Abstract
Triple negative breast cancers (TNBC) lacking estrogen, progesterone and HER2 receptors account for 10–20% of breast cancer and are indicative of poor prognosis. The development of effective treatment strategies therefore represents a pressing unmet clinical need. We previously identified a molecularly-targeted approach to target aberrant epigenetics of TNBC using a peptide corresponding to the SIN3 interaction domain (SID) of MAD. SID peptide selectively blocked binding of SID-containing proteins to the paired α-helix (PAH2) domain of SIN3, resulting in epigenetic and transcriptional modulation of genes associated with epithelial-mesenchymal transition (EMT). To find small molecule inhibitor (SMI) mimetics of SID peptide we performed an in silico screen for PAH2 domain-binding compounds. This led to the identification of the avermectin macrocyclic lactone derivatives selamectin and ivermectin (Mectizan) as candidate compounds. Both selamectin and ivermectin phenocopied the effects of SID peptide to block SIN3-PAH2 interaction with MAD, induce expression of CDH1 and ESR1 and restore tamoxifen sensitivity in MDA-MB-231 human and MMTV-Myc mouse TNBC cells in vitro. Treatment with selamectin or ivermectin led to transcriptional modulation of genes associated with EMT and maintenance of a cancer stem cell phenotype in TNBC cells. This resulted in impairment of clonogenic self-renewal in vitro and inhibition of tumor growth and metastasis in vivo. Underlining the potential of avermectins in TNBC, pathway analysis revealed that selamectin also modulated the expression of therapeutically-targetable genes. Consistent with this, an unbiased drug screen in TNBC cells identified selamectin-induced sensitization to a number of drugs, including those targeting modulated genes.
Keywords: selamectin, ivermectin, SIN3A, triple-negative breast cancer
Introduction
Triple negative breast cancer (TNBC), as defined by absence of estrogen receptor (ER) and progesterone receptor (PR) and lack of overexpression of HER2, is an aggressive subtype comprising 10–20% of breast cancer incidences (1). TNBC patients have a shorter median survival time after relapse (~18 month) and more readily develop chemoresistant disease (2). In contrast to advances in the treatment of ER-positive and HER2-positive tumors, TNBC remains a disease with poor prognosis and limited treatment options (3). Molecular profiling suggests that while ~75% of TNBC tumors exhibit a basal-like phenotype (4) and commonly harbor BRCA1 and TP53 mutations (5) the subtype can nevertheless also be considered a heterogeneous entity (6).
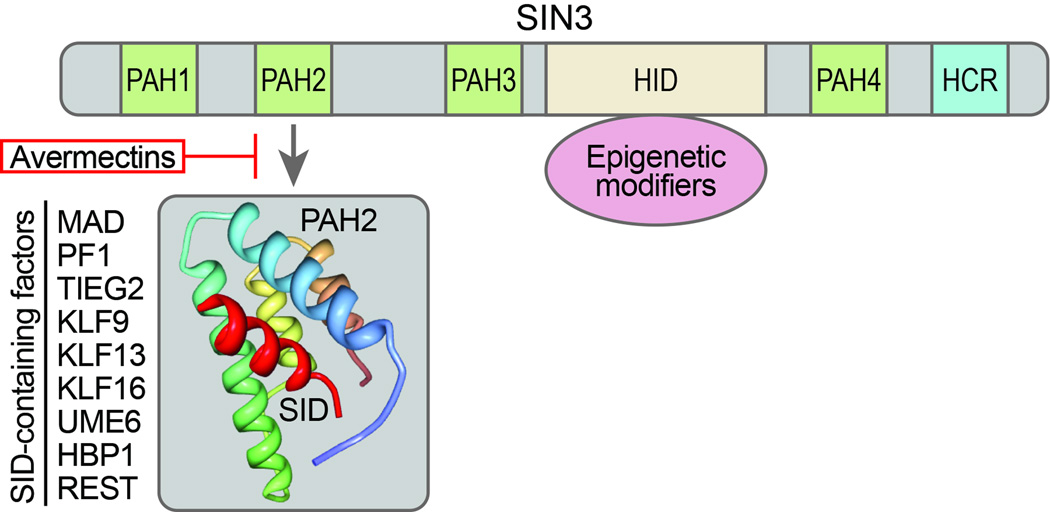
In addition to underlying genetic factors, epigenetics are increasingly recognized as playing an important role in the phenotypic and molecular heterogeneity of TNBC. Aberrant DNA promoter methylation and histone modification can lead to the deregulated expression of key TNBC-associated genes (7, 8). Deregulated epigenetics have also been functionally linked to processes critical to breast cancer tumorigenesis such as epithelial-mesenchymal transition (EMT) that is necessary for the tumor invasion-metastasis cascade and acquisition and maintenance of a cancer stem cell phenotype (9). Epigenetic reconfiguration in cancer cells is brought about by aberrant recruitment of chromatin-modifying complexes that perform diverse activities. Important facilitators of epigenetic deregulation are the SIN3A, which has been implicated breast cancer pathogenesis (10–12), and SIN3B multidomain adaptor proteins. SIN3A and SIN3B, which lack intrinsic DNA binding activity, serve as molecular scaffolds that bridge interactions between chromatin regulators and sequence-specific DNA binding transcription factors. The multiprotein repressor complexes that are generated control cell proliferation and differentiation (13–15). Both SIN3A and SIN3B are characterized by a unique arrangement of four paired amphipathic α-helix (PAH1–PAH4) motifs (Fig. 1). Despite sharing sequence homology, the different PAH domains mediate specific interactions with SIN3A and SIN3B, with the second PAH repeat (PAH2) reported to bind a functionally diverse group of proteins, including the Mad family of repressors, that contain a motif known as a SIN3 interaction domain (SID, Fig. 1) (13).
Figure 1. Schematic representation of SIN3 PAH2 and SID-containing factors.
SIN3A and SIN3B interact with numerous transcriptional factors via highly conserved domains comprising four paired amphipathic α-helices (PAH 1–4), a HDAC interaction domain (HID), bound by many core corepressor components that mediate epigenetic modifications, and a C-terminal highly conserved region (HCR). The molecular structure PAH2 is highlighted together with the SIN3 interaction domain (α-helix SID, red). A list of SID-containing factors is also shown. Interactions between PAH2 and SID-containing factors may be disrupted using SID decoy peptides or avermectins.
We previously reported that inhibition of SIN3A to interact with partner proteins via its PAH2 domain induced differentiation and inhibited tumorigenicity in TNBC (10). This was achieved through the use of a peptide corresponding to the SID domain of MAD, which binds to SIN3-PAH2 and competes with PAH2 partner proteins. In this study we sought to identify small molecule inhibitor (SMI) mimetics of SID. We describe an in silico screen of small molecule drugs, leading to the identification of two members avermectin family of macrocyclic lactones, selamectin and ivermectin as clinical-candidate compounds for the treatment of TNBC.
Materials and Methods
Cell lines and reagents
The mouse metastatic mammary 4T1 tumor cell line (Cat# CRL-2539) and human MDA-MB-231 breast adenocarcinoma cell line (Cat# HTB-26) were purchased from the American Type Culture Collection (ATCC). The MDA-MB-231-Luc-D3H2LN Bioware (D3H2LN) cell line (16) was purchased from PerkinElmer (Cat# 119369). The mouse mammary tumor MMTV-Myc cell line has been previously reported (17, 18). Cell lines were authenticated by short tandem repeat (STR) profiling in accordance with the standard ASN-0002-2011 in April 2015 (DDC Medical, Fairfield, OH). 4T1 cells were maintained in RPMI supplemented with 10% fetal bovine serum (FBS) and 1% Antibiotic-Antimycotic solution (Invitrogen). D3H2LN and MDA-MB-231 cell lines were maintained in DMEM supplemented with 10% FBS, 1% GlutaMAX (Invitrogen), 10mM HEPES, 1mM sodium pyruvate, non-essential amino acids and 1% antibiotic-antimycotic solution. MMTV-Myc cells were cultured in DMEM/F12 medium supplemented with 5% FBS, 1% GlutaMAX, 10mM HEPES, and 1% antibiotic-antimycotic solution. Ivermectin was purchased from Sigma. Selamectin was synthesized in-house.
NMR Spectroscopy
The PAH2 domain of mouse SIN3A was expressed in Escherichia coli BL21 (DE3) cells in the pET22b vector (Novagen) as previously described (19). Isotope-labeled protein was prepared from cells grown on a minimal medium containing 15NH4Cl with or without 13C-glucose in H2O. The protein was purified by nickel-IDA affinity chromatography, followed by thrombin cleavage to remove an N-terminal poly-His-tag. The protein solution for NMR study contained the SIN3A PAH2 domain at 0.1 mM in 100 mM phosphate buffer, pH 6.5, with 5 mM perdeuterated DTT and 10% 2H2O. All NMR spectra were collected at 25oC in a buffer consisting of 50 mM Tris-HCl at pH 8.0, containing 150 mM NaCl, 20% DMSO and 10% 2H2O on NMR spectrometers of 800 or 600 MHz. The 1H, 13C and 15N resonances of the protein were assigned by triple-resonance NMR spectra collected with a 13C/15N-labeled SIN3A PAH2 domain (20).
In Silico Chemical Screening
Computational screening of chemical compounds was conducted using an ensemble of 20 NMR structures of the SIN3A PAH2 domain (19). For the virtual screening, three of these structures were selected: the one whose root-mean-square derivations (RMSDs) with the rest are the smallest (i.e., the structure in the middle of the ensemble) and a pair of structures with RMSDs that were the largest among all pairs (i.e., the two extremes). The screening was performed on a collection of 2000 FDA-approved small molecule drugs (Microsource Discovery Systems). Two programs were used for virtual screening: Autodock-4, combined with AutoDockTools to set up the target structure file (21) and eHiTS (22). The screening and the analysis of the results were driven by the script set Full-screen and the program Dockres (23). Docking was focused on the surface region of the PAH2 domain where the SID-containing protein binds, between the two longer helices near the end of the two shorter helices. The twenty top scoring ligands were tested for their binding to the SIN3A PAH2 domain experimentally by NMR spectroscopy.
GST-pull down assay
The experimental procedures were performed as reported (10). Briefly, GST and GST-SIN3A PAH2 were prepared in Escherichia coli according to the standard procedures. MAD was immunoprecipitated from MDA-MB-231 cell lysates using rabbit polyclonal anti-MAD (C19) antibody (Santa Cruz Biotechnology). Following vehicle, compound or peptide treatment, bait protein (GST or GST-PAH2) and prey protein (immunoprecipitated MAD protein) were pre-incubated separately for 1 hour at 4°C and subjected to GST-pull down assay procedures. The protein was visualized by immunoblotting using mouse monoclonal anti-MAD (F-1) antibody (Santa Cruz Biotechnology). Tat-SID peptide consists the amino acid sequence 5–24 of MAD protein and HIV type I Tat arginine-rich RNA-binding motif as a leader sequence which has been mutated (RRR>GGG) to improve nuclear entry (10).
Immunoblot analysis
Immunoblotting was performed according to standard procedures and visualized using ECL plus Western Blotting detection reagent (Life Technologies). Blots were probed with either anti-ERα antibody (Cell Signaling Technology, #8644) or E-cadherin antibody (Cell signaling technology, #3195). Anti-GAPDH antibody (Ambion, #4300) was used as a loading control.
Mammalian two-hybrid assay
Experiments were performed as previously described (10). Cells were treated with 15 µM SID peptides or 10 µM compounds 24 hour post-transfection. pSID and pSID2MUT, GAL4, pVP16-MAD, pGALO- SIN3B, GAL4UASX5-Tk-luc have been previously described (10).
Duolink Proximity ligation assay
MDA-MB-231 cells were plated onto coverslips in 12 well plates and treated with vehicle, Tat-SID or compound. Cells were stained with monoclonal SIN3A (sc-5299) 1:100 and polyclonal MAD (sc-222) 1:1000 according to the manufacturer’s instructions (Olink Bioscience) except utilizing 1% BSA in PBS as a blocking reagent and carrying out initial washes in PBS. Cells were counterstained in To-pro-3-iodide in PBS, 3×5 min washes at RT and mounted in Vectashield mounting medium (Vector Labs). Images were collected on a Zeiss LSM700 confocal microscope and Duolink software was utilized to quantitate signals.
Immunofluorescence
MDA-MB-231 cells were plated onto coverslips in 12 well plates and treated with vehicle or compound. Cells were stained with monoclonal E-cadherin (67A4) antibody (sc-21791) 1:50 overnight at 4°C according to standard protocols. Secondary antibodies (dilution 1:200 in 1% normal goat serum/PBS) were added for 1 h and then washed. Cells were counterstained in To-pro-3-iodide in PBS, 3×5 min washes at RT and mounted in Vectashield mounting medium (vector labs). Images were collected on a Zeiss LSM700 confocal microscope.
Real-time quantitative PCR (qPCR
RNA was prepared using RNeasy Plus Mini kit (Qiagen). For real-time qPCR, 1µg RNA was reverse-transcribed with SuperScript First-Strand Synthesis System (Invitrogen). 50–250 ng of resulting cDNA were amplified and analyzed in real time with QuantiTect SYBR Green PCR Kit (Qiagen). Results are represented after values were normalized to housekeeping genes (RPL30 or GAPDH) and are presented as fold-differences over control using the ΔΔCt method for relative quantifications. Each comparison was made using triplicate reactions and in at least 3 experiments. Primer sequences are detailed in Supplemental Table S1.
Modified Boyden chamber invasion assay
5×104 cells were seeded onto the top well of a 24-well matrigel-coated filter insert (BD biosciences) and 0.1% FBS was used as chemoattractant in the lower chamber. The numbers of invaded cells were counted after 24 hours, stained with HEMA 3 stain set (Fisher Scientific), and the percentage of invasive cells calculated. A total 5 fields were counted per filter under 400× magnification using a Nikon Eclipse E600 microscope.
Estrogen Receptor Reporter Luciferase assay
Luciferase reporter activities were measured using the Dual Luciferase reporter assay system (Promega) according to the manufacturer’s instructions. To determine ERα transcriptional activity, cells grown in phenol-red free DMEM/F12 growth medium containing 5% charcoal-stripped FBS were pre-treated with 1 µM selamectin (or ivermectin) for 4 days. Cells were split, re-plated and co-transfected with 0.3 µg pGL3-ERE responsive reporter plasmids and 0.1 µg pRL-Tk expressing renilla luciferase (24) prior to vehicle or drug treatment in phenol-red free DMEM/F12 containing 1% charcoal-stripped FBS for 48hrs. Firefly luciferase activity was normalized to renilla luciferase activity and demonstrated as ERα reporter activity.
Tumorsphere formation assay
Cells were pre-treated for 7 days in 2D culture in growth medium. For tumorsphere cultures, cells (10,000 cells per 6 well) were replated in ultra-low attachment plates (Corning Costar) for a further 7 days without treatment. Tumorsphere media contained DMEM/F12 supplemented with B27 growth supplement minus Vitamin A (Invitrogen), recombinant EGF (20ng/ml, Invitrogen) and 1% antibiotic-antimycotic solution (Invitrogen). Cells were counted and spherical colonies with a diameter of greater than 50 µm considered tumorspheres (25). Other cell aggregates were excluded.
Aldehyde dehydrogenase (ALDH) assay
D3H2LN cells were cultured and treated with vehicle or drugs as described for the tumorsphere formation assay. Tumorspheres were dissociated using cell dissociation buffer (Invitrogen) and tested for ALDH activity (2×105 cells/sample), using the Aldefluor assay (Aldegen) according to the manufacturer’s instructions.
In vivo animal studies
All procedures were approved by and performed according to Institutional Animal Care and Use Committee (IACUC) procedures of the Icahn School of Medicine at Mount Sinai. For ex vivo treatments MMTV-Myc cells were pre-treated daily with selamectin (1µM) in vitro for 7 days. After pre-treatment, 200,000 cells were injected into inguinal mammary fat pads of six to eight week-old female transgenic FVB/N mice (Jackson laboratory, n = 10). Tumor diameters were measured with a caliper every other day and volumes (mm3) were calculated according to the formula X = d2 × D/2 (mm3), d = short diameter, D = long diameter. Mice were sacrificed 12 days after cell inoculation. For in vivo treatments, MMTV-Myc cells (50,000 cells per mouse) were injected into the left flank. From next day, mice were treated daily with 1.6mg/kg/day selamectin by intraperitoneal injection for 15 days when tumor volume in the vehicle group reached 1000 mm3. Mice were euthanized using CO2. Tumor volume was calculated as indicated and dissected tumors were weighed. The lungs were removed from mice, fixed with Bouin’s fixative solution so that the number of lung metastases at the surface was counted.
Lung metastasis dissemination studies
BALB/c mice were inoculated subcutaneously with 5×103 4T1 cells (triple negative breast cancer mouse model) (ATCC) into the interscapular space. When the tumors reached ~300 mm3 in volume, established primary tumors were surgically resected under anesthesia/analgesia (ketamine/xylazine) following IACUC guidelines. The day after surgery the mice received vehicle (DMSO) or 3.2 mg/kg/day selamectin by intraperitoneal injection for 30 days. The mice were monitored for signs of cachexia (changes in weight, temperature, fur condition, activity, lethargy, respiratory distress). When signs of cachexia were detected, the mice were euthanized and the lungs were fixed in Bouin’s fixative solution and overt lung metastasis counted.
Expression microarray analysis
Sub-confluent cultures of MDA-MB-231 cells were treated with vehicle or selamectin for 24h. Total RNA was isolated using the ZR RNA MiniPrep Kit (Zymo Research). The concentration and quality of the total RNA was assessed on an Agilent 2100 BioAnalyzer (Agilent Technologies). All samples were normalized to 200ng and processed according to standard Affymetrix protocols using GeneChip WT Terminal Labeling and Controls Kit (Affymetrix) and WT Expression Kit (Ambion). The quality and quantity of labeled cRNA was checked and 750 ng of labeled cRNA were hybridized to a GeneChip Human Gene 1.0 ST Arrays using GeneChip Hybridization, Wash, and Stain Kit (Affymetrix). The arrays were scanned on a GeneChip Scanner 3000 7G. Array data were analyzed by Chipinspector 2.1 (Genomatix) and transcripts were considered to be significantly regulated if at least 3 significant probes mapped to them and the log2 fold change of the transcript calculated from these probes was above 1 or below -1. For all subsequent analyses, the median expression values of two independent biological replicates were used. Replicates were combined exhaustively, i.e. mean fold changes were calculated by comparing each replicate from the treatment group to each replicate from the control group. Log2 fold change values for genes were calculated as the average of the log2 fold change values of the corresponding significantly regulated transcripts and a False Discovery Rate (FDR) was set as 5%. Ingenuity pathway analysis (IPA) software (Qiagen) was used to identify significantly overrepresented pathways, cellular functions and upstream transcription factor analysis in the list of identified proteins. Vehicle versus selamectin treatment expression data were imported into IPA and core analysis was performed to identify the most significantly regulated proteins and associated cellular functions. Expression microarray analysis was performed according to Minimum Information About a Microarray Gene Experiment (MIAME) guidelines and data have deposited on the Gene Expression Ontology (GEO) database with accession number GSE67438.
Drug sensitivity assay
MDA-MB-231 cells (500 cells per well) were plated in 384 well plates and exposed to small molecule inhibitors selamectin (0.5 µM) for five consecutive days after which cell viability in each well was estimated by the use of CellTitre Glo assay (Promega). Luminescence data were log2 transformed, plate median centered and then Z score standardized according to the library median effect and the library median absolute deviation (26). Drug sensitization Z-score values of <−1.5 were considered as representing selamectin sensitization effects.
Statistical analyses
Statistical analyses were performed with GraphPad Prism software (version 5.0). The experiments were conducted with at least three independent experiments unless otherwise mentioned. Where shown, p values were calculated using the unpaired Student’s t-test, Mann-Whitney or one-way ANOVA as indicated.
Results
Avermectins are small molecule mimetics of SID peptide
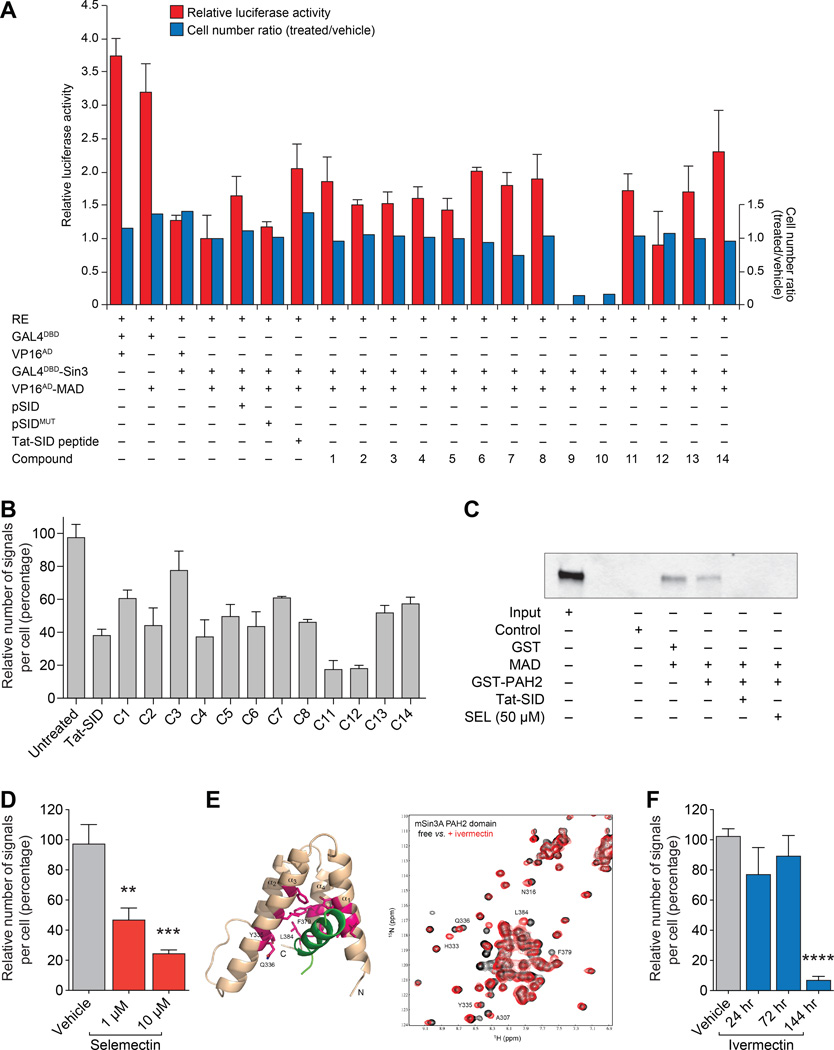
We previously selectively targeted the PAH2 domain of SIN3B corepressor through introduction of a 20-mer peptide fragment comprising amino acids 5–24 of MAD SID. The peptide effectively bound the PAH2 domain and prevented interactions with SID-containing partner proteins, leading to epigenetic reprogramming, re-expression of CDH1 and ESR1, and induction of differentiation in TNBC breast cancer cells (10). To identify small molecule inhibitor (SMI) mimetics of the SID motif, we selected 20 domain structures identified by NMR (19) and performed an in silico screen against the structures of 2,000 FDA-approved drugs. This structure-guided analysis identified 14 candidate compounds (Supplementary Fig. S1), which were screened using two complementary techniques to identify whether they could interfere with binding of MAD to SIN3 PAH2 (Fig. 2A and B). First, we used a mammalian two-hybrid assay as previously described (10) and consistent with previous data (10, 27), co-expression of GAL4DBD-Sin3B and VP16AD-MAD led a 3.7-fold decrease in luciferase activity compared to co-expression of GAL4DBD and VP16AD empty vectors on a reporter under the control of GAL4UAS elements (Fig. 2A). Treatment with Tat-SID peptide led to a 2-fold increase in luciferase activity compared to that found with GAL4DBD-Sin3B and VP16AD-MAD co-expression and the majority of candidate compounds were also found to increase luciferase activity, with the exception of compounds 9 and 10, which were cytotoxic.
Figure 2. Small molecule mimetics of MAD SID block interaction with SIN3 PAH2.
A, Mammalian two-hybrid analysis of 293T cells transfected with GAL4UASX5-Tk-Luc reporter (RE), GAL4DBD-SIN3B and VP16AD-MAD. Cells were treated for 24 hours with 15 µM SID peptide or compounds (10 µM) as indicated. Cells were also co-transfected with pSID or pSIDMUT expressing wild-type or mutated SID peptide, respectively, as indicated. Post-treatment cells were counted and measured. Shown is cell number ratio (treated/vehicle, blue) and luciferase activity relative to renilla control (red). Cell number ratio and relative luciferase activity values were normalized to the value for untreated GAL4DBD-SIN3B and VP16AD-MAD.
B, Proximity ligation assay (PLA) analyzing SIN3A-MAD interactions following treatment for 24 hours with 15 µM Tat-SID peptide or compounds (10 µM) as indicated. Shown are numbers of signals (red dots) per cell relative to untreated control representing SIN3A-MAD interactions (see Supplementary Fig. S2A). Signal generation is dependent on close proximity (< 40 nm) of antibody conjugated PLA probes that have been ligated, amplified and detected with complementary fluorescent probes.
CIn vitro GST pull-down assay of GST-tagged SIN3A-PAH2 (amino acids 306–450) with MAD protein immunoprecipitated from MDA-MB-231 cell lysates in the presence of Tat-SID peptide (15µM) and C14 (selamectin, SEL 50µM). Input is 10% amount used for immunoprecipitation.
D, PLA was performed in MDA-MB-231 cells treated with indicated concentrations of selamectin for 24 hours (1 µM, P = 0.0046; 10 µM, P = 0.0007). See also Supplementary Fig. S2B.
E, Structure-guided discovery of ivermectin (IVM) as a chemical ligand for the SIN3A PAH2 domain. Left, ribbon diagram of the NMR structure of SIN3A PAH2 domain bound to a MAD-SID (green) (PDB: 1G1E). The protein residues with major backbone amide chemical shift perturbations with ivermectin are in magenta. Right2D 15N-HSQC spectra of the 15N-labeled SIN3A PAH2 domain (0.1 mM) in the free form (black) and in the presence of ivermectin (0.2 mM) (red).
F, PLA was performed in MDA-MB-231 cells treated with ivermectin (1 µM) for 24, 72 or 144 hrs as indicated (144 hrs, P < 0.0001). See also Supplementary Fig. S2B.
Error bars represent mean ± SD. P, unpaired t-test.
We also used the proximity ligation assay (PLA) (28) to confirm the ability of the candidate compounds to block SIN3A PAH2-SID interactions. Consistent with the mammalian two-hybrid results, the 12 compounds tested were able to interfere with SIN3A PAH2-SID interactions (Fig. 2B and Supplementary Fig. S2A). Compound 14 (selamectin) is a member of the avermectin family of macrocyclic lactones (29) and another member of this group, ivermectin, has been approved by Food and Drug Administration (FDA) to treat human parasitic diseases, including scabies (30). Selamectin and ivermectin share the same macrocyclic ring and differ only in the number of sugar moieties (31). Based on the activity of selamectin in our SIN3A and SIN3B PAH2-SID interference assays and favorable drug-like qualities of avermectins, these compounds were selected for further study. Consistent with the result from our initial focused screens, selamectin blocked interactions between SIN3A PAH2 and in vitro-translated MAD (Fig. 2C). Moreover, the degree of inhibition of interaction between SIN3A and MAD SID was dose-dependent (Fig. 2D and Supplementary Fig. S2B).
The NMR structure of SIN3A-PAH2 domain bound to MAD SID peptide (green) has previously been resolved (PDB: 1G1E) (19) (Fig. 2E, left panel) and we performed a detailed NMR titration to establish whether ivermectin interacts with PAH2 through residues critical for the PAH2-SID interaction. NMR titration of 15N-labeled PAH2 with ivermectin resulted in shift of signals for a group of amino acids (including Y335, Q336, F379, L384) consistent with the interaction between SIN3A PAH2 and the MAD SID domain (Fig. 2E, right panel). Similar to selamectin, PLA analysis revealed that ivermectin efficiently blocked PAH2-SID interactions, with treatment promoting a 7-fold reduction (Fig. 2F and Supplementary Fig. S2B).
Ivermectin and selamectin upregulate CDH1 and ESR1, inhibit invasion and confer tamoxifen sensitivity in TNBC
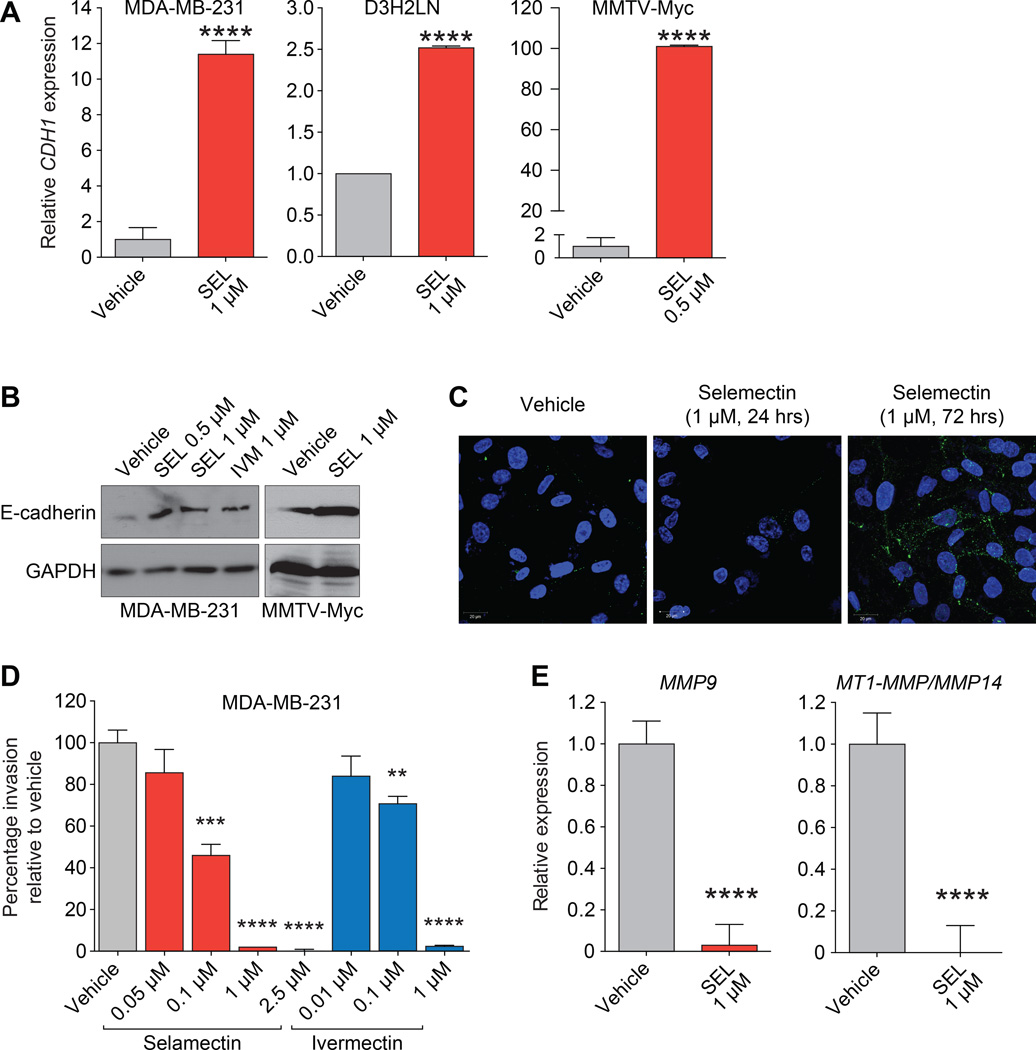
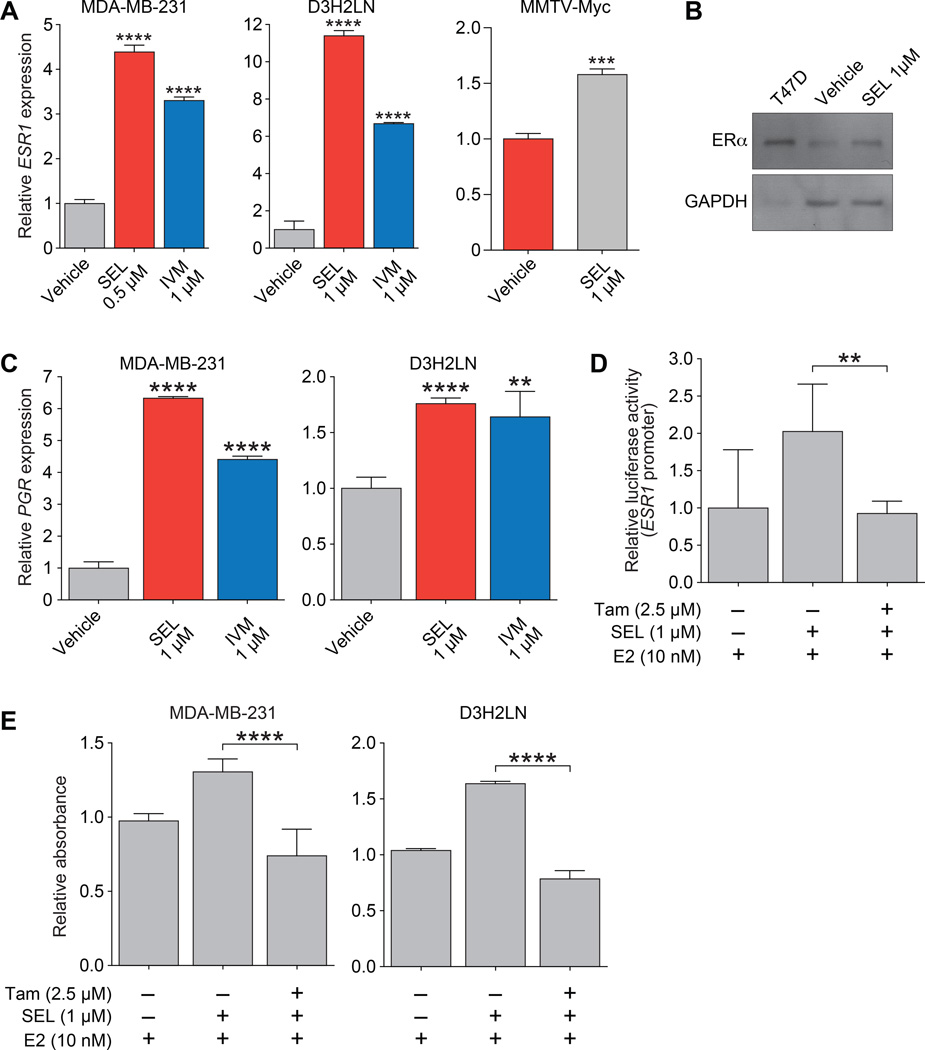
We tested the effect of selamectin and ivermectin on CDH1 and ESR1 expression in the wild-type MDA-MB-231 TNBC cell line as well as the mouse MMTV-Myc cell line (18). We also used the MDA-MB-231-derived reporter cell line (MDA-MB-231-luc-D3H2LN), which possesses greater metastatic potential compared to parental MDA-MB-231 cells (16). Consistent with our previous results with SID peptide (10), treatment with selamectin induced CDH1 mRNA expression by 2.5 to 12-fold in MDA-MB-231 and D3H2LN cell lines, respectively, and by 100-fold in MMTV-Myc cells (Fig. 3A). Upregulation of cytoplasmic and membrane-associated E-cadherin protein was also observed after selamectin or ivermectin treatment in MDA-MB-231 (Fig. 3B and 3C). We previously demonstrated that stable expression of SID inhibited the formation of large and invasive colonies in 3D culture in Matrigel (10) and consistent with these results, both selamectin and ivermectin inhibited invasion in dose-dependent manner (Fig. 3D). Selamectin treatment also resulted in the downregulation in MDA-MB-231 cells of the MMP9 and MT1-MMP/MMP14 genes that are associated with invasion and metastasis (32, 33), (Fig. 3E). Treatment with selamectin also induced ESR1 mRNA expression by 4 to 11-fold, respectively, in MDA-MB-231 and D3H2LN cell lines, and by 1.5-fold in MMTV-Myc cells (Fig. 4A). Ivermectin was also found to increase ESR1 expression in MDA-MB-231 and D3H2LN cell lines by 3-fold and 6-fold, respectively. Consistent with these results, selamectin treatment also increased levels of ERα protein (Fig. 4B). Expression of progesterone receptor (PGR), a direct transcriptional target of ERα, was also increased by between 1.5-fold and 6-fold following treatment with selamectin or ivermectin (Fig. 4C). We next investigated the impact of re-expression of ESR1 on ERα function. Co-treatment with selamectin in the presence of 17β-estradiol (E2), led to a 2-fold increase in ERα activity in a reporter assay, whereas co-treatment with ERα antagonist tamoxifen (Tam) led to a 2-fold reduction ERα activity (Fig. 4D). Again, these results mirror those obtained with SID peptide (10). Additionally, re-expression of ERα in response to selamectin treatment sensitized MDA-MB-231 and D3H2LN cells to the growth promoting or inhibiting effects of E2 or Tam, respectively (Fig. 4E).
Figure 3. Treatment with selamectin or ivermectin leads to upregulation of CDH1 expression in TNBC.
A, qPCR of CDH1 mRNA in MDA-MB-231 (P < 0.0001), D3H2LN (P < 0.0001), and MMTV-Myc (P < 0.0001) cell lines following treatment with selamectin (5 days) as indicated.
B, MDA-MB-231 and MMTV-Myc cells were treated with selamectin or ivermectin as indicated (5 days) and subjected to immunoblot analysis to determine expression of E-cadherin protein.
C, MDA-MB-231 cells were treated with selamectin as indicated and subjected to immunofluorescence analysis to determine expression of E-cadherin protein.
D, MDA-MB-231 cells were plated in transwell invasion filters and treated with indicated concentrations of selamectin or ivermectin. Shown are percentage of cells that traversed Matrigel-coated filters relative to vehicle control (SEL: 0.1 µM, P = 0.0003; SEL: 1 µM, P < 0.0001; 2.5 µM, P < 0.0001. IVM: 0.1 µM, P = 0.0019; SEL: 1 µM, P < 0.0001).
E, qPCR of MMP9 (leftP < 0.0001) and MT1-MMP/MMP14 (rightP < 0.0001) of MDA-MB-231 cells in 3D culture following treatment with selamectin (24 hrs) as indicated.
Error bars represent mean ± SD. P, unpaired t-test.
Figure 4. Ivermectin or selamectin treatment increases ESR1 and PGR expression leading to restoration of tamoxifen and estrogen sensitivity in TNBC cells.
A, qPCR of ESR1 in MDA-MB-231 cells (SEL, P < 0.0001; IVM, P < 0.0001) and D3H2LN (SEL, P < 0.0001; IVM, P < 0.0001) following treatment with selamectin or ivermectin (5 days) as indicated.
B, MDA-MB-231 cells were treated with selamectin (5 days) and subjected to immunoblot analysis to determine expression of ERα protein. Expression of ERα in T47D breast cancer cells is shown as a positive control.
C, qPCR of PGR in MDA-MB-231 cells (SEL, P < 0.0001; IVM, P < 0.0001) and D3H2LN (SEL, P < 0.0001; IVM, P = 0.0021) following treatment with selamectin or ivermectin (5 days) as indicated.
D, MDA-MB-231 cells were pre-treated with selamectin (1µM) for 4 days. Cells were transfected with a luciferase reporter under the control of an estrogen response element, and further treated with selamectin or selamectin plus tamoxifen (Tam) in the presence of 17β-estradiol (E2) for 48 hours (E2 vs. Tam, P = 0.0056). Luciferase activity relative to renilla control (red) is shown.
E, MDA-MB-231 or D3H2LN cells were pre-treated with selamectin (1µM) for 4 days and then further treated with selamectin or selamectin plus Tam in the presence of E2 for 48 hours. Cell viability was determined by MTS tetrazolium assay (E2 vs. Tam: MDA-MB-231, P < 0.0001; D3H2LN, P < 0.0001).
Error bars represent mean ± SD. P, unpaired t-test.
Ivermectin and selamectin target TNBC clonogenic self-renewal to inhibit growth and metastasis
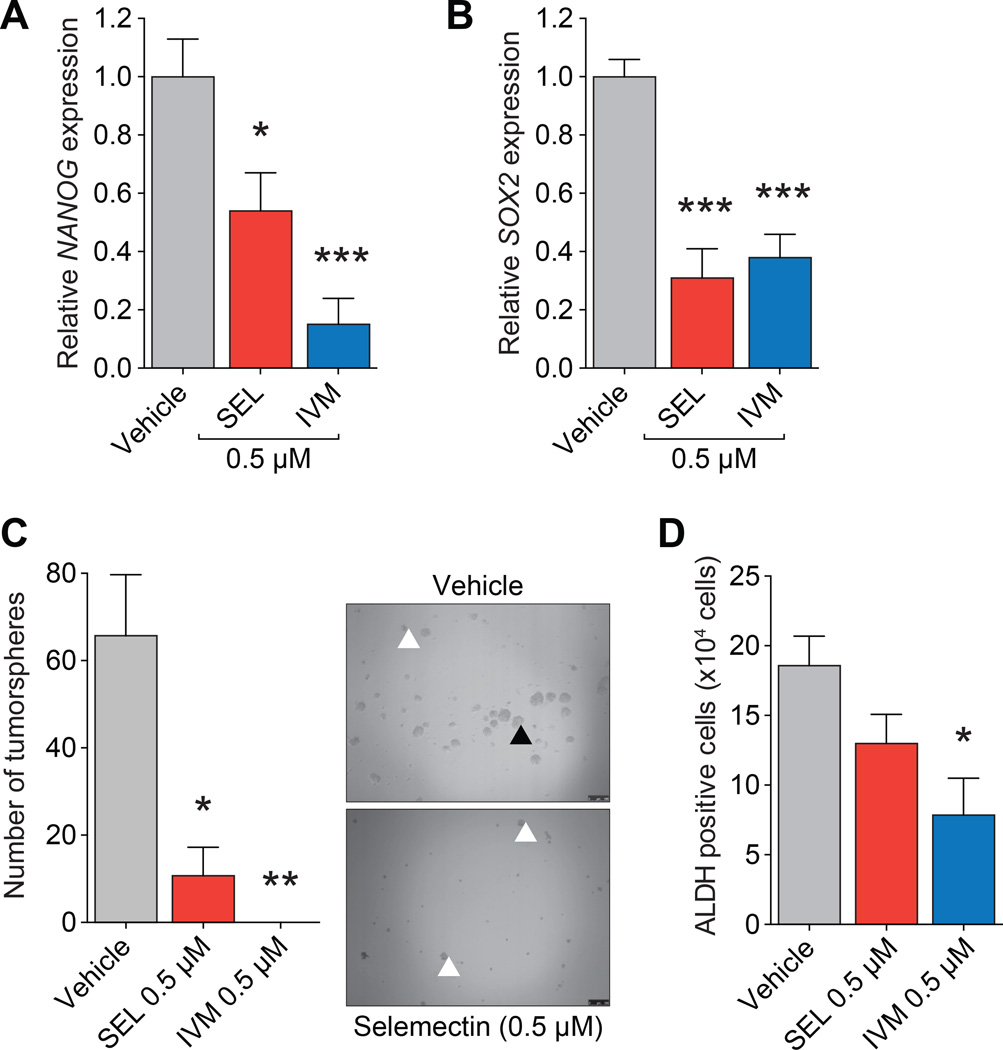
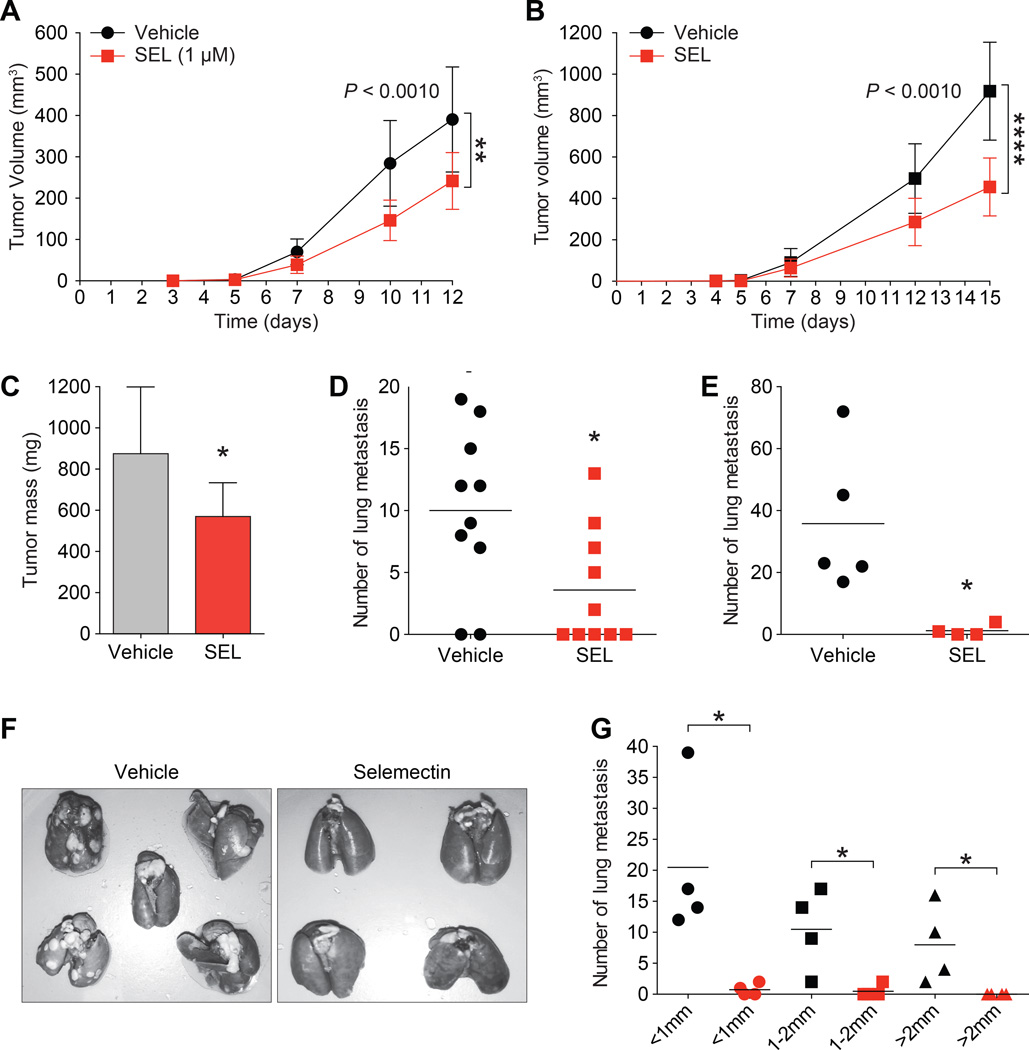
In embryonic stem (ES) cells, SIN3A is a part of a complex that positively regulates NANOG and SOX2, expression of which are hallmarks of stem cell pluripotency and self-renewal (34). Furthermore, both NANOG and SOX2 interact with SIN3A-containing complexes to repress gene targets in ES cells (35). Consistent with this, treatment of D3H2LN cells with 0.5µM of selamectin or ivermectin reduced NANOG (Fig. 5A) and SOX2 (Fig. 5B) gene expression by 50 to 80%. Furthermore, clonogenic self-renewal dependent (32, 33) tumorsphere growth by was diminished by 90–100% (Fig. 5C). Basal-B sub-type cell lines such as MDA-MB-231 have increased ALDH activity, which is another marker of normal and malignant breast stem cells (36). Compared with vehicle-treated cells (18.6×104 ALDH+ cells), reduced ALDH activity was observed after treatment with selamectin (13×104 ALDH+ cells) or ivermectin (7.8×104 ALDH+ cells, P = 0.0149) (Fig. 5D). Our in vitro results could also be recapitulated in vivo. Both in vitro pre-treatment of MMTV-Myc cells with selamectin (Fig. 6A) and in vivo administration of the drug (Fig. 6B and C) led to a significant reduction in volume and mass of orthotopically-implanted tumors. Selamectin treated tumors were also observed to be locally less invasive (not shown). In vivo treatment with selamectin also lead to a dramatic reduction in the number of lung metastases observed (Fig. 6D). Lastly, the effect of selamectin on tumor cell dissemination and metastasis was examined following surgery to remove established tumors arising from implantation of 4T1 metastatic mammary tumor cells and was found to greatly reduce the number of lung metastases under these conditions (Fig. 6E–G).
Figure 5. Ivermectin or selamectin decrease expression and activity of CSC markers in TNBC cells.
A, qPCR of NANOG in D3H2LN cells treated with selamectin or ivermectin (7 days) as indicated (SEL, P = 0.0123; IVM, P = 0.0007).
B, qPCR of SOX2 in D3H2LN cells treated with selamectin or ivermectin (7 days) as indicated (SEL, P = 0.0005; IVM, P = 0.0004).
C, D3H2LN cells were pre-treated with selamectin or ivermectin (7 days) at the indicated concentrations, replated in ultra-low attachment plates and cultured without treatment for a further 7 days. Cells were counted and spherical colonies with a diameter of greater than 50 µm considered to represent tumorspheres (▲). Other cell aggregates (indicated by white arrowheads) were excluded (SEL, P = 0.0117; IVM, P = 0.0033).
D, D3H2LN cells were pre-treated with selamectin or ivermectin (7 days) at the indicated concentrations, replated in ultra-low attachment plates and cultured without treatment for a further 7 days. Cells were collected to determine ALDH activity and results are quantified as number of ALDH+ cells (P = 0.0149).
Error bars represent mean ± SD. P, unpaired t-test.
Figure 6. Treatment with selamectin inhibits TNBC growth and metastasis in vivo.
A, MMTV-Myc mouse mammary tumor cells were pre-treated in vitro with vehicle or selamectin for 7 days. 200,000 cells were inoculated into the inguinal mammary fat pads of FVB/N mice (n = 10 per arm). Tumor volume (mm3) was calculated on the days indicated (P = 0.0017).
B–D, 50,000 MMTV-Myc cells were inoculated into the flanks of FVB/N mice on day 0 and treated with selamectin (1.6 mg/kg/day) for 15 days (n = 10 mice per arm). Tumor volume (B) (P < 0.0001), tumor mass (C) (P = 0.0161), and number of lung metastases (D) (P = 0.0224) in each group were measured.
E–G, 4T1 cells (10,000 per mouse) were inoculated into the flanks of BALB/c mice. Tumors were allowed to grow for 10 days before surgical removal. Treatment was then initiated with selamectin (3.2 mg/kg/day for 30 days, n = 4) or vehicle (n = 5). Animals were sacrificed after 30 days and the total number of lung metastases were measured (E, F) (P = 0.0017). Also measured were number of metastases according to size in each group (G) (< 1mm, P = 0.0198; 1–2mm, P = 0.0235; > 2mm, P = 0.0447).
Error bars represent mean ± SD. P = 0.0224, unpaired t test.
Selamectin treatment alters the expression of genes involved in clinically targetable TNBC pathways, leading to drug sensitization
To further investigate the mechanism of action of selamectin in TNBC, we performed expression microarray analysis at 24 hours to identify genes and pathways upstream of E-cadherin and ERα. Pathway analysis identified downregulation of important markers of epithelial to mesenchymal transition: FGFR2-4, SMAD2, ID2, PIK3CA and WNT5A (Table 1 and Supplementary Table S2) (37–40). Genes involved in estrogen-dependent signaling were modulated (Table 2 and Supplementary Table S3) and upstream regulator analysis also identified activation of ER receptor signaling (Z score, 0.433; P = 1.36E-05) and the β-estradiol pathway (Z score, 0.577; P = 3.77E-04). Other associated pathways that were significantly negatively regulated included fibroblast growth factor (FGF), integrin-linked kinase (ILK) signaling (which plays an important role in silencing of E-cadherin (41)), growth hormone, and HGF cytokine signaling (Table 2 and Supplementary Table S3). Analysis of the genes differentially expressed in the pathways modulated by selamectin treatment revealed four additional genes that have been identified as playing critical roles in TNBC (IGF1R, −1.58 fold; IRS1, −1.52 fold; ITGB3, −1.48 fold; see Supplementary Table S2). Importantly, together with those identified as markers of EMT, these genes encode factors that either sit at the top of clinically relevant receptor tyrosine kinase (RTK) pathways or, as is the case with PIK3CA (encoding PI3 kinase subunit p100α), occupy a central node of control. PI3K is a key player in TNBC pathogenesis and represents a major therapeutic target, and a number of the receptor tyrosine kinases downregulated by selamectin, including IGF1-R (42) and FGF (40), signal through PI3K via RAS (43). The finding that selamectin also downregulates ITGB3 is also significant given the role of integrin αvβ3 in EMT, maintaining a cancer stem cell phenotype and conferring resistance to RTK inhibitors (44, 45).
Table 1.
Genes involved in EMT that are downregulated in MDA-MB-231 cells treated with selamectin
| Gene | Fold change | P value |
|---|---|---|
| SMAD2 | −2.11 | 1.76E-05 |
| ID2 | −1.75 | 1.15E-06 |
| FGFR2 | −1.69 | 4.69E-06 |
| FGFR4 | −1.46 | 6.39E-05 |
| PIK3CA | −1.37 | 8.24E-04 |
| FGFR3 | −1.32 | 9.46E-04 |
| WNT5A | −1.28 | 7.47E-04 |
Table 2.
Pathway analysis for genes differentially expressed in MDA-MB-231 cells treated with selamectin
| Canonical Pathway | Z- score | P value |
|---|---|---|
| Integrin-linked kinase (ILK) signaling | −0.229 | 4.61E-05 |
| PTEN signalling | 0.577 | 2.36E-03 |
| Growth hormone signaling | −1.134 | 5.54E-03 |
| Role of p14/p19ARF in tumor supression | 0.447 | 5.84E-03 |
| Estrogen-dependent breast cancer signaling | NA | 1.06E-02 |
| Hepatocyte growth factor (HGF) signaling | −0.707 | 2.25E-02 |
| Fibroblast growth factor (FGF) signaling | −1.134 | 4.98E-02 |
In parallel, we carried out a sensitivity screen in MDA-MB-231 cells to identify compound to which selamectin treatment conferred sensitization, using a focused library of 80 drugs that are either currently used in the treatment of cancer or are in late stage clinical development. We measured viability in cells exposed to compound alone versus compound in addition to selamectin. Drug sensitization Z scores of less than −1.5 for were considered significant and 23 compounds passed this threshold (Table 3). Consistent with our results, and validating this approach, selamectin caused sensitization to 4-hydroxytamoxifen (4–OHT), an active metabolite of tamoxifen (Z score: −2.93 at 50 nM 4-OHT). In the context of TNBC therapy, other drugs with significant Z-scores included the FGFR inhibitor AZD4547 (Z score: −1.65), RTK inhibitor Sunitinib (Z score: −2.76), PI3K/mTOR inhibitor PF-04691502 (Z score: −2.78) and the IGF-1R inhibitor GSK1904529A (Z score: −2.18). It is notable that many of the drugs to which selamectin conferred sensitivity target genes and pathways modulated by selamectin treatment.
Table 3.
Drug sensitivity screen in MDA-MB-231 cells treated with selamectin
| Drug | Mode of Action | Concentration | DMSO Z | Median DE Z |
|---|---|---|---|---|
| 4-OHT | ERi | 50 nM | 1.15 | −2.93 |
| AZ4547 | FGFRi | 10 nM | 2.26 | −3.67 |
| PD173074 | FGFRi | 10 nM | 1.34 | −2.14 |
| Sunitinib | FGFRi, VEGFR1-3i,PDGFRi, Kiti, CSF1Ri | 100 nM | 1.03 | −2.76 |
| MK2206 | AKTi | 10 nM | 0.64 | −1.57 |
| MLN-4924 | NEDDi | 100 nM | 0.27 | −3.01 |
| PF-04691502 | PI3Ki/mTORi | 1 nM | 2.55 | −2.78 |
| PLX-4720 | BRAFi | 50 nM | −0.19 | −2.92 |
| Sotrastaurin/AEB071 | PKCi | 5 nM | 0.78 | −2.15 |
| PF-00477736 | CHK1i | 0.5 nM | 1.70 | −2.74 |
| Rucaparib/AG-014699 | PARPi | 500 nM | −0.10 | −3.24 |
| GDC-0449 | SMOi | 50 nM | −0.14 | −3.46 |
| XAV-939 | TNKSi/Wnti | 10 nM | 1.53 | −2.30 |
| Resveratrol | NSAID | 10 nM | 0.59 | −1.53 |
| GSK1904529A | IGF-1Ri, IRi | 0.5 nM | 1.36 | −2.18 |
| BMS-911543 | JAK2i | 0.5 nM | 0.70 | −1.88 |
| Crizotonib | c-Meti, ALKi | 5 nM | 2.10 | −2.63 |
| Nilotinib | BCR-ABLi | 50 nM | 0.75 | −2.62 |
Discussion
In this study, we have identified the macrocyclic lactones selamectin and ivermectin as small molecule mimetics of a therapeutic peptide corresponding to the MAD SID motif. We demonstrated that the effects of these drugs on TNBC phenocopy SID peptide and that selamectin treatment as a single agent exhibits inhibitory activity against TNBC tumor growth and metastasis. Work to identify the molecular mechanisms underlying the activity of selamectin or ivermectin in TNBC is ongoing but candidate SIN3A/SIN3B-PAH2 interacting proteins include PF1 (PHF12) and TIEG1 (KLF10). PF1 recruits a complex containing two known breast cancer factors, EMSY and JARID1B (46–48). JARID1B is a H3K4 demethylase and inhibition of this complex could explain, at least in part, the epigenetic remodeling we have observed with SID peptide treatment (10). PF1 also interacts with SIN3B to modify chromatin downstream of transcriptional start sites and mitigate RNA polymerase II progression within transcribed loci (49), although it is unclear whether this plays a role in aberrant gene expression in TNBC. As its name implies, TIEG1 (TGFβ inducible early gene), plays a role in the TGFβ/SMAD signal transduction pathway where it drives expression and activation of SMAD2 (50, 51). A direct link between TIEG1 and TGFβ pathway-associated EMT in TNBC remains to be established but it is notable that expression of SMAD2 is downregulated following selamectin treatment. The identities of DNA or chromatin binding factors that recruit SIN3-containing repressive complexes also remain to be established but candidates include ZEB1 (via CTBP and/or SMARCA4) (52) and components of the DNA methylation machinery such as MECP2, MBD2 or DNMT3B (53–55).
While up to 33% of TNBC patients achieve a pathological complete response following neoadjuvant chemotherapy and have a good prognosis, overall survival for those TNBC patients with residual disease is dramatically worse compared with non-TNBC patients with residual disease (56, 57). A major reason for this discrepancy lies with the inability of TNBC patients to respond to conventional hormone or HER2-directed therapies that improve overall survival in other breast cancer subtypes. Thus, our results demonstrating that selamectin and ivermectin treatment leads to re-expression of ERα and induction of tamoxifen responsiveness have clinical significance. These findings are consistent with HDAC and DNMT inhibition where has been reported that tamoxifen-bound, reactivated ERα recruited the Mi2/NuRD corepressor complex but not the Sin3A corepressor complex to ERα-responsive genes (58). The inactivation of specific Sin3A repressor functions is therefore not predicted to interfere with tamoxifen-induced downstream activities of ERα. Furthermore, given that TNBC patients with metastaic disease relapse very quickly when on chemotherapy (59), the ability of selamectin to inhibit metastasis is particulary exciting. With conventional treatment options delivering poor results in TNBC, there has been much focus in recent years on the development of molecularly-targeted strategies to combat this disease. However thus far, with a few notable exceptions, molecularly-targeted drugs have not been curative or even effective in cancer in general and rational combinatorial approaches are needed to improve outcome (60, 61). Thus, our results showing that selamectin modulates expression of therapeutically relevant genes to confer sensitization to molecularly-targeted drugs in clinical development warrants further investigation.
The results of this study are in line with recent research demonstrating that avermectin treatment at concentrations comparable to those used here inhibited the WNT/TCF pathway, leading to inhibition of colon and lung tumor growth in vitro and in vivo without non-specific cell toxicity (62). Oral ivermectin is already extensively used to treat parasite infections in humans including river blindness (63) with few side effects observed at clinical doses (64, 65). Selamectin and ivermectin do not readily cross the blood–brain barrier (66) and thus far only dogs with mutated ABCB1 (MDR1) and knockout mice for this gene have exhibited neurotoxicity (67) with these drugs. In summary, our results strongly suggests that selective inhibition of SIN3 function using selamectin or ivermectin should be investigated clinically in combinatorial adjuvant and neoadjuvant therapy, both with existing agents such as tamoxifen as well as molecularly-targeted drugs.
Supplementary Material
Acknowledgments
Financial support
This work was funded by National Cancer Institute 1R01CA158121-01, Samuel Waxman Cancer Research Foundation, and Chemotherapy Foundation grants. K. Petrie and L. Howell were supported by a Leukaemia and Lymphoma Research (LLR) UK Specialist Programme Grant (11046). Y. Sbirkov was supported a LLR Gordon Piller Fellowship (10053). V. Gil was supported by a CRUK Project Grant (A12747).
Footnotes
Disclosure of potential conflicts of interest
The authors disclose no potential conflicts of interest.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. The New England journal of medicine. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. doi: 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- 3.Tomao F, Papa A, Zaccarelli E, Rossi L, Caruso D, Minozzi M, et al. Triple-negative breast cancer: new perspectives for targeted therapies. OncoTargets and therapy. 2015;8:177–193. doi: 10.2147/OTT.S67673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2011;16(Suppl 1):61–70. doi: 10.1634/theoncologist.2011-S1-61. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. The Journal of pathology. 2010;220:263–280. doi: 10.1002/path.2648. [DOI] [PubMed] [Google Scholar]
- 7.Roll JD, Rivenbark AG, Sandhu R, Parker JS, Jones WD, Carey LA, et al. Dysregulation of the epigenome in triple-negative breast cancers: basal-like and claudin-low breast cancers express aberrant DNA hypermethylation. Exp Mol Pathol. 2013;95:276–287. doi: 10.1016/j.yexmp.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Kagara N, Huynh KT, Kuo C, Okano H, Sim MS, Elashoff D, et al. Epigenetic regulation of cancer stem cell genes in triple-negative breast cancer. The American journal of pathology. 2012;181:257–267. doi: 10.1016/j.ajpath.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 9.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farias EF, Petrie K, Leibovitch B, Murtagh J, Chornet MB, Schenk T, et al. Interference with Sin3 function induces epigenetic reprogramming and differentiation in breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11811–11816. doi: 10.1073/pnas.1006737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellison-Zelski SJ, Solodin NM, Alarid ET. Repression of ESR1 through actions of estrogen receptor alpha and Sin3A at the proximal promoter. Mol Cell Biol. 2009;29:4949–4958. doi: 10.1128/MCB.00383-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellison-Zelski SJ, Alarid ET. Maximum growth and survival of estrogen receptor-alpha positive breast cancer cells requires the Sin3A transcriptional repressor. Molecular cancer. 2010;9:263. doi: 10.1186/1476-4598-9-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Current genetics. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 14.Grzenda A, Lomberk G, Zhang JS, Urrutia R. Sin3: master scaffold and transcriptional corepressor. Biochimica et biophysica acta. 2009;1789:443–450. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadamb R, Mittal S, Bansal N, Batra H, Saluja D. Sin3: insight into its transcription regulatory functions. Eur J Cell Biol. 2013;92:237–246. doi: 10.1016/j.ejcb.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins DE, Hornig YS, Oei Y, Dusich J, Purchio T. Bioluminescent human breast cancer cell lines that permit rapid and sensitive in vivo detection of mammary tumors and multiple metastases in immune deficient mice. Breast cancer research. 2005;7:R444–R454. doi: 10.1186/bcr1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosch A, Bertran SP, Lu Y, Garcia A, Jones AM, Dawson MI, et al. Reversal by RARalpha agonist Am580 of c-Myc-induced imbalance in RARalpha/RARgamma expression during MMTV-Myc tumorigenesis. Breast cancer research. 2012;14:R121. doi: 10.1186/bcr3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38:627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- 19.Brubaker K, Cowley SM, Huang K, Loo L, Yochum GS, Ayer DE, et al. Solution structure of the interacting domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex. Cell. 2000;103:655–665. doi: 10.1016/s0092-8674(00)00168-9. [DOI] [PubMed] [Google Scholar]
- 20.Clore GM, Gronenborn AM. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods in enzymology. 1994;239:349–363. doi: 10.1016/s0076-6879(94)39013-4. [DOI] [PubMed] [Google Scholar]
- 21.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of computational chemistry. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zsoldos Z, Reid D, Simon A, Sadjad SB, Johnson AP. eHiTS: a new fast, exhaustive flexible ligand docking system. Journal of molecular graphics & modelling. 2007;26:198–212. doi: 10.1016/j.jmgm.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Mezei M, Zhou MM. Dockres: a computer program that analyzes the output of virtual screening of small molecules. Source code for biology and medicine. 2010;5:2. doi: 10.1186/1751-0473-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii Y, Pirkmaier A, Alvarez JV, Frank DA, Keselman I, Logothetis D, et al. Cyclin D1 overexpression and response to bortezomib treatment in a breast cancer model. Journal of the National Cancer Institute. 2006;98:1238–1247. doi: 10.1093/jnci/djj334. [DOI] [PubMed] [Google Scholar]
- 25.Shaw FL, Harrison H, Spence K, Ablett MP, Simoes BM, Farnie G, et al. A detailed mammosphere assay protocol for the quantification of breast stem cell activity. Journal of mammary gland biology and neoplasia. 2012;17:111–117. doi: 10.1007/s10911-012-9255-3. [DOI] [PubMed] [Google Scholar]
- 26.Bajrami I, Kigozi A, Van Weverwijk A, Brough R, Frankum J, Lord CJ, et al. Synthetic lethality of PARP and NAMPT inhibition in triple-negative breast cancer cells. EMBO molecular medicine. 2012;4:1087–1096. doi: 10.1002/emmm.201201250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayer DE, Laherty CD, Lawrence QA, Armstrong AP, Eisenman RN. Mad proteins contain a dominant transcription repression domain. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nature methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 29.Griffin J, Fletcher N, Clemence R, Blanchflower S, Brayden DJ. Selamectin is a potent substrate and inhibitor of human and canine P-glycoprotein. Journal of veterinary pharmacology and therapeutics. 2005;28:257–265. doi: 10.1111/j.1365-2885.2005.00655.x. [DOI] [PubMed] [Google Scholar]
- 30.Nolan TJ, Lok JB. Macrocyclic lactones in the treatment and control of parasitism in small companion animals. Current pharmaceutical biotechnology. 2012;13:1078–1094. doi: 10.2174/138920112800399167. [DOI] [PubMed] [Google Scholar]
- 31.Lespine A, Dupuy J, Alvinerie M, Comera C, Nagy T, Krajcsi P, et al. Interaction of macrocyclic lactones with the multidrug transporters: the bases of the pharmacokinetics of lipid-like drugs. Current drug metabolism. 2009;10:272–288. doi: 10.2174/138920009787846297. [DOI] [PubMed] [Google Scholar]
- 32.Monteiro P, Rosse C, Castro-Castro A, Irondelle M, Lagoutte E, Paul-Gilloteaux P, et al. Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. The Journal of cell biology. 2013;203:1063–1079. doi: 10.1083/jcb.201306162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacob A, Jing J, Lee J, Schedin P, Gilbert SM, Peden AA, et al. Rab40b regulates trafficking of MMP2 and MMP9 during invadopodia formation and invasion of breast cancer cells. Journal of cell science. 2013;126:4647–4658. doi: 10.1242/jcs.126573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baltus GA, Kowalski MP, Tutter AV, Kadam S. A positive regulatory role for the mSin3A-HDAC complex in pluripotency through Nanog and Sox2. The Journal of biological chemistry. 2009;284:6998–7006. doi: 10.1074/jbc.M807670200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nature cell biology. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 36.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell stem cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li K, Yao L, Chen L, Cao ZG, Yu SJ, Kuang XY, et al. ID2 predicts poor prognosis in breast cancer, especially in triple-negative breast cancer, and inhibits E-cadherin expression. OncoTargets and therapy. 2014;7:1083–1094. doi: 10.2147/OTT.S64759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larue L, Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3' kinase/AKT pathways. Oncogene. 2005;24:7443–7454. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 39.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nature reviews Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 40.Katoh M, Nakagama H. FGF receptors: cancer biology and therapeutics. Medicinal research reviews. 2014;34:280–300. doi: 10.1002/med.21288. [DOI] [PubMed] [Google Scholar]
- 41.Hannigan G, Troussard AA, Dedhar S. Integrin-linked kinase: a cancer therapeutic target unique among its ILK. Nature reviews Cancer. 2005;5:51–63. doi: 10.1038/nrc1524. [DOI] [PubMed] [Google Scholar]
- 42.Davison Z, de Blacquiere GE, Westley BR, May FE. Insulin-like growth factor-dependent proliferation and survival of triple-negative breast cancer cells: implications for therapy. Neoplasia. 2011;13:504–515. doi: 10.1593/neo.101590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordon V, Banerji S. Molecular pathways: PI3K pathway targets in triple-negative breast cancers. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:3738–3744. doi: 10.1158/1078-0432.CCR-12-0274. [DOI] [PubMed] [Google Scholar]
- 44.Galliher AJ, Schiemann WP. Beta3 integrin and Src facilitate transforming growth factor-beta mediated induction of epithelial-mesenchymal transition in mammary epithelial cells. Breast cancer research. 2006;8:R42. doi: 10.1186/bcr1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seguin L, Kato S, Franovic A, Camargo MF, Lesperance J, Elliott KC, et al. An integrin beta(3)-KRAS-RalB complex drives tumour stemness and resistance to EGFR inhibition. Nature cell biology. 2014;16:457–468. doi: 10.1038/ncb2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitra D, Das PM, Huynh FC, Jones FE. Jumonji/ARID1 B (JARID1B) protein promotes breast tumor cell cycle progression through epigenetic repression of microRNA let-7e. The Journal of biological chemistry. 2011;286:40531–40535. doi: 10.1074/jbc.M111.304865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vire E, Curtis C, Davalos V, Git A, Robson S, Villanueva A, et al. The breast cancer oncogene EMSY represses transcription of antimetastatic microRNA miR-31. Molecular cell. 2014;53:806–818. doi: 10.1016/j.molcel.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto S, Wu Z, Russnes HG, Takagi S, Peluffo G, Vaske C, et al. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer cell. 2014;25:762–777. doi: 10.1016/j.ccr.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jelinic P, Pellegrino J, David G. A novel mammalian complex containing Sin3B mitigates histone acetylation and RNA polymerase II progression within transcribed loci. Mol Cell Biol. 2011;31:54–62. doi: 10.1128/MCB.00840-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnsen SA, Subramaniam M, Janknecht R, Spelsberg TC. TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene. 2002;21:5783–5790. doi: 10.1038/sj.onc.1205681. [DOI] [PubMed] [Google Scholar]
- 51.Johnsen SA, Subramaniam M, Katagiri T, Janknecht R, Spelsberg TC. Transcriptional regulation of Smad2 is required for enhancement of TGFbeta/Smad signaling by TGFbeta inducible early gene. Journal of cellular biochemistry. 2002;87:233–241. doi: 10.1002/jcb.10299. [DOI] [PubMed] [Google Scholar]
- 52.Sanchez-Tillo E, Lazaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29:3490–3500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 53.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 54.Boeke J, Ammerpohl O, Kegel S, Moehren U, Renkawitz R. The minimal repression domain of MBD2b overlaps with the methyl-CpG-binding domain and binds directly to Sin3A. The Journal of biological chemistry. 2000;275:34963–34967. doi: 10.1074/jbc.M005929200. [DOI] [PubMed] [Google Scholar]
- 55.Geiman TM, Sankpal UT, Robertson AK, Chen Y, Mazumdar M, Heale JT, et al. Isolation and characterization of a novel DNA methyltransferase complex linking DNMT3B with components of the mitotic chromosome condensation machinery. Nucleic acids research. 2004;32:2716–2729. doi: 10.1093/nar/gkh589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384:164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
- 57.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 58.Sharma D, Saxena NK, Davidson NE, Vertino PM. Restoration of tamoxifen sensitivity in estrogen receptor-negative breast cancer cells: tamoxifen-bound reactivated ER recruits distinctive corepressor complexes. Cancer research. 2006;66:6370–6378. doi: 10.1158/0008-5472.CAN-06-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kassam F, Enright K, Dent R, Dranitsaris G, Myers J, Flynn C, et al. Survival outcomes for patients with metastatic triple-negative breast cancer: implications for clinical practice and trial design. Clinical breast cancer. 2009;9:29–33. doi: 10.3816/CBC.2009.n.005. [DOI] [PubMed] [Google Scholar]
- 60.Al-Lazikani B, Banerji U, Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nature biotechnology. 2012;30:679–692. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 61.Balch C, Nephew KP. Epigenetic targeting therapies to overcome chemotherapy resistance. Advances in experimental medicine and biology. 2013;754:285–311. doi: 10.1007/978-1-4419-9967-2_14. [DOI] [PubMed] [Google Scholar]
- 62.Melotti A, Mas C, Kuciak M, Lorente-Trigos A, Borges I, Ruiz IAA. The river blindness drug Ivermectin and related macrocyclic lactones inhibit WNT-TCF pathway responses in human cancer. EMBO molecular medicine. 2014 doi: 10.15252/emmm.201404084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thylefors B. The Mectizan Donation Program (MDP) . Annals of tropical medicine and parasitology. 2008;102(Suppl 1):39–44. doi: 10.1179/136485908X337481. [DOI] [PubMed] [Google Scholar]
- 64.Yang CC. Acute human toxicity of macrocyclic lactones. Current pharmaceutical biotechnology. 2012;13:999–1003. doi: 10.2174/138920112800399059. [DOI] [PubMed] [Google Scholar]
- 65.Pacque M, Munoz B, Poetschke G, Foose J, Greene BM, Taylor HR. Pregnancy outcome after inadvertent ivermectin treatment during community-based distribution. Lancet. 1990;336:1486–1489. doi: 10.1016/0140-6736(90)93187-t. [DOI] [PubMed] [Google Scholar]
- 66.Kokoz YM, Tsyganova VG, Korystova AF, Grichenko AS, Zenchenko KI, Drinyaev VA, et al. Selective cytostatic and neurotoxic effects of avermectins and activation of the GABAalpha receptors. Bioscience reports. 1999;19:535–546. doi: 10.1023/a:1020262811459. [DOI] [PubMed] [Google Scholar]
- 67.Mealey KL, Bentjen SA, Gay JM, Cantor GH. Ivermectin sensitivity in collies is associated with a deletion mutation of the mdr1 gene. Pharmacogenetics. 2001;11:727–733. doi: 10.1097/00008571-200111000-00012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.