Abstract
Streptomyces are aerobic Gram-positive bacteria characterized by a complex life cycle that includes hyphae differentiation and spore formation. Morphological differentiation is triggered by stressful conditions and takes place in a pro-oxidant environment, which sets the basis for an involvement of the oxidative stress response in this cellular process. Characterization of the phenotypic traits of Streptomyces natalensis ΔkatA1 (mono-functional catalase) and ΔcatR (Fur-like repressor of katA1 expression) strains in solid medium revealed that both mutants had an impaired morphological development process. The sub-lethal oxidative stress caused by the absence of KatA1 resulted in the formation of a highly proliferative and undifferentiated vegetative mycelium, whereas de-repression of CatR regulon, from which KatA1 is the only known representative, resulted in the formation of scarce aerial mycelium. Both mutant strains had the transcription of genes associated with aerial mycelium formation and biosynthesis of the hyphae hydrophobic layer down-regulated. The first round of the programmed cell death (PCD) was inhibited in both strains which caused the prevalence of the transient primary mycelium (MI) over secondary mycelium (MII). Our data shows that the first round of PCD and morphological differentiation in S. natalensis is dependent on oxidative stress in the right amount at the right time.
Streptomyces is the largest genus of the Actinobacteria phylum, which constitutes a very robust phylogenetic group of Gram-positive bacteria with high G + C content1. Streptomyces display an aerobic lifestyle and undergo a complex life cycle that comprises hyphae differentiation and formation of resistant unigenomic spores2. Furthermore, streptomycetes are most noticeable for their ability to produce a plethora of secondary metabolites with a wide array of biological activities, e.g. antifungals, anticancer agents or immunosuppressants3.
During the last decade, several studies have demonstrated that the Streptomyces typical life cycle on solid medium is more complex than initially thought. A previously unsuspected highly compartmentalized mycelium was shown to be formed after spore germination. This primary mycelium (MI) displays an active primary metabolism and undergoes an early event of programmed cell death (PCD) at certain hyphae segments4,5. A multinucleated secondary mycelium (MII) arises from the viable segments of MI and starts to grow into and above the solid medium. The passage into this developmental stage is accompanied with the metabolic shift from primary to secondary metabolism and with the triggering of the development program5,6. As a consequence, biosynthetic gene clusters of secondary metabolites are activated and proteins involved in the onset of aerial mycelium are expressed, namely the components of the hydrophobic layer: chaplins, rodlins and the lantipeptide SapB5. In parallel with aerial mycelium formation, the innermost mycelium suffers a second round of PCD, possibly to increase nutrient availability6. Finally, spores are formed through septation of aerial hyphae.
The development program in streptomycetes is orchestrated by BldD that avoids a premature morphological differentiation by repressing the transcription of many development related genes in a manner that depends on cyclic di-GMP-mediated dimerization7,8. Among BldD targets, BldH and BldN play a pivotal role in aerial mycelium formation since they activate the transcription of genes coding for the lantipeptide SapB9,10, chaplins (Chp) and rodlins (Rdl)11.
Aerial mycelium formation occurs in an environment with a high oxygen pressure, which promotes the generation of reactive oxygen species (ROS). Interestingly, it has been demonstrated that ROS may display multiple functions in the development of different microorganisms12. Moreover, the development program in streptomycetes is triggered by environmental stressful conditions (e.g. nutrient scarcity) that are usually accompanied with oxidative stress13,14,15. Thus, although largely uncomprehended, there are evidences pointing out to an active involvement of oxidative stress in Streptomyces development.
Streptomyces possess a fine-tuned oxidative stress response, which likely evolved as a response to its aerobic lifestyle. Typically, streptomycetes possess several H2O2-sensitive transcription regulators, namely CatR (Fur-like), OxyR (LysR-family) and the sigma/anti-sigma factors SigR/RsrA that control the expression of the catalase KatA1, the alkylhydroperoxide reductase system AhpCD and the thioredoxin system TrxAB, respectively16,17,18; an organic hydroperoxide-sensitive transcription regulator OhrR (MarR-family) that controls the organic peroxide resistance proteins OhrABC19; and the nickel-responsive transcription regulator Nur (Fur-like) that governs the expression of iron- and nickel-superoxide dismutases SodF and SodN20.
Previously we have shown that the deletion of genes encoding enzymes involved in the oxidative stress response, namely the alkylhydroperoxide reductase system (ahpCD), the mono-functional catalase (katA1) and the superoxide anion scavenging enzyme superoxide dismutase (sodF), led to an effective modulation of intracellular ROS levels in Streptomyces natalensis that resulted in a modulation of secondary metabolism in liquid medium21. Unlike the other mutant strains, ΔkatA1 presented an impaired aerial mycelium formation, i.e. it displayed a bald phenotype. In this work we generated a S. natalensis defective mutant on the katA1 transcription repressor encoding gene catR that displayed a severe, but not complete, impairment of aerial mycelium formation. Characterization of S. natalensis ΔcatR and ΔkatA1 strains on solid medium provided, for the first time, evidences regarding the influence of oxidative stress over Streptomyces morphological differentiation.
Results
Genes associated with morphological development and oxidative stress response in S. natalensis ATCC 27448
The S. natalensis ATCC 27448 genome sequence (GenBank Accession Number: JRKI01000000) was analysed for the presence of genes associated with Streptomyces morphological differentiation (reviewed in Claessen, et al.22, Flardh and Buttner2 and McCormick and Flardh23). Orthologues for the most important development genes of the model organism S. coelicolor are present in the S. natalensis genome (summarized in Supplementary Table S1).
S. natalensis ATCC 27448 harbours homologues to genes related to the formation of aerial mycelium (e.g. bldD, bldH and bldN), biosynthesis of the aerial hyphae hydrophobic layer, cell division and chromosome segregation (e.g. whiA, whiB and ftsZ) and spore maturation (e.g. the whiE locus). Regarding proteins related to aerial mycelium hydrophobic layer, S. natalensis possesses two long chaplins (SNA_06310 and SNA_01235), seven short chaplins (SNA_00490, SNA_12255, SNA_11245, SNA_11250, SNA_00485, SNA_05975 and SNA_12040) and one rodlin encoding gene (SNA_27905), differing from S. coelicolor that presents three long chaplins, five short-chaplins and two rodlin encoding genes. S. natalensis lantipeptide SapB biosynthetic genes (ram genes) are arranged into a cluster, similarly to what is observed in S. coelicolor and other streptomycetes. Finally, it is noteworthy that the whiE locus coding for the grey polyketide spore pigment in S. natalensis displays a high degree of synteny with its S. coelicolor counterpart.
Regarding genes associated with the oxidative stress response, particularly those related to H2O2 detoxification, in addition to the previously described H2O2-inducible mono-functional catalase katA1 (SNA_34325), its Fur-like repressor catR (SNA_34330), the alkyl hydroperoxide reductase system (ahpCD, SNA_35115 and SNA_35120) and its H2O2 responsive regulator oxyR (SNA_35110)21, we have identified an additional Clade 3 mono-functional catalase katA3 (SNA_29075) and a bi-functional catalase-peroxidase encoding gene cpx (SNA_19710) in S. natalensis.
The knock-out mutants for catalase and Fur-like repressor CatR present impaired aerial mycelium formation
In a previous study, we generated a S. natalensis mutant defective on the mono-functional catalase KatA1 that displayed an impaired morphological differentiation in solid medium21. To investigate the effect of KatA1 activity in the development program of S. natalensis, we further analysed the ΔkatA1 strain and a knock-out mutant on the katA1 transcription repressor, CatR. S. natalensis ΔcatR::aac(3)IV-oriT mutant strain was generated by conjugation using a PCR targeting procedure that replaced the native catR locus with an apramycin resistance cassette. Several exconjugants were obtained that displayed the same phenotype. Three exconjugants were randomly selected and their identity confirmed by PCR and Southern blot hybridization; one exconjugant was randomly selected for subsequent studies and named S. natalensis ΔcatR (Supplementary Fig. S1).
Morphological development of the S. natalensis wild-type, ΔkatA1 and ΔcatR strains was characterized in R5 medium growing as mycelium lawns and as isolated colonies (Fig. 1). In the wild-type mycelium lawn, aerial mycelium was first observed at 48 h after inoculation (data not shown) and at 72 h was fully formed (Fig. 1A). ΔcatR strain development was delayed when compared to the wild-type forming a scarce aerial mycelium at 72 h, which did not further develop in subsequent days (data not shown). Moreover, ΔcatR accumulated a dark-blue pigment that might correspond to the blue-coloured pigment previously observed in S. natalensis ΔsngA liquid cultures24. ΔkatA1 did not form aerial mycelium at any time point, i.e. it displayed a bald phenotype (Fig. 1A). The phenotypes of isolated colonies were similar to those observed in mycelium lawns. However, isolated colonies allowed us to notice that the mutant strains vegetative mycelium presented a higher growth rate, leading to the formation of larger colonies than in the wild-type strain (notice the differences in the scale bars in Fig. 1B). This was particularly visible in ΔkatA1, which completely lacked the typical wild-type colony structure (Fig. 1B).
Figure 1. Morphological phenotypes and mycelium proliferation assay of S. natalensis wild-type, ΔkatA1 and ΔcatR strains grown in R5 solid medium.
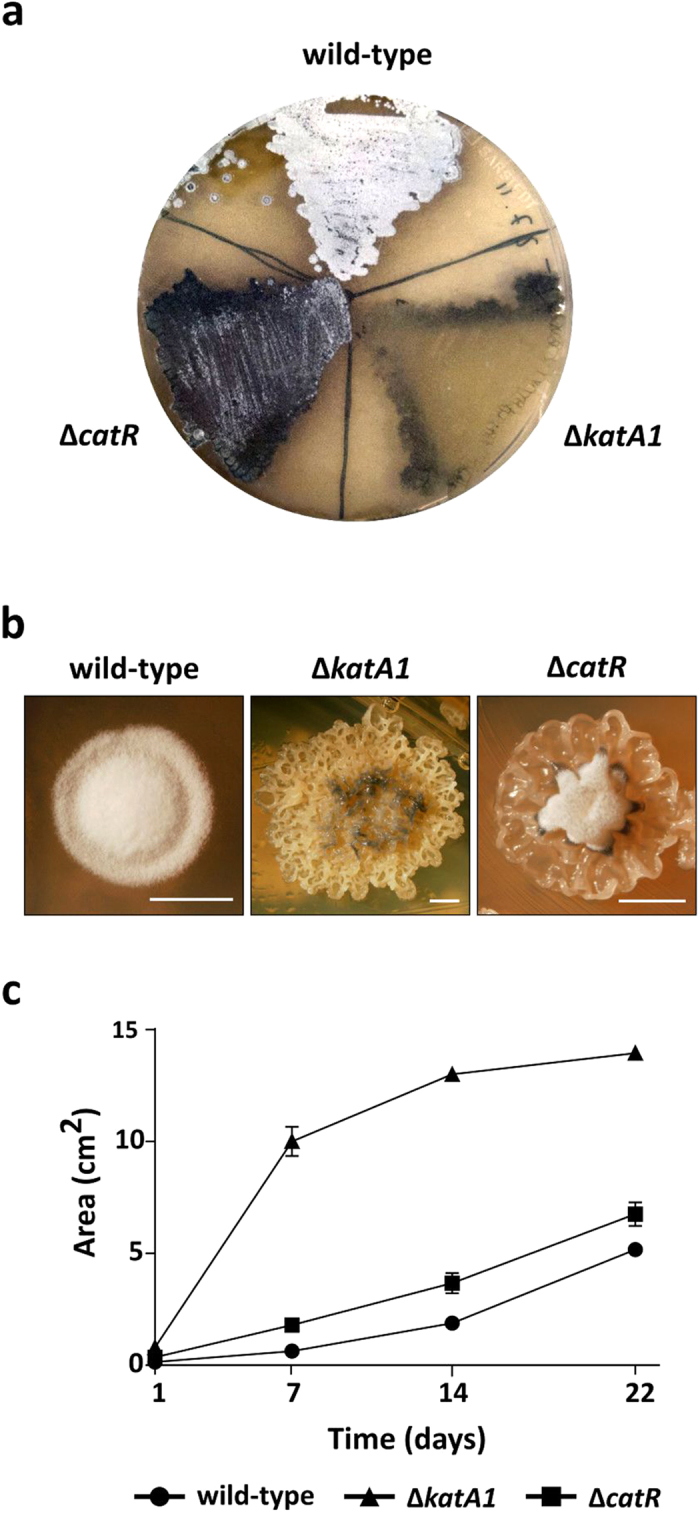
(a) Photographs of mycelium lawns at 72 h. (b) Photographs of isolated colonies at 72 h. Scale bar: 2,50 mm. Photographs by Tiago Beites. (c) Mycelium proliferation rate in R5 solid medium. 10 μl drops of liquid cultures grown to an OD600nm of 4–5 were placed in the middle of R5 plates and mycelium proliferation was measured for 22 days. R5 plates were photographed and the mycelium area was determined using the measure function of the ImageJ software. Results are representative of three independent experiments.
To characterize the vegetative mycelium growth rate, we assessed the rate of mycelium proliferation (Fig. 1C). With this purpose, a 10 μl drop of YEME liquid cultures grown to an OD600nm of 4–5 was inoculated in the centre of R5 plates and mycelium proliferation was followed for 22 days by measuring the vegetative mycelium area. Both mutant strains presented a higher rate of mycelium proliferation when compared with the wild-type, in particular ΔkatA1 strain that at day 22 occupied a 2.7-fold larger area than the wild-type (Fig. 1C).
Key development genes are down-regulated in ΔkatA1 and ΔcatR
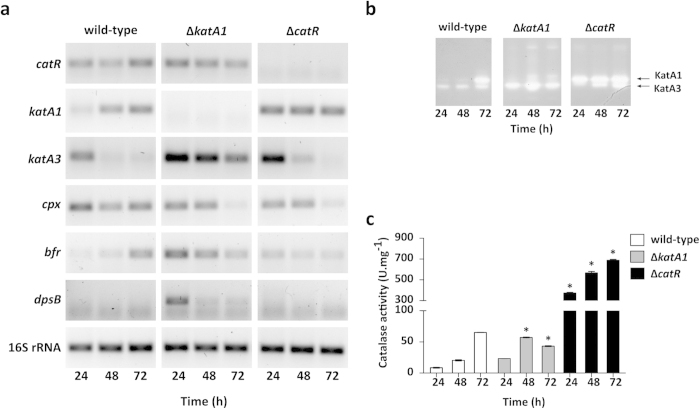
To further characterize the morphological impairment phenotype displayed by the mutant strains in solid medium we examined the transcription of genes associated with the onset of aerial mycelium (bldD, bldH and bldN) and formation of the aerial hyphae hydrophobic coat (ramS, ramC, chpC, and rdlA) at 24 h (vegetative growth), 48 h and 72 h (formation of aerial mycelium) (Fig. 2). No major differences were observed for bldD expression under the conditions tested. However, ΔkatA1 strain displayed a progressive decrease of bldH transcript levels in contrast with the wild-type and ΔcatR strains that presented a constitutive expression. Interestingly, the transcription of ramC and ramS was down-regulated in both mutant strains when compared to the wild-type, pointing out to a deficient SapB production, particularly in ΔkatA1 strain. In addition, the poor correlation between the transcription of bldH and ram genes, especially in ΔcatR strain, suggests the presence of additional players in the regulation of SapB biosynthesis in S. natalensis.
Figure 2. Transcription analysis by RT-PCR of genes associated with the development program in S. natalensis wild-type, ΔkatA1 and ΔcatR strains.

The used pair of primers and annealing temperatures are presented in Supplementary Table S2. The amplicons are the result of 30 PCR cycles. Transcription profiles are representative of three independent experiments.
The transcription of bldN, chpC and rdlA was down-regulated in ΔkatA1 and, to a lesser extent, in ΔcatR when compared to the wild-type. These results suggest a defective synthesis of chaplins and rodlins in the mutant strains.
The oxidative stress defences are activated at early developmental stages in ΔkatA1 and ΔcatR
We also examined the transcription of genes associated with the oxidative stress response in particular the catalase encoding genes (katA1, katA3 and cpx) and the Fur-like repressor catR at 24 h (vegetative mycelium), 48 h and 72 h (formation of aerial mycelium) (Fig. 3A).
Figure 3. Characterization of oxidative stress response in S. natalensis wild-type, ΔkatA1 and ΔcatR strains.
(a) Transcription analysis of genes associated with the oxidative stress response by RT-PCR. The used pair of primers and annealing temperatures are presented in Supplementary Table S2. The amplicons are the result of 30 PCR cycles. The transcription profiles are representative of three independent experiments. (b) Catalase activity determined by native-PAGE. 50 μg of total protein per lane were loaded and separated by electrophoresis before in-gel zymography. Arrows indicate bands displaying enzymatic activity. (c) Total catalase activity in protein crude extracts. Results (average of triplicates and standard deviation) are representative of three independent experiments. Statistically significant differences to the wild-type strain at each time point were determined by one-way ANOVA followed by post hoc test (Tukey test; GraphPad Prism). * p < 0.01.
The wild-type strain displayed a growth stage-dependent transcription of the two mono-functional catalases: while katA3 was preferably expressed before the onset of aerial hyphae (24 h), katA1 transcription was temporally correlated with aerial mycelium formation (48 h and 72 h). The increasing expression of KatA1 reflected in a progressive increase in the KatA1 activity band as assessed by native-PAGE (Fig. 3B) and total catalase activity (Fig. 3C). In ΔkatA1 strain, katA3 transcription was up-regulated at all time points when compared to the wild-type (Fig. 3A). This profile was reflected in a high intensity of KatA3 activity band (Fig. 3B) and a peak at 48 h of total catalase activity levels (Fig. 3C). Regarding ΔcatR strain, mutated in the repressor of katA1 (catR), the constitutive over-expression of katA1 (Fig. 3A) and the consequent induction of KatA1 activity (Fig. 3B,C) was an expected outcome. Regarding the transcription pattern of the catalase-peroxidase encoding gene cpx, there was no clear correlation with morphological development. Nevertheless, both mutants presented a down-regulation of cpx transcription at 72 h.
Toxicity of H2O2 is intimately associated to the availability of free ferrous iron (Fe2 + ) due to the formation of hydroxyl radicals in the so-called Fenton reaction. Iron storage proteins such as bacterioferritins and Dps, play an important role on the oxidative stress response due to their ability to chelate intracellular ferrous iron and circumvent free iron-driven intracellular toxicity25. The in silico analysis of S. natalensis genome revealed two proteins harbouring a ferritin domain (Pfam domain PF00210), SNA_09350 and SNA_32055, orthologues to the S. coelicolor bacterioferritin Bfr and DpsB protein, respectively. Analysis of bfr expression revealed a clear up-regulation in ΔkatA1 at 24 h and 48 h. In addition, ΔkatA1 was the only strain in which transcripts of dpsB could be detected under the conditions tested (24 h).
These results show that ΔkatA1 and ΔcatR mutant strains have elements of the oxidative stress response system induced at earlier stages of the development program when compared to the wild-type. The expression of iron storage proteins in S. natalensis ΔkatA1 indicates that KatA3 activity is not sufficient to counteract the lack of KatA1 concerning H2O2 detoxification and it is safe to assume that ΔkatA1 endures a sub-lethal oxidative stress. Regarding ΔcatR strain, induction of catalase activity is the consequence of catR deletion and the consequent derepression of katA1 transcription.
ΔkatA1 and ΔcatR presented an extended primary metabolism and inhibition of development-related PCD
Total protein extracts of the wild-type, ΔkatA1 and ΔcatR strains grown in solid medium for 72 h were analysed by 2D-PAGE (Supplementary Fig. S2). Analysis of the 2D gels revealed a total of 353 valid protein spots. The mutant strains ΔkatA1 and ΔcatR presented 61 and 33 spots with significant differences when compared with the wild-type, respectively (P > 0.05; two-fold change). From these, 41 well-individualized spots were analysed by mass spectrometry (PMF + MSMS). Excluding the spots that were identified as isoforms of the same protein and spots with mixture of proteins, we have successfully identified 26 individual proteins (Table 1; Supplementary Fig. S2).
Table 1. List of identified proteins in the 2D-PAGE proteome comparison between the S. natalensis mutant strains ∆katA1 and ∆catR with the wild-type.
| SNA code (spot ID§) | Description | ΔkatA1 vs wt (fold change) | ΔcatR vs wt (fold change) |
|---|---|---|---|
| Ribosome constituents | |||
| SNA_38815 (4) | 50S ribosomal protein L7/L12 | Up (*) | Absent |
| Chaperones | |||
| SNA_03365 (6) | molecular chaperone DnaK | Down (0.5) | Down (# 0.5) |
| Protection responses | |||
| SNA_07730 (11) | organic hydroperoxide resistance reductase B | Up (*) | Up (*) |
| SNA_31980 (37) | recombinase A | Up (*) | Absent |
| SNA_34325 (28) | catalase | Absent | Up (27) |
| Macromolecule degradation | |||
| SNA_02830 (39) | aminopeptidase | Up (*) | Absent |
| SNA_12425 (34) | serine protease 2 | Absent | Up (*) |
| SNA_32135 (30) | RNA-metabolising metallo-beta-lactamase | Up (*) | Absent |
| SNA_32175 (10) | guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase | Absent | Absent |
| Aminoacid biosynthesis | |||
| SNA_33095 (21) | acetolactate synthase | Up (8.8) | Absent |
| Biosynthesis of cofactors, carriers | |||
| SNA_36475 (24) | bifunctional 5,10-methylene-tetrahydrofolate dehydrogenase/5,10-methylene-tetrahydrofolate cyclohydrolase | Up (3.8) | Down (# 0.4) |
| Central intermediary metabolism | |||
| SNA_09155 (8) | ribokinase | Down (0.4) | Down (# 0.7) |
| Degradation of small molecules | |||
| SNA_17045 (40) | glyoxylate carboligase | Up (*) | Absent |
| SNA_33060 (12) | 1-pyrroline-5-carboxylate dehydrogenase | Down (0.3) | Down (0.2) |
| Energy metabolism, carbon | |||
| SNA_07865 (33) | pyruvate dehydrogenase E1 | Up (*) | Absent |
| SNA_09035 (35) | dihydrolipoamide acyltransferase | Absent | Up (*) |
| SNA_09040 (27) | dihydrolipoamide dehydrogenase | Up (*) | Up (*) |
| SNA_10650 (22) | glucose 6-phosphate dehydrogenase | Up (*) | Absent |
| SNA_12565 (25) | alanine dehydrogenase | Up (*) | Absent |
| SNA_33550 (19) | pyruvate kinase | Up (5.7) | Up (# 1.5) |
| Nucleotide biosynthesis | |||
| SNA_12580 (31) | CTP synthetase | Up (*) | Absent |
| SNA_13390 (38) | carbamoyl phosphate synthase small subunit | Up (*) | Absent |
| SNA_36490 (18) | purine biosynthesis protein | Up (*) | Absent |
| Unknown function | |||
| SNA_01165 (17) | Hypothetical protein | Up (*) | Up (*) |
| SNA_17125 (36) | superfamily DUF2263 | Up (# 3.8) | Up (5.6) |
| SNA_27255 (23) | Hypothetical protein | Down (# 0.7) | Down (0.4) |
Proteins were grouped according to the functional categorization available for S. coelicolor genome (ftp://ftp.sanger.ac.uk/pub/ project/pathogens/S_coelicolor).
*Not detected in the wild-type.
#Not statistically significant different (P < 0.05).
§Spot ID refers to protein spot numbers in Supplementary Fig. S2.
The set of over-expressed proteins in ΔkatA1 when compared to the wild-type, was majorly associated with energy metabolism (SNA_07865, SNA_09040, SNA_10650, SNA_12565 and SNA_33550) and anabolic processes, such as the biosynthesis of co-factors (SNA_36475), amino acids (SNA_33095) and nucleotides (SNA_12580, SNA_13390 and SNA_36490), as well as protein synthesis (SNA_38815). These findings suggest that, at 72 h, ΔkatA1 maintained active metabolic pathways associated with anabolic processes and energy production. Although at a less extent, we have also identified over-expressed proteins in ΔcatR related with energy metabolism (SNA_09035, SNA_09040 and SNA_33550).
The set of proteins identified in the wild-type strain that were down-regulated or even not detected in the ΔkatA1 and ΔcatR protein extracts included the molecular chaperon DnaK (SNA_03365), the proline catabolism enzyme pyrroline-5-carboxylate dehydrogenase (SNA_33060), the pentose phosphate pathway (PPP) enzyme ribokinase (SNA_09155) and the polyribonucleotide nucleotidyltransferase PNPase (SNA_32175). Also, it is noteworthy the absence of PNPase in the mutant strains proteomes; this nuclease was previously shown to be expressed during the developmentally related programmed cell death (PCD) process in S. coelicolor26.
Finally, stress-related over-expressed proteins were also identified in the protein extracts of ΔkatA1 and ΔcatR namely the organic hydroperoxide resistance reductase B (SNA_07730) in both mutant strains.
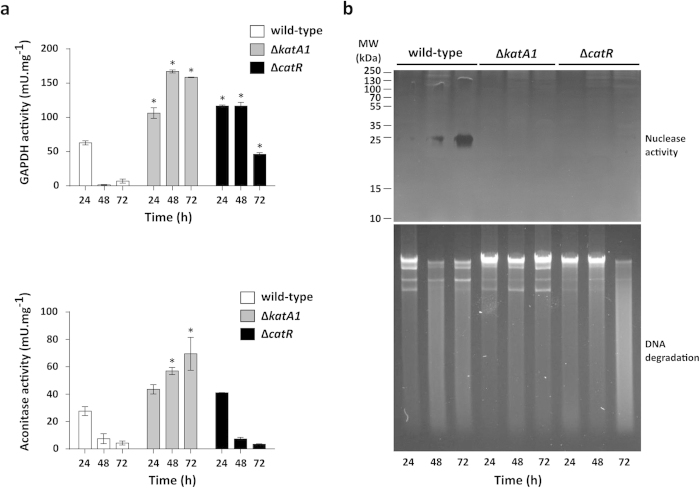
Overall, this proteome analysis suggested that the mutant strains, particularly ΔkatA1 strain, displayed an active primary metabolism at 72 h and that the PCD process was inhibited. To further confirm these results we determined the enzymatic activities of central carbon metabolism proteins throughout growth in solid medium, namely the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and the tricarboxylic acid (TCA) cycle enzyme aconitase (Acn) (Fig. 4A). In addition, we have also characterized two PCD markers: intracellular nuclease activity and DNA degradation (Fig. 4B).
Figure 4. Assessment of primary metabolism enzyme activities and programmed cell death markers in S. natalensis wild-type, ΔkatA1 and ΔcatR strains.
(a) Determination of GAPDH (top panel) and aconitase (bottom panel) enzymatic activities in protein crude extracts. Results (average of triplicates and standard deviation) are representative of three independent experiments. Statistically significant differences between strains at each time point were assessed by one-way ANOVA followed by post hoc test (Tukey test; GraphPad Prism). * p < 0.01. (b) The upper panel depicts nuclease activity in protein crude extracts (20 μg of protein per lane); the SDS-PAGE performed with the protein extracts was used as loading control (Supplementary Fig. S3). The lower panel shows the integrity of total genomic DNA in a 0.8% (w/v) agarose gel (1 μg of total DNA per lane).
The GAPDH and Acn activity profiles in the wild-type presented a pronounced decrease from 24 h to 48 h, which overlapped with the onset of aerial hyphae formation (Fig. 4A). Conversely, the enzymatic activities of GAPDH and Acn in ΔkatA1 protein crude extracts were significantly higher than the wild-type throughout growth (Fig. 4A). This profile further supports the idea that ΔkatA1 experiences a highly active primary metabolism throughout growth. Finally, S. natalensis ΔcatR presented significantly higher GAPDH activity levels when compared with the wild-type during all growth stages, albeit a down-shift on its activity was also observed upon aerial mycelium emergence (72 h). Acn activity profile was similar to that of the wild-type (Fig. 4A).
Regarding PCD markers, results have shown that the wild-type protein extracts displayed a single nuclease activity band (approx. 25 kDa) with increasing intensity throughout time (Fig. 4B). Noteworthy, S. coelicolor protein extracts display a nuclease activity band associated with PCD that presents a similar molecular weight26. No nuclease activity was detected in the mutant strains protein extracts (Fig. 4B). Accordingly, DNA was degraded at 48 h and 72 h in the wild-type whilst DNA integrity was maintained throughout growth in the ΔkatA1. For ΔcatR strain, DNA degradation was detected at 72 h (Fig. 4B).
ΔkatA1 development is arrested at MI stage and ΔcatR presents an extended life-span of MI
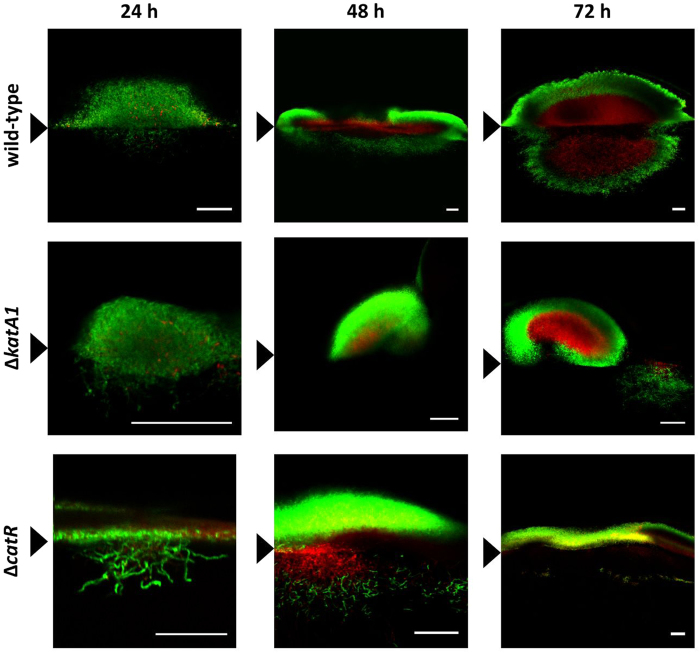
Morphological differentiation of the ΔkatA1 and ΔcatR mutants and S. natalensis wild-type strain was characterized in solid cultures by confocal microscopy, making use of a live/dead cell specific staining protocol (Figs 5 and 6A).
Figure 5. S. natalensis wild-type, ΔkatA1 and ΔcatR strains colony development by confocal microscopy and using a cell viability assay.
The presented micrographs are the result of merging the red (dead cells) and green (live cells) channels with ImageJ software. Black arrowheads indicate the medium-air interface. The results are representative of three independent experiments. Scale bar: 40 μm.
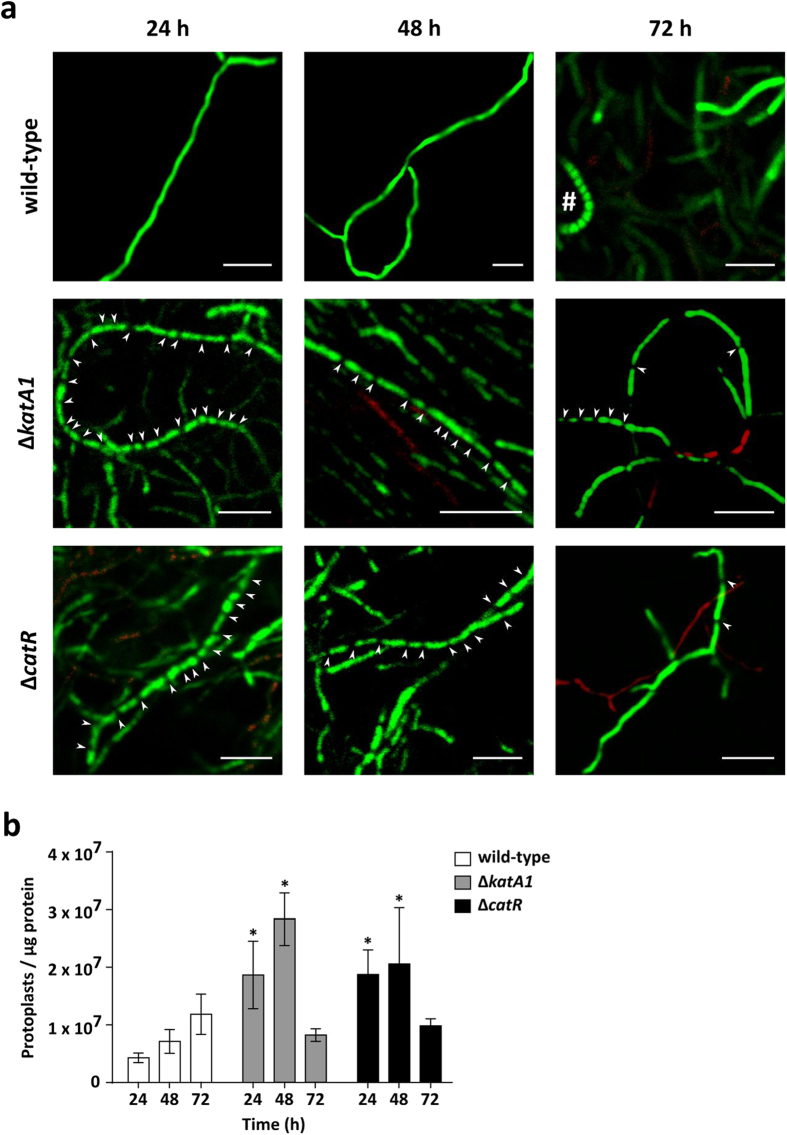
Figure 6. Assessment of the presence of compartmentalized (MI) and multinucleated (MII) mycelia in S. natalensis wild-type, ΔkatA1 and ΔcatR strains.
(a) Characterization of isolated hyphae through confocal microscopy using a cell viability assay. The presented micrographs are the result of merging the red (dead cells) and green (live cells) channels with ImageJ software. White arrowheads indicate the visible septa in the hyphae. # - aerial hypha forming a chain of spores. These results are representative of three independent experiments. Scale bar: 5 μm. (b) Quantification of protoplast formation through cytometry. Results are the average of biological duplicates and standard deviation. Statistically significant differences between strains at each time point were assessed by a one-way ANOVA followed by post hoc test (Tukey test; GraphPad Prism). * p < 0.01.
At 24 h, the wild-type colonies were majorly composed by live hyphae that accumulated at the medium surface. Mycelium continued to proliferate and at 48 h the centre of the colony started to present cell death, while the most peripheral mycelium remained viable. At 72 h, colonies maintained this defined mycelium stratification, albeit the area occupied by dead mycelium was much larger than at 48 h (Fig. 5). This pattern showed that the development of streptomycetes colonies is a dynamic process where the “old hyphae” (colony centre) undergo PCD and the “young hyphae” (colony periphery) remain viable and differentiate. Detailed hyphae images of the wild-type (Fig. 6A) showed a low-septation pattern or absence of septa throughout the different time points, indicating a differentiation into the MII stage prior to 24 h. At 72 h spore chains were observed, showing that the wild-type is able to complete its development cycle in R5 medium.
Regarding ΔkatA1, an accumulation of mycelium at the medium surface was also observed at 24 h. However, in the subsequent time points, mycelium presented an apparently independent growth above and within the medium: growth into the medium was restricted to small live hyphae aggregates, whereas the growth above the medium surface consisted in mycelium clumps, which were mostly alive, only presenting some cell death in the centre (Fig. 5). Hyphae of the mutant strain ΔkatA1 (Fig. 6A) showed a high-septation pattern at all time points (24 h, 48 and 72 h), demonstrating that the compartmentalized mycelium MI survives until later growth stages in this strain. Nevertheless, at 72 h some hyphae presented less frequent septa, pointing out to a transition from MI into MII (Fig. 6A).
As observed in the other strains, ΔcatR presented an accumulation of mycelium at the medium surface at 24 h. Strikingly, ΔcatR colonies showed a desynchronized behaviour at 48 h: the mycelium immersed in the medium presented cell death in the centre and live mycelium in the periphery, similar to the wild-type, whereas the mycelium located above the medium surface was mostly alive, lacking the stratification observed in the wild-type. At 72 h, the mycelium immersed in the medium degenerated; however, the mycelium grown above the medium-air interface was very heterogeneous: completely alive in some colonies, whereas others presented the wild-type dead/live stratification (Fig. 5). Observing hyphae in detail, ΔcatR presented a high number of septa at 24 h and 48 h, demonstrating that its mycelium was in MI at these time points. At 72 h MII seemed to be fully formed, due to the predominance of low-septated hyphae and no MI was observed (Fig. 6A).
To quantify hyphae septation in S. natalensis mutant strains, we have measured the capacity of protoplasts formation in S. natalensis strains using the methodology developed recently by the research group of A. Manteca (unpublished results) (Fig. 6B). Manteca, et al.27 demonstrated that protoplast formation correlates with the septation of the mycelium, since MII multinucleated hyphae form big protoplast that are highly unstable, while MI compartmentalized hyphae form large numbers of stable protoplasts. The number of protoplasts generated by the mycelia from ΔkatA1 and ΔcatR mutants at 24 h and 48 h was significantly higher than the wild-type. These results are consistent with the high-septation (MI) observed in the confocal micrographs of both mutant strains at these time points. At 72 h there were no significant differences between the mutants and the wild-type strain. Also interesting was the tendency of an increasing number of protoplasts formed by the wild-type mycelium throughout time. This happens also in other streptomycetes (Manteca et al. personal communication) and is consequence of sporulation, which led to an overestimation of the number of events counted by cytometry.
Discussion
Streptomycetes morphological differentiation takes place in a pro-oxidant environment. Furthermore, it is well established that Streptomyces development program is triggered by physiological stresses13,14,15. Curiously, the construction of a S. natalensis knock-out mutant strain for an important antioxidant enzyme - the H2O2 inducible mono-functional catalase KatA1 - resulted in a blockage of aerial mycelium formation (bald phenotype) and to a complete lack of colony organization. In addition, deletion of its transcriptional repressor encoding gene catR resulted in a delayed but not completely impaired, aerial mycelium formation (Fig. 1). The lack of sporulation in both mutant strains hampered strain manipulation and consequently we were not able to perform mutant complementation. DNA delivery methodologies such as transformation or intergeneric conjugation using mycelium have been unfruitful for S. natalensis28. However, considering the genomic context of catR and katA1 it is not likely that the mutations had a polar effect on the transcription of downstream genes.
Previously we have shown that KatA1 is the preferred antioxidant defence against H2O2-induced oxidative stress in S. natalensis21; the katA1 expression profile and the induction of total catalase activity suggests that the onset of aerial mycelium is associated with H2O2 induced oxidative stress (Fig. 3). In addition, the expression of the bacterioferritin encoding gene bfr in the wild-type at 72 h emphasized the need to control the levels of free-iron in the cell, which is also a common response to oxidative stress25. The results suggest that the formation of aerial mycelium in S. natalensis is accompanied by the emergence of intracellular oxidative stress.
Given the important role played by KatA1 in the S. natalensis antioxidant background, it was not surprising to observe that the deletion of its encoding gene was accompanied with an adaptive response that comprised the activation of an array of defence mechanisms from early growth stages, namely the alternative mono-functional catalase KatA3, the ferritins Bfr and DpsB and the organic hydroperoperoxide resistance protein OhrB (Fig. 3; Table 1). The up-regulation of the iron-chelating ferritin encoding genes would likely lead to the decrease of intracellular iron availability in order to reduce iron-dependent H2O2 toxicity. Interestingly, it is known that the transcription of key development related genes in S. coelicolor is responsive to iron availability, in particular bldN transcription is down-regulated in iron limited conditions29. We hypothesize that the limited iron availability caused by the adaptive response to an early intracellular oxidative stress endured by ΔkatA1 mutant contributes to the down-regulation of bldN transcription and its regulatory cascade (chaplin and rodlin proteins), contributing to the observed bald phenotype. It should be also highlighted the induction of dpsB transcription (the only Dps encoding gene found in S. natalensis genome) in the ΔkatA1 mutant. Our results suggest that contrary to S. coelicolor30, dpsB transcription in S. natalensis might be induced by oxidative stress. Finally, it is noteworthy that despite the vast array of mechanisms that bacteria use to counteract redox imbalance, no proteins related with other redox systems (e.g. mycothiol) were identified by 2D-PAGE as presenting altered expression levels. However, for a complete characterization of the oxidative stress response a genome-wide approach would be needed.
Deletion of catR forced a constitutive over-expression of katA1. It would be safe to assume that the high levels of total catalase activity observed in ΔcatR mutant prevent this strain to undergo H2O2 induced oxidative stress. Moreover, no major differences were observed in the transcription of ferritin encoding genes when compared to the wild-type strain. Nevertheless a down regulation of key development related genes is also observed in ΔcatR. Although katA1 is the only known target of CatR in Streptomyces sp.16,21, one cannot exclude the hypothesis that CatR orchestrates a wider molecular response, as the appearance of the blue-pigmented compound in ΔcatR might suggest. In fact, the CatR homolog in Bacillus subtilis - PerR - was shown to coordinate the expression of several antioxidant defences encoding genes31.
Although no significant differences were observed in the transcription of the gene encoding the master regulator BldD, we cannot rule out its involvement in the arrest of morphological differentiation in both mutants. Cyclic di-GMP (c-di-GMP) is a bacterial second messenger that has been implicated in the regulation of numerous physiological processes, including oxygen sensing32 and the oxidative stress response33. Recently, it was also identified as a key mediator of BldD activity7. Thus, the intracellular redox status of the mutant strains might be modulating the cyclic di-GMP turnover and consequently BldD activity. Nevertheless, activation of the oxidative stress response machinery either by the generation of a sub-lethal oxidative stress (ΔkatA1) or by de-repressing katA1 expression (ΔcatR), was associated with an active primary metabolism until later time points and with an increased vegetative mycelium proliferation rate. Moreover, the developmentally related PCD was shown to be inhibited (ΔkatA1) or delayed (ΔcatR) in the mutant strains, whereas in the wild-type, PCD was observed during aerial mycelium formation. Overall, these characteristics suggested an extended life-span of a mycelium in an early stage of development - the highly compartmentalized and transient mycelium MI.
The novelty of these observations, specifically regarding the prevalence of MI, required more conclusive data. The analysis of confocal micrographs showed that the wild-type was in the multinucleated stage MII at all time points (Fig. 6A). In addition the centre of the wild-type colonies was composed by dying hyphae that undergo PCD (Fig. 5), and aerial mycelium was being formed. It is noteworthy that this pattern followed the same trend observed in S. coelicolor6. Conversely, ΔkatA1 strain confocal micrographs and the analysis of the availability of these hyphae to form protoplasts, unequivocally demonstrated that a highly compartmentalized mycelium was present until 72 h, confirming the development arrest at MI. As a consequence, ΔkatA1 grew as a highly proliferative and undifferentiated mycelium with no defined colony organization (Fig. 1B and Fig. 5). Interestingly, this phenotype suggests that colony organization in the S. natalensis wild-type strain is dependent on the formation of the multinucleated mycelium MII. Regarding ΔcatR, a differentiation blockage at MI was observed until 48 h. Nevertheless, at 72 h the great majority of the mycelium was already in MII (Fig. 6A), which was consistent with the appearance of colonies with the wild-type live/dead mycelium (Fig. 5) stratification and with the formation of a scarce aerial mycelium (Fig. 1B).
The delayed morphological differentiation of MI into MII displayed by ΔkatA1 and ΔcatR mutant strains resembles the oxidative stress hormesis phenomenon described for other organisms, where an initial exposure to sub-lethal ROS dosages leads to the activation of an adaptive response and a concomitant increase in cell survival34. Ultimately, this hormetic effect implicates oxidative stress as a trigger of PCD, which has been previously demonstrated in Escherichia coli35. Taking into consideration this context, the phenotypes of ΔkatA1 and ΔcatR strains demonstrate that the observed early activation of the oxidative stress response system avoided or delayed the first round of PCD, which blocked MI differentiation to MII. Importantly, this development arrest fully supports the mutant strains morphological phenotypes, since the emergence of MII is preceded by a PCD, and only MII can differentiate into aerial mycelium. In light of our results, we propose that the oxidative stress generated as a consequence of nutrient scarcity - main signal for the development program onset – and other physiological stresses is an important trigger of the developmentally related PCD process.
In conclusion, to the best of our knowledge, this work presents the first documented cases of Streptomyces strains arrested at the MI phase and show compelling evidences that place oxidative stress as an important player in the transition between developmental stages, namely in the differentiation of MI to MII. Additionally, it brings relevant knowledge for the industrial production of added-value compounds by Streptomyces strains, due to the strict correlation of MI/MII transition with the onset of secondary metabolism36.
Methods
Streptomyces natalensis genome sequencing
Whole-genome shotgun sequencing of S. natalensis strain ATCC 27448 was determined using an Illumina HiSeq 2000 sequencing system at BGI (www.genomics.cn). Two paired-end libraries of 0,5-kbp (10 000 000 reads) and 2-kbp (5 000 000 reads) were used for sequencing that yielded 900 and 450 Mbp of clean data, respectively. The reads were assembled using the SOAPdenovo software (version 1.05) of the SOAP tool package (http://soap.genomics.org.cn/soapdenovo.html). Scaffolds were submitted to GenBank as a whole shotgun sequence and annotated through the Prokaryotic Genome Annotation Pipeline (PGAP). This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession JRKI00000000. The version described in this paper is JRKI01000000.
Bacterial strains and growth conditions
Escherichia coli strains were routinely grown in LB medium at 37 °C. S. natalensis strains were routinely cultivated in TBO37 solid medium at 30 °C. Differentiation studies were performed in R538 solid medium at 30 °C; 500 μl of YEME39 liquid cultures grown to late exponential phase were used as inoculum for S. natalensis solid cultures. To harvest mycelium for nucleic acids or protein extractions, S. natalensis strains were grown in a nitrocellulose membrane with a 0.45 μm pore (Merck Millipore) placed on top of R5 solid medium. For the mycelium proliferation assays, a drop of 10 μl of YEME liquid cultures grown to OD600nm of 4–5 was inoculated in the centre of R5 plates. Plates were photographed and the mycelium area was determined using the ImageJ software.
Construction of ΔcatR knock-out mutant strain
The coding sequence of catR from S. natalensis ATCC 27448 was replaced by the aac(3)IV/oriT cassette from plasmid pIJ773 using a PCR-targeting approach40. The primers used for the amplification of the cassette were D_catR_F and D_catR_R (Supplementary Table S2). The native catR gene was replaced by the resistance cassette through homologous recombination resulting in the knock-out mutant strain S. natalensis ΔcatR::aac(3)IV/oriT. The genetic identity of this strain was confirmed by Southern hybridization and PCR (Supplementary Fig. S1).
Nucleic acids and protein procedures
S. natalensis genomic DNA was isolated from 1 ml of YEME liquid cultures using the MasterPureTM Gram Positive DNA Purification Kit (Epicentre). Southern hybridization was performed with probes labelled with digoxigenin using the DIG DNA labelling kit (Roche). Intergeneric conjugation between E. coli ET12567 [pUZ8002] and S. natalensis ATCC 27448 was performed as previously described28.
For total RNA isolation from mycelium grown in solid medium, a portion of mycelium (approx. 2 cm2) was scrapped from the membrane and resuspended in 500 ml of K-phosphate buffer 50 mM pH 6.8. Two volumes of RNA Protect Bacteria Reagent (Qiagen) were added to the suspension; mycelium was harvested by centrifugation and immediately frozen by immersion in liquid nitrogen. RNA extraction was performed using RNeasy® Mini kit (Quiagen) and following the manufacturer instructions with modifications described elsewhere21. DNase treatments, RNA concentration determination and RNA quality and integrity evaluation was performed as previously described21.
Protein crude extracts were obtained from a section (approx. 2 cm2) of scrapped mycelium. This mycelium was washed and resuspended in the appropriate buffer containing 25% (v/v) of protease inhibitor (Roche). Mycelium disruption was performed through sonication (Branson Sonifier, Model B-15), the lysate was centrifuged and the pellet discarded. Protein content of cellular extracts was determined by the BCATM Protein Assay Kit (Pierce) or Bradford reagent (Bio-rad) when buffers contained reducing agents. Bovine serum albumin was used to determine standard curves. Unidimensional protein profiles were determined by SDS_PAGE in 12.5% (w/v) gels and using the coomassie coloration (Bio-Rad).
Enzymatic activity assays
Catalase activity was quantified spectrophotometrically by following the rate of decrease in absorbance at 240 nm caused by the disappearance of H2O241 as described elsewhere21. 1 unit of enzyme activity is defined as the amount required for the conversion of 1 μmol substrate into product per min. The specific activity of glyceraldeheyde 3-phosphate dehydrogenase (GAPDH) was measured as previously described42. 1 unit of GAPDH activity corresponds to the amount of enzyme necessary to produce 1 μmol of NADH per min, at 25 °C. The glucose 6-phosphate dehydrogenase (G6PDH) specific activity was determined spectrophotometrically by following the reduction rate of NADP+ into NADPH at 340 nm43. 1 unit of G6PDH activity is equivalent to the amount of enzyme necessary to produce 1 μmol of NADPH per min, at 25 °C. Aconitase specific activity was measured as previously described44. The rate of isocitrate formation from citrate was followed at 240 nm, using an absorption coefficient of 3.6 mM−1. 1 unit of activity transformed 1 nmol of substrate per minute, at 25 °C.
In gel determination of catalase activity was performed in 7.5% (w/v) non-denaturing polyacrylamide gels as previously described45. For nuclease activity, protein extracts were separated in 12.5% (w/v) polyacrylamide gels in denaturing conditions containing DNA from herring testes type XIV (SIGMA) 10 μg/ml. Protein renaturation, induction of nuclease activity and gel visualization were performed as previously described46.
Two-dimensional polyacrylamide gel electrophoresis
The two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) technique was performed as previously described47. Total protein extracts, isoelectric focusing and SDS-PAGE gels were prepared as described elsewhere21. Gels were silver stained through a mass spectrometry (MS) compatible protocol48. PageRulerTM (Fermentas) was used as size marker. In silico analysis of the 2D gels was performed in PDQuest 2-D (Bio-Rad). Spots with a significant statistical difference between strains (biological duplicates) were considered using the following criteria: P > 0.05 and two-fold change.
Protein spots were excised from gels and digested with trypsin. Samples were analysed using the 4700 Proteomics Analyzer MALDI-TOF/TOF (Applied Biosystems) as previously described49. Data were analysed using GPS Explorer (Version 3.6; Applied Biosystems). Protein identification was performed combining data from PMF (Peptide Mass Fingerprint) and tandem mass (MS/MS) spectra. The Mascot (Matrix Science, UK) algorithm was used to determine the statistical significance (score > 52) of protein identification against the genome database of S. natalensis ATCC 27448.
PCR and RT-PCR
DNA amplification by PCR was performed with GoTaq Flexi DNA Polymerase (Promega) or Pfu DNA polymerase (Fermentas) according to manufacturer instructions.
For gene expression studies 1 μg of DNase I-treated (DNA-free Kit, Ambion) total RNA was transcribed with the iScriptTM Select cDNA Synthesis Kit (Bio-Rad), using the random primers supplied, and following the manufacturer instructions. RT-PCR amplifications were performed using the primer pairs listed in Supplementary Table S2 and using GoTaq Flexi DNA Polymerase (Promega). PCR products were separated on 2% (w/v) agarose gels.
Confocal microscopy
To prepare samples for confocal microscopy analysis, 4 cm2 blocks of R5 solid cultures were cut out using a scalpel and sliced using a microtome (sections with a width of approx. 0.15 mm). To characterize isolated hyphae, mycelium was scrapped from R5 solid cultures and resuspended in NaCl 0,9% (w/v). 10 μl of the mycelium resuspension were used for staining. Viability staining was performed using the “LIVE/DEAD BacLight Bacterial Viability Kit” (Molecular Probes) with a mixture of SYTO9 and propidium iodide in a 1:1 proportion and following the manufacturer instructions. Samples were observed under a Leica SP2 AOBS SE confocal laser-scanning microscope using 488 nm and 568 nm as excitation wavelengths and emitting between 520–545 nm (green) or 620–640 nm (red), as previously described6. Micrographs were treated using the software ImageJ.
Mycelium protoplasting and protoplast quantification
Protoplast were obtained and quantified according to the methodology developed recently by Manteca et al. (unpublished results). These authors made the control experiments necessary to warrant that the protocol used for protoplast formation had an efficiency close to 100%, which is necessary for reproducibility of the results, and to warrant the correlation between hypha compartmentalization and protoplast formation. Briefly, 60 mg of mycelia (fresh weight) were resuspended in 1.4 ml of modified buffer P (0.6% TES buffer pH 7.2, 103% sucrose; filtered through a 0.2 μm filter) into a 2-ml Eppendorf tube. Lysozyme was added from a freshly prepared stock at a final concentration of 2 mg/ml and incubated for 30 minutes at 600 rpm and 37 °C. Protoplasts were drawn in and out in a 1-ml pipette two times, incubated for an additional 30 minutes, washed two times by sedimentation (1000 g) and resuspended in buffer P. After the last washing step, protoplasts were resuspended in 500 μl of buffer P.
Samples were stained with SYTO9 (Invitrogen, 6 μM) and quantified with a flow cytometer (Cytomics FC500, Beckman-Coulter Inc., USA). Measurements were performed using BD Trucount Tubes®, with 500 μl of protoplasts stained with SYTO9. The trigger signal was established with an FL1 detector (530/540 nm) with the adequate negative control (buffer P) and biological control (BD Plasma Count®). The absolute quantifications were performed counting 10,000 of the standard beads included in the BD Trucount Tubes®. Protoplast dilutions used for cytometry quantifications were those reporting absolute protoplast numbers within the 5,000–10,000 interval, close to the number of beads used as standard. The number of protoplasts per μl was calculated based on the number of standard beads, and the number of protoplasts per mg of protein was calculated based on the protein estimated for the mycelium processed for protoplast formation. Protoplasts from two biological replicates were quantified.
Additional Information
How to cite this article: Beites, T. et al. Streptomyces natalensis programmed cell death and morphological differentiation are dependent on oxidative stress. Sci. Rep. 5, 12887; doi: 10.1038/srep12887 (2015).
Supplementary Material
Acknowledgments
This work was funded by: “NORTE-07-0124-FEDER-000003 – Cell homeostasis tissue organization and organism biology” project co-funded by FEDER funds through the Operational North Region Programme (ON.2 – O Novo Norte) under National Strategic Reference Framework (QREN) and by National funds through FCT – Fundação para a Ciência e Tecnologia/MEC – Ministério da Educação e Ciência and when applicable co-funded by FEDER funds within the partnership agreement PT2020 related with the research unit number 4293. TB was supported by a post-doctoral fellowship under the PEst-C/SAU/LA0002/2013 (FCOMP-01-0124-FEDER-037277) project; PO, MVM and SDSP were supported by the FCT fellowships SFRH/BPD/74894/2010, SFRH/BPD/95683/2013 and SFRH/BD/66367/2009, respectively. We thank Rui Fernandes, Hugo Osório and Catarina Santos for excellent technical assistance in the preparation of samples for confocal microscopy, protein identification and in silico analysis of the S. natalensis genome, respectively.
Footnotes
Author Contributions T.B., P.O., B.R. and A.M. performed the experiments; S.D., S.P., R.O., P.T. and P.M.-F. assisted with data analysis; all authors contributed to the critical discussion of the manuscript; T.B. and M.V.M. conceived, designed the work and wrote the manuscript. All authors read and approved the final manuscript.
References
- Garrity G., Bell J. & Lilburn T. in Bergey’s Manual of Systematic Bacteriology (Bergey’s Manual Trust, Springer-Verlag, 2004). [Google Scholar]
- Flardh K. & Buttner M. J. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7, 36–49 (2009). [DOI] [PubMed] [Google Scholar]
- Berdy J. Bioactive microbial metabolites. J. Antibiot (Tokyo) 58, 1–26 (2005). [DOI] [PubMed] [Google Scholar]
- Manteca A., Fernandez M. & Sanchez J. A death round affecting a young compartmentalized mycelium precedes aerial mycelium dismantling in confluent surface cultures of Streptomyces antibioticus. Microbiology 151, 3689–3697 (2005). [DOI] [PubMed] [Google Scholar]
- Yague P. et al. Transcriptomic analysis of Streptomyces coelicolor differentiation in solid sporulating cultures: first compartmentalized and second multinucleated mycelia have different and distinctive transcriptomes. PLoS One 8, e60665 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteca A., Claessen D., Lopez-Iglesias C. & Sanchez J. Aerial hyphae in surface cultures of Streptomyces lividans and Streptomyces coelicolor originate from viable segments surviving an early programmed cell death event. FEMS Microbiol. Lett. 274, 118–125 (2007). [DOI] [PubMed] [Google Scholar]
- Tschowri N. et al. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158, 1136–1147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Hengst C. D. et al. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 78, 361–379 (2010). [DOI] [PubMed] [Google Scholar]
- Yamazaki H., Takano Y., Ohnishi Y. & Horinouchi S. amfR, an essential gene for aerial mycelium formation, is a member of the AdpA regulon in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 50, 1173–1187 (2003). [DOI] [PubMed] [Google Scholar]
- Wolanski M. et al. The level of AdpA directly affects expression of developmental genes in Streptomyces coelicolor. J. Bacteriol. 193, 6358–6365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Domonkos A., Chandra G. & Buttner M. J. Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by sigma(BldN) and a cognate anti-sigma factor, RsbN. Mol. Microbiol. 84, 1033–1049 (2012). [DOI] [PubMed] [Google Scholar]
- Cap M., Vachova L. & Palkova Z. Reactive oxygen species in the signaling and adaptation of multicellular microbial communities. Oxid. Med. Cell. Longev. 2012, 976753 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera T. S., Csonka L. N. & Paliy O. Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J. Bacteriol. 190, 3712–3720 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakimova M., Ahlgren H. G., Harrison J. J., English A. M. & Nguyen D. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J. Bacteriol. 195, 2011–2020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-García A., Barreiro C., Santos-Beneit F., Sola-Landa A. Martín J. F. Genome-wide transcriptomic and proteomic analysis of the primary response to phosphate limitation in Streptomyces coelicolor M145 and in a ΔphoP mutant. Proteomics 7, 2410–2429 (2007). [DOI] [PubMed] [Google Scholar]
- Hahn J. S., Oh S. Y., Chater K. F., Cho Y. H. & Roe J. H. H2O2-sensitive fur-like repressor CatR regulating the major catalase gene in Streptomyces coelicolor. J. Biol. Chem. 275, 38254–38260 (2000). [DOI] [PubMed] [Google Scholar]
- Hahn J. S., Oh S. Y. & Roe J. H. Role of OxyR as a peroxide-sensing positive regulator in Streptomyces coelicolor A3(2). J. Bacteriol. 184, 5214–5222 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallifidas D., Thomas D., Doughty P. & Paget M. S. B. The σR regulon of Streptomyces coelicolor A3(2) reveals a key role in protein quality control during disulphide stress. Microbiology 156, 1661–1672 (2010). [DOI] [PubMed] [Google Scholar]
- Oh S.-Y., Shin J.-H. & Roe J.-H. Dual role of OhrR as a repressor and an activator in response to organic hydroperoxides in Streptomyces coelicolor. J. Bacteriol. 189, 6284–6292 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn B.-E. et al. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 59, 1848–1858 (2006). [DOI] [PubMed] [Google Scholar]
- Beites T. et al. Crosstalk between ROS homeostasis and secondary metabolism in S. natalensis ATCC 27448: modulation of pimaricin production by intracellular ROS. PLoS One 6, e27472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D., de Jong W., Dijkhuizen L. & Wösten H. A. Regulation of Streptomyces development: reach for the sky! Trends Microbiol. 14, 313–319 (2006). [DOI] [PubMed] [Google Scholar]
- McCormick J. R. & Flardh K. Signals and regulators that govern Streptomyces development. FEMS Microbiol. Rev. 36, 206–231 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-M., Lee C.-K., Choi S.-U., Park H.-R. & Hwang Y.-I. Functional analysis of a BarX homologue (SngA) as a pleiotropic regulator in Streptomyces natalensis. Arch. Microbiol. 189, 569–577 (2008). [DOI] [PubMed] [Google Scholar]
- Andrews S. C., Robinson A. K. & Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237 (2003). [DOI] [PubMed] [Google Scholar]
- Manteca A., Mäder U., Connolly B. A. & Sanchez J. A proteomic analysis of Streptomyces coelicolor programmed cell death. Proteomics 6, 6008–6022 (2006). [DOI] [PubMed] [Google Scholar]
- Manteca A., Fernandez M. & Sanchez J. Cytological and biochemical evidence for an early cell dismantling event in surface cultures of Streptomyces antibioticus. Res. Microbiol. 157, 143–152 (2006). [DOI] [PubMed] [Google Scholar]
- Enriquez L. et al. An efficient gene transfer system for the pimaricin producer Streptomyces natalensis. FEMS Microbiol. Lett. 257, 312–318 (2006). [DOI] [PubMed] [Google Scholar]
- Traxler M. F., Seyedsayamdost M. R., Clardy J. & Kolter R. Interspecies modulation of bacterial development through iron competition and siderophore piracy. Mol. Microbiol. 86, 628–644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facey P. D. et al. Streptomyces coelicolor Dps-like proteins: differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol. Microbiol. 73, 1186–1202 (2009). [DOI] [PubMed] [Google Scholar]
- Faulkner M. J. & Helmann J. D. Peroxide stress elicits adaptive changes in bacterial metal ion homeostasis. Antioxid. Redox Signal. 15, 175–189 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckerman J. R. et al. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 48, 9764–9774 (2009). [DOI] [PubMed] [Google Scholar]
- Lacey M. M., Partridge J. D. & Green J. Escherichia coli K-12 YfgF is an anaerobic cyclic di-GMP phosphodiesterase with roles in cell surface remodelling and the oxidative stress response. Microbiology 156, 2873–2886 (2010). [DOI] [PubMed] [Google Scholar]
- Ristow M. & Schmeisser S. Extending life span by increasing oxidative stress. Free Radic. Biol. Med. 51, 327–336 (2011). [DOI] [PubMed] [Google Scholar]
- Hazan R., Sat B. & Engelberg-Kulka H. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186, 3663–3669 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioseras B., Lopez-Garcia M. T., Yague P., Sanchez J. & Manteca A. Mycelium differentiation and development of Streptomyces coelicolor in lab-scale bioreactors: programmed cell death, differentiation, and lysis are closely linked to undecylprodigiosin and actinorhodin production. Bioresour. Technol. 151, 191–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgens C. E., Hamill R. L., Sands T. H., Hoehn M. M. & Davis N. E. The occurrence of deacetoxycephalosporin C in fungi and streptomycetes. J. Antibiot. 27, 298–300 (1974). [DOI] [PubMed] [Google Scholar]
- Kieser T., Bibb M., Buttner M., Chater K. & Hopwood D. A. Practical Streptomyces Genetics. (John Innes Centre, Norwich, United Kigdom, 2000). [Google Scholar]
- Chater K. F., Hopwood D. A., Kieser T. & Thompson C. J. Gene cloning in Streptomyces. Curr. Top. Microbiol. Immunol. 96, 69–95 (1982). [DOI] [PubMed] [Google Scholar]
- Gust B., Challis G. L., Fowler K., Kieser T. & Chater K. F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100, 1541–1546 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers R. F. & Sizer I. W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195, 133–140 (1952). [PubMed] [Google Scholar]
- Holland M. J. & Westhead E. W. Chemical reactivity at the catalytic sites of aspartic β-semialdehyde dehydrogenase and glyceraldehyde 3-phosphate dehydrogenase. Biochemistry 12, 2276–2281 (1973). [DOI] [PubMed] [Google Scholar]
- Rudack D., Chisholm E. M. & Holten D. Rat Liver glucose 6-phosphate dehydrogenase. J. Biol. Chem. 246, 1249–1254 (1971). [PubMed] [Google Scholar]
- Muschko K., Kienzlen G., Fiedler H.-P., Wohlleben W. & Schwartz D. Tricarboxylic acid cycle aconitase activity during the life cycle of Streptomyces viridochromogenes Tü494. Arch. Microbiol. 178, 499–505 (2002). [DOI] [PubMed] [Google Scholar]
- Clare D. A., Duong M. N., Darr D., Archibald F. & Fridovich I. Effects of molecular-oxygen on detection of superoxide radical with nitroblue tetrazolium and on activity stains for catalase. Anal. Biochem. 140, 532–537 (1984). [DOI] [PubMed] [Google Scholar]
- Fernandez M. & Sanchez J. Nuclease activities and cell death processes associated with the development of surface cultures of Streptomyces antibioticus ETH 7451. Microbiology 148, 405–412 (2002). [DOI] [PubMed] [Google Scholar]
- Gorg A. et al. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis 21, 1037–1053 (2000). [DOI] [PubMed] [Google Scholar]
- Goromova I. & Celis J. E. in Cell Biology: A Laboratory Handbook. 3rd Edition. (ed Celis J.E., Carter N., Hunter T., Simons K., Small J.V., Shotton D. ) (Elsevier. Academic Press., 2006). [Google Scholar]
- Pinho S. S. et al. Role of E-cadherin N-glycosylation profile in a mammary tumor model. Biochem. Biophys. Res. Commun. 379, 1091–1096 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.