after disaggregation and delayed reaggregation without inducer. Cell Differ Dev. 1989;28:211–217. doi: 10.1016/0922-3371(89)90006-3. [DOI] [PubMed] [Google Scholar]
- 106.Di-Gregorio A, Sancho M, Stuckey DW, Crompton LA, Godwin J, Mishina Y, Rodriguez TA. BMP signalling inhibits premature neural differentiation in the mouse embryo. Development. 2007;134:3359–3369. doi: 10.1242/dev.005967. 10.1242/dev.005967. [DOI] [PubMed] [Google Scholar]
- 107.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kiecker C, Lumsden A. The role of organizers in patterning the nervous system. Annu Rev Neurosci. 2012;35:347–367. doi: 10.1146/annurev-neuro-062111-150543. 10.1146/annurev-neuro-062111-150543. [DOI] [PubMed] [Google Scholar]
- 109.Cayuso J, Marti E. Morphogens in motion: growth control of the neural tube. J Neurobiol. 2005;64:376–387. doi: 10.1002/neu.20169. 10.1002/neu.20169. [DOI] [PubMed] [Google Scholar]
- 110.Rubenstein JLR, Rakic P. Comprehensive developmental neuroscience : patterning and cell type specification in the developing CNS and PNS. First edition Elsevier/Academic Press; Amsterdam: 2013. [Google Scholar]
- 111.Maden M. Retinoid signalling in the development of the central nervous system. Nat Rev Neurosci. 2002;3:843–853. doi: 10.1038/nrn963. 10.1038/nrn963. [DOI] [PubMed] [Google Scholar]
- 112.O’Leary DD, Chou SJ, Sahara S. Area patterning of the mammalian cortex. Neuron. 2007;56:252–269. doi: 10.1016/j.neuron.2007.10.010. 10.1016/j.neuron.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 113.Crossley PH, Martinez S, Ohkubo Y, Rubenstein JL. Coordinate expression of Fgf8, Otx2, Bmp4, and Shh in the rostral prosencephalon during development of the telencephalic and optic vesicles. Neuroscience. 2001;108:183–206. doi: 10.1016/s0306-4522(01)00411-0. [DOI] [PubMed] [Google Scholar]
- 114.Ericson J, Muhr J, Placzek M, Lints T, Jessell TM, Edlund T. Sonic hedgehog induces the differentiation of ventral forebrain neurons: a common signal for ventral patterning within the neural tube. Cell. 1995;81:747–756. doi: 10.1016/0092-8674(95)90536-7. [DOI] [PubMed] [Google Scholar]
- 115.Gunhaga L, Marklund M, Sjodal M, Hsieh JC, Jessell TM, Edlund T. Specification of dorsal telencephalic character by sequential Wnt and FGF signaling. Nat Neurosci. 2003;6:701–707. doi: 10.1038/nn1068. 10.1038/nn1068. [DOI] [PubMed] [Google Scholar]
- 116.Hansen DV, Rubenstein JL, Kriegstein AR. Deriving excitatory neurons of the neocortex from pluripotent stem cells. Neuron. 2011;70:645–660. doi: 10.1016/j.neuron.2011.05.006. 10.1016/j.neuron.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, van den Ameele J, Espuny-Camacho I, Herpoel A, Passante L, Schiffmann SN, et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455:351–357. doi: 10.1038/nature07287. 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- 118.Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- 119.Espuny-Camacho I, Michelsen KA, Gall D, Linaro D, Hasche A, Bonnefont J, Bali C, Orduz D, Bilheu A, Herpoel A, et al. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 120.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 121.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mariani J, Simonini MV, Palejev D, Tomasini L, Coppola G, Szekely AM, Horvath TL, Vaccarino FM. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2012;109:12770–12775. doi: 10.1073/pnas.1202944109. 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci U S A. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Krencik R, Zhang SC. Directed differentiation of functional astroglial subtypes from human pluripotent stem cells. Nat Protoc. 2011;6:1710–1717. doi: 10.1038/nprot.2011.405. 10.1038/nprot.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Krencik R, Weick JP, Liu Y, Zhang ZJ, Zhang SC. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat Biotechnol. 2011;29:528–534. doi: 10.1038/nbt.1877. 10.1038/nbt.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maroof AM, Keros S, Tyson JA, Ying SW, Ganat YM, Merkle FT, Liu B, Goulburn A, Stanley EG, Elefanty AG, et al. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu Y, Liu H, Sauvey C, Yao L, Zarnowska ED, Zhang SC. Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nat Protoc. 2013;8:1670–1679. doi: 10.1038/nprot.2013.106. 10.1038/nprot.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D, Arnold CM, Chen YJ, Stanley EG, Elefanty AG, et al. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013;12:573–586. doi: 10.1016/j.stem.2013.04.005. 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu Y, Weick JP, Liu H, Krencik R, Zhang X, Ma L, Zhou GM, Ayala M, Zhang SC. Medial ganglionic eminence-like cells derived from human embryonic stem cells correct learning and memory deficits. Nat Biotechnol. 2013;31:440–447. doi: 10.1038/nbt.2565. 10.1038/nbt.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cambray S, Arber C, Little G, Dougalis AG, de Paola V, Ungless MA, Li M, Rodriguez TA. Activin induces cortical interneuron identity and differentiation in embryonic stem cell-derived telencephalic neural precursors. Nat Commun. 2012;3:841. doi: 10.1038/ncomms1817. 10.1038/ncomms1817. [DOI] [PubMed] [Google Scholar]
- 131.Stacpoole SR, Spitzer S, Bilican B, Compston A, Karadottir R, Chandran S, Franklin RJ. High yields of oligodendrocyte lineage cells from human embryonic stem cells at physiological oxygen tensions for evaluation of translational biology. Stem Cell Reports. 2013;1:437–450. doi: 10.1016/j.stemcr.2013.09.006. 10.1016/j.stemcr.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Billon N, Jolicoeur C, Tokumoto Y, Vennstrom B, Raff M. Normal timing of oligodendrocyte development depends on thyroid hormone receptor alpha 1 (TRalpha1) EMBO J. 2002;21:6452–6460. doi: 10.1093/emboj/cdf662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Brustle O, Jones KN, Learish RD, Karram K, Choudhary K, Wiestler OD, Duncan ID, McKay RD. Embryonic stem cell-derived glial precursors: a source of myelinating transplants. Science. 1999;285:754–756. doi: 10.1126/science.285.5428.754. [DOI] [PubMed] [Google Scholar]
- 134.Amamoto R, Arlotta P. Development-inspired reprogramming of the mammalian central nervous system. Science. 2014;343:1239882. doi: 10.1126/science.1239882. 10.1126/science.1239882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, Watanabe Y, Mizuseki K, Sasai Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat Neurosci. 2005;8:288–296. doi: 10.1038/nn1402. 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- 136.Zeisel A, Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015 doi: 10.1126/science.aaa1934. 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 137.Rossier J, Bernard A, Cabungcal JH, Perrenoud Q, Savoye A, Gallopin T, Hawrylycz M, Cuenod M, Do K, Urban A, et al. Cortical fast-spiking parvalbumin interneurons enwrapped in the perineuronal net express the metallopeptidases Adamts8, Adamts15 and Neprilysin. Mol Psychiatry. 2015;20:154–161. doi: 10.1038/mp.2014.162. 10.1038/mp.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rubin AN, Kessaris N. PROX1: a lineage tracer for cortical interneurons originating in the lateral/caudal ganglionic eminence and preoptic area. PLoS One. 2013;8:e77339. doi: 10.1371/journal.pone.0077339. 10.1371/journal.pone.0077339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26:10967–10983. doi: 10.1523/JNEUROSCI.2572-06.2006. 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ahmed Z, Shaw G, Sharma VP, Yang C, McGowan E, Dickson DW. Actin-binding proteins coronin-1a and IBA-1 are effective microglial markers for immunohistochemistry. J Histochem Cytochem. 2007;55:687–700. doi: 10.1369/jhc.6A7156.2007. 10.1369/jhc.6A7156.2007. [DOI] [PubMed] [Google Scholar]