Abstract
The emergence of drug-resistant strains of influenza virus, makes exploring new classes of inhibitors that target universally conserved viral targets a highly important goal. The influenza A viral genome is made up of 8 single-stranded RNA negative segments. The RNA promoter, consisting of the conserved sequences at the 3′ and 5′ end of each RNA genomic segment, is universally conserved among influenza A virus strains and in all segments. Previously we reported on the identification and NMR structure of DPQ (6,7-dimethoxy-2-(1-piperazinyl)-4-quinazolinamine) (compound 1) in complex with the RNA promoter. Here we report on additional screening and SAR studies with compound 1, including ex vivo anti-influenza activity assays, resulted in improved cellular activity against influenza A virus in the micromolar range.
The influenza A RNA promoter is universally conserved among influenza A virus strains, making it potentially an ideal drug target for novel antiviral agents. Using an NMR-based approach, we report on the characterization and initial SAR studies of RNA binding compounds, including ex vivo anti-influenza activity assays.
Introduction
Influenza A viral infection poses great threat to public health worldwide, affecting lives of thousands of individuals every year. There are currently two classes of medications available to treat influenza. The first class includes amantadine and rimantadine, both are M2 ion channel inhibitors (1) which block the release of the viral genome into the cytoplasm and the maturation of the viral proteins.(2) However, H1N1 and H3N2 influenza strains circulating in the North American continent carry a S31N mutation of the M2 protein, which renders them resistant to amantadine (http://www.cdc.gov/flu/about/qa/antiviralresistance for 2013–2014 flu season).(1) The second class of anti-influenza drugs includes neuraminidase inhibitors, oseltamivir phosphate (Tamiflu) (3) and zanamivir (Relenza),(4) which abolish the release of new virions from the host cell. Unfortunately, neuraminidase resistant strains are also emerging. In light of the rising resistance of influenza virus strains to current anti-influenza medications, finding other druggable targets that are as conserved and less prone to the development of drug resistance due to mutations is of critical importance.(5, 6)
The influenza A virus is a member of orthomyxoviridae family and its genome consists of eight single negative-stranded RNA segments.(7) The genome of influenza A virus is organized into eight separated ribonucleoprotein complexes (RNP complexes), where the hetero-trimeric RNA dependent RNA polymerase (RdRp) and multiple copies of nucleoproteins bind to single stranded viral RNA.(6) The RdRp is composed of three subunits, PA, PB1 and PB2, arranged in a head to tail fashion.(8) The RdRp binds to the 3′ and 5′ ends of a viral RNA segment, triggering both transcription initiation of complementary RNA (cRNA) and replication of the viral genome (vRNA).
The 13 nucleotides on the 5′ terminus and 12 nucleotides on the 3′ terminus of each viral RNA segment are conserved throughout various human influenza A virus strains and come together to form a so-called panhandle-like structure.(7) Mutations in these conserved sequences negatively affect viral replication efficiency.(9) We will refer to this region (13 nucleotides of the 5′ terminus and 12 nucleotides the 3′ terminus) of the viral RNA as the influenza RNA promoter throughout this work. The structure of this RNA promoter has been solved by solution NMR spectroscopy using a designed RNA segment containing all conserved nucleotides underlined in the following sequence: 5′-GAGUAGAAACAAGGCUUCGGCCUGCUUUUGCU - 3′.(10) We found that the RNA promoter adopts an A-form helix with an interloop containing an AA-U motif and a C-A mismatch pair. In addition, there was a 46 ± 10° bend close to the initiation site of the viral RNA, that has been postulated to be important for RNA polymerase recognition.(7)
Armed with the knowledge of the RNA promoter structure and encouraged by the fact that promoter sequences are highly conserved and intolerant of mutations,(7) we carried out a small molecule screen against the RNA promoter in the hope to find hits that possess anti-influenza replication activity.(10) A small molecule, 6,7-dimethoxy-2-(1-piperazinyl)-4-quinazolinamine (DPQ, compound 1), was identified in a NMR-based screen of 4279 compounds to bind to influenza RNA promoter close to the AA-U interloop region (Figure 1A).(10) However, compound 1 showed only a modest anti-influenza activity in a cell-based viral replication assay with an IC50 value of 549 μM (Table 1).(10) Upon examination of the compound 1-RNA promoter complex, we reported that the binding of compound 1 straightened the bend at the A5-U29 base pair and widened the major groove near base pairs G13-C22 and G14-C21 (numbering from the 5′ end in the above listed RNA sequence).(10) We hypothesized that this change in structure would interfere with the binding of RdRp and subsequently the ability of the virus to replicate.
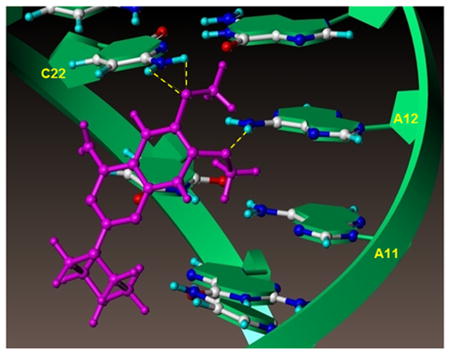
Figure 1. Influenza RNA promoter in complex with compound 1.
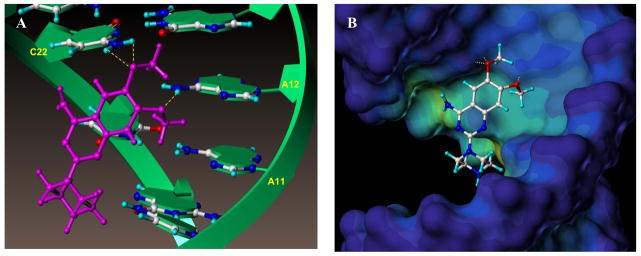
(A) Detail of the interactions between compound 1 and the RNA promoter from the NMR structure of the complex (PDB ID 2LWK).(10) The RNA helix is shown as a green ribbon and the nucleoside atoms and compound 1 are shown as balls and sticks. The yellow dotted lines indicated hydrogen bonds between the adenine 12 H61/H62 hydrogens and 6-methoxy oxygen atom of compound 1, and between cytosine 22 H41/H42 hydrogens and the 7-methoxy oxygen of compound 1.
(B) The RNA promoter was depicted in surface representation and color coded according to cavity depth (blue indicates the most outer surface and yellow indicated a deeper cavity relative to the surface). Compound 1 binds in the major groove of the RNA helix and the amine on the piperazine ring extents toward the cavity but does not interact significantly with the RNA.
Table 1.
Summary of IC50, CC50, IC50/CC50 and Kd values of compounds
| ID | Structure | IC50 (μM) | CC50 (μM) | IC50/CC50 | Kd (μM) |
|---|---|---|---|---|---|
| 1 |
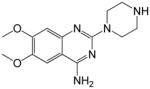
|
549*1 | >1000*1 | <0.55 | 61.45 |
| 3 |
|
>250*2 | >250*2 | >1.00 | 131.9 |
| 4 |
|
>250 | >250 | >1.00 | + |
| 5 |
|
>250 | >250 | >1.00 | + |
| 6 |
|
39.67 ± 6.49 | >250 | <0.16 | 114.2 |
| 7 |
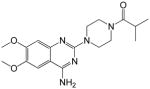
|
33.89 ± 23.2 | >250 | <0.14 | 158.5 |
| 8 |
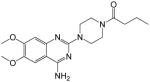
|
44.18 ± 11.28 | >250 | <0.18 | 44.35 |
| 9 |
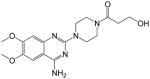
|
>250 | >250 | >1.00 | + |
| 10 |
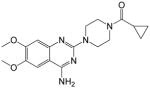
|
34.18 ± 17.23 | >250 | <0.14 | 127.2 |
| 11 |
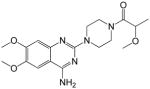
|
>250 | >250 | >1.00 | + |
| 12 |
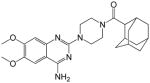
|
>250 | 16.4 | >15.24 | + |
| 13 |
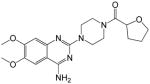
|
54.06 ± 9.5 | >250 | <0.22 | 331.6 |
| 14 |
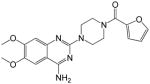
|
27.88 ± 10.2 | 78.23 | 0.36 | + |
| 15 |
|
16.77 ± 3.99 | 66.97 | 0.25 | 221.1 |
| 16 |
|
107.55 ± 49.09 | >250 | <0.43 | 320.7 |
| 17 |
|
126 ± 24.65 | >250 | <0.50 | 149.9 |
Values were determined and reported in published paper.(10)
Due to compound solubility properties, the IC50 and CC50 values were determined with serial titration concentrations ranging from 250μM to 38nM.
+ indicates that the analogue binds to the influenza RNA promoter and results in decreased intensities of imino protons of U26 and G13 yet the Kd value was not determined.
When studying the complex between compound 1 and the RNA promoter more closely, we found that the interaction is mainly mediated by contacts involving the methoxy groups on compound 1 and the bases of adenosine 12 and cytosine 22 (Figure 1A).(10) Moreover, the primary amine of compound 1, likely protonated under physiological conditions, is located across from the phosphate backbone, presumably stabilizing the binding via electrostatic interactions (Figure 1B). However, the secondary amine on the piperazine seems to be pointing away from the target and is not making significant contacts with the RNA promoter. Based on these observations, we hypothesized that modifying and/or extending the secondary amine on the piperazine could result in analogues with either improved affinity and/or pharmacological properties (Figure 1B). Our studies resulted in improved compound 1 derivatives with cellular activity against influenza A virus in the micromolar range.
In a parallel approach, we also tested a combinatorial peptide library against the RNA promoter by NMR, in an attempt to identify possible peptide binding motifs that may recapitulate the recognition by the RdRp. However, no suitable peptide sequences were identified, corroborating the hypothesis that the RNA promoter functions as an anchoring ligand for the RdRp and that changes in its confirmation, as induced by our compounds, are sufficient to antagonize its function.
Methods and Materials
Compound 1 analogues synthesis (compounds 3 to 16)
General
Unless otherwise indicated, all anhydrous solvents were commercially obtained and stored in Sure-seal bottles under nitrogen. All other reagents and solvents were purchased as the highest grade available and used without further purification. Thin-layer chromatography (TLC) analysis of reaction mixtures was performed using Merck silica gel 60 F254 TLC plates, and visualized using ultraviolet light. NMR spectra were recorded on JEOL 400 MHz instruments. Chemical shifts (δ) are reported in parts per million (ppm) referenced to 1H (Me4Si at 0.00). Coupling constants (J) are reported in Hz throughout. Mass spectral data were acquired on Shimadzu LCMS-2010EV for low resolution, and on an Agilent ESI-TOF for either high or low resolution; or Bruker Datonics Autoflex II MALDI TOF/TOF. Purity of all compounds was obtained in a HPLC Breeze from Waters Co. using an Atlantis T3 3 μm 4.6×150 mm reverse phase column. The eluant was a linear gradient with a flow rate of 1 ml/min from 95% A and 5% B to 5% A and 95% B in 15 min followed by 5 min at 100% B (Solvent A: H2O with 0.1% TFA; Solvent B: acetonitrile with 0.1% TFA). The compounds were detected at λ=254 nm. Purity of key compounds was established by HPLC and/or elemental analysis as performed on a Perkin Elmer series II-2400. Combustion analysis was performed by NuMega Resonance Labs, San Diego, CA, USA.
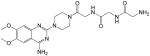
2-amino-N-(2-((2-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)-2-oxoethyl)amino)-2-oxoethyl)acetamide (3)
Yield: 35%; 1H NMR (400 MHz, DMSO-d6) δ 3.60–3.66 (m, 4 H), 3.75–3.80 (m, 4 H), 3.82 (s, 2 H), 3.88 (s, 2 H), 4.05 (br s, 2 H), 7.19 (s, 1 H), 7.67 (s, 1 H), 7.99 (br s, 2 H, NH2), 8.14 (br s, 1 H), 8.62 (br s, 1 H, NH), 8.67 (br s, 2 H, NH2); MS (MALDI) m/z 461 (M+H)+, 483 (M+Na)+.
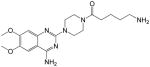
5-amino-1-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)pentan-1-one (4)
Yield: 41%; 1H NMR (400 MHz, DMSO-d6) δ 1.52–1.56 (m, 4 H), 2.41–2.49 (m, 2 H), 2.78–2.81 (m, 2 H), 3.61–3.64 (m, 4 H), 3.75–3.81 (m, 4 H), 3.83 (s, 3 H), 3.89 (s, 3 H), 7.21 (s, 1 H), 7.67 (s, 1 H), 7.72 (br s, 2 H, NH2), 8.69 (s, 1 H, NH), 8.82 (s, 1 H, NH); MS (MALDI) m/z 389 (M+H)+, 411 (M+Na)+.
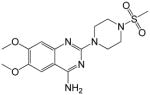
6,7-dimethoxy-2-(4-(methylsulfonyl)piperazin-1-yl)quinazolin-4-amine (5)
Yield: 69%; 1H NMR (400 MHz, DMSO-d6) δ 2.92 (s, 3 H), 3.26–3.30 (m, 4 H), 3.83 (s, 3 H), 3.88 (s, 3 H), 3.91–3.95 (m, 4 H), 7.30 (s, 1 H), 7.69 (s, 1 H), 8.73 (br s, 1 H), 8.87 (br s, 1 H); MS (MALDI) m/z 368 (M+H)+, 390 (M+Na)+.
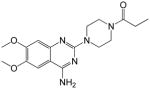
Synthesis of 1-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)propan-1-one (6)
A mixture of 1 (62 mg, 0.21 mmol, 1 eqv), propionic acid (16 mg, 0.21 mmol, 1 eqv), Oxyma pure (31 mg, 0.25 mmol, 1.2 eqv), N,N′-Diisopropylcarbodiimide (DIC, 32 mg, 0.25 mmol, 1.2 eqv), DIEA (40 mg, 0.31 mmol, 1.5 eqv) in DMF (2 mL) was stirred for 16 h at room temperature. After completion, DMF was removed in vacuo followed by chromatographic purification using 5–10% MeOH in CH2Cl2 to afford a pure product, 6 (43 mg, 60%).
1H NMR (400 MHz, DMSO-d6) δ 0.96 (t, J = 7.2 Hz, 3 H), 2.33 (q, J = 6.8 Hz, 2 H), 3.59–3.62 (m, 4 H), 3.65–3.71 (m, 4 H), 3.74 (s, 3 H, OMe), 3.79 (s, 3 H, OMe), 6.76 (s, 1 H), 7.35 (br s, 2 H, NH2), 7.41 (s, 1 H); MS (MALDI): m/z 346 (M+H)+, 368 (M+Na)+.
Similarly, compounds 7 to 16 were synthesized; characterization data were as following:
1-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)-2-methylpropan-1-one (7)
Yield: 67%; 1H NMR (400 MHz, DMSO-d6) δ 0.97 (d, J = 6.8 Hz, 6 H), 2.84–2.89 (m, 1 H), 3.59–3.63 (m, 4 H), 3.67–3.71 (m, 4 H), 3.73 (s, 3 H, OMe), 3.78 (s, 3 H, OMe), 6.70 (s, 1 H), 7.14 (br s, 2 H, NH2), 7.37 (s, 1 H); MS (MALDI): m/z 360 (M+H)+, 382 (M+Na)+.
1-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)butan-1-one (8)
Yield: 56%; 1H NMR (400 MHz, DMSO-d6) δ 0.86 (t, J = 7.2 Hz, 3 H), 2.30–2.34 (m, 2 H), 2.45 (t, J = 6.8 Hz, 2 H), 3.46–3.50 (m, 4 H), 3.56–3.61 (m, 4 H), 3.78 (s, 3 H, OMe), 3.86 (s, 3 H, OMe), 6.90 (s, 1 H), 7.35 (br s, 2 H, NH2), 7.41 (s, 1 H); MS (MALDI) m/z 360 (M+H)+, 382 (M+Na)+.
1-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)-3-hydroxypropan-1-one (9)
Yield: 61%; 1H NMR (400 MHz, DMSO-d6) δ 2.54 (br s, 1 H), 3.45–3.57 (m, 4 H), 3.65 (q, J = 5.5 Hz, 2 H), 3.67–3.75 (m, 4 H), 3.79 (s, 3 H), 3.82 (s, 3 H), 4.55 (t, J = 6.2 H, 2 H), 6.80 (br s, 2 H, NH2), 7.47 (s, 2 H); MS (MALDI) m/z 362 (M+H)+, 384 (M+Na)+.
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(cyclopropyl)methanone (10)
Yield: 59%; 1H NMR (400 MHz, DMSO-d6) δ 0.70–0.72 (m, 4 H), 1.91–2.01 (m, 1 H), 3.41–3.54 (m, 4 H), 3.65–3.71 (m 4 H), 3.74 (s, 3 H), 3.79 (s, 3 H), 6.76 (s, 1 H), 7.30 (br s, 2 H, NH2), 7.41 (s, 1 H); MS (MALDI) m/z 358 (M+H)+, 380 (M+Na)+.
1-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)-2-methoxypropan-1-one (11)
Yield: 52%; 1H NMR (400 MHz, DMSO-d6) δ 1.20 (d, J = 4.5 Hz, 3 H, Me), 3.16 (s, 3 H), 3.38–3.45 (m, 4 H), 3.58–3.69 (m, 4 H), 3.78 (s, 3 H), 3.79 (s, 3 H), 4.20–4.24 (m, 1 H), 6.69 (s, 1 H), 7.10 (br s, 2 H, NH2), 7.37 (s, 1 H); MS (MALDI) m/z 384 (M+H)+, 406 (M+Na)+.
(1r,3R,5S)-adamantan-1-yl(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)methanone (12)
Yield: 52%; 1H NMR (400 MHz, DMSO-d6) δ 1.21 (t, J = 7.6 H, 2 H), 1.60–1.67 (m, 6 H), 1.86–1.96 (m, 7 H), 3.41–3.48 (m, 4 H), 3.62–3.66 (m, 4 H), 3.75 (s, 3 H), 3.80 (s, 3 H), 6.81 (s, 1 H), 7.45 (s, 1 H), 7.55 (br s, 2 H, NH2); MS (MALDI) m/z 452 (M+H)+, 474 (M+Na)+.
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(tetrahydrofuran-2-yl)methanone (13)
Yield: 65%; 1H NMR (400 MHz, DMSO-d6) δ 1.80–1.88 (m, 2 H), 1.96–2.10 (m, 2 H), 3.57–3.71 (m, 8 H), 3.72–3.80 (m, 2 H), 3.83 (s, 3 H), 3.86 (s, 3 H), 4.72–4.76 (m, 1 H), 7.56 (br s, 2 H, NH2), 7.75 (s, 2 H); MS (MALDI) m/z 388 (M+H)+, 410 (M+Na)+.
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(furan-2-yl)methanone (14)
Yield: 61%; 1H NMR (400 MHz, DMSO-d6) δ 3.42–3.55 (m, 4 H), 3.63–3.68 (m, 4 H), 3.83 (s, 3 H), 3.99 (s, 3 H), 6.67 (s, 1 H), 7.10 (t, J = 3.6 Hz, 1 H), 7.24 (s, 1H), 7.41 (s, 1 H), 7.35 (br s, 2 H, NH2), 7.43 (d, J = 3.8 Hz, 1 H); MS (MALDI) m/z 384 (M+H)+, 406 (M+Na)+.
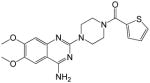
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(thiophen-2-yl)methanone (15)
Yield: 60%; 1H NMR (400 MHz, DMSO-d6) δ 3.45–3.56 (m, 4 H), 3.67–3.70 (m, 4 H), 3.76 (s, 3 H), 3.79 (s, 3 H), 6.75 (s, 1 H), 7.11 (t, J = 3.8 Hz, 1 H), 7.30 (br s, 2 H, NH2), 7.41 (s, 1 H), 7.43 (d, J = 3.8 Hz, 1 H), 7.72 (d, J = 3.8 Hz, 1 H); MS (MALDI) m/z 400 (M+H)+, 422 (M+Na)+.
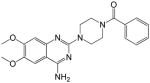
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(phenyl)methanone (16)
Yield: 59%; 1H NMR (400 MHz, DMSO-d6) δ 3.39–3.45 (m, 4 H), 3.62–3.72 (m, 4 H), 3.77 9s, 3 H), 3.88 (s, 3 H), 6.73 (s, 1 H), 7.17 (br s, 2H, NH2), 7.42 (s, 1 H), 7.43–7.48 (m, 5 H); MS (MALDI) m/z 394 (M+H)+, 416 (M+Na)+.
Binding assays and Kd determinations using NMR spectroscopy
Compound stocks (in DMSO-d6) were diluted in binding buffer (10 mM potassium phosphate, 150 mM NaCl, pH 6.0) and added to a solution of either 30 or 50 μM influenza RNA promoter at the indicated final concentrations (Supplementary Figure 1). Initial binding was assessed by observing the peak intensities of the imino protons of U26, G13 and G24. The reduction of peak intensity of U26 and G13 was measured and the values were used to calculate Kd values using GraphPad PRISM 6. The chemical shift displacement at the ribose region at peak ~5.714 ppm corresponding to the H1′ of adenosine 12(10) was also monitored to confirm binding of the compounds and for Kd determinations. Increasing concentrations of compounds 1, 3, 6, 7, 8, 10, 13, 15, 16 and 17 was titrated into 50 μM influenza RNA at the concentrations of 10, 20, 40, 60, 80, 100, 200 and 370 μM in buffer composed of 10 mM potassium phosphate, 50 mM NaCl, (pH = 6.0). NMR spectra were obtained on 600 MHz Bruker Avance spectrometer equipped with TCI cryoprobe. The NMR data was processed and analyzed using TOPSPIN2.1 (Bruker Biospin, MA). The Kd values were calculated with GraphPad PRISM 6.
Peptide library screening and guanidine and amidino containing compounds screening
A combinatorial tetra-peptide library consisted of 19 natural amino acids excluding cysteine was obtained (Pepscan, Zuidersluisweg, The Netherlands). The tetra-peptide library was assembled in a positional scanning format. (11, 12) The samples are grouped into mixtures where one amino acid is fixed at a certain position while the other 3 positions contain all possible combinations for the 19 amino acids. For example, a mixture of OXXX, where O is one of the 19 natural amino and X represents a combination of all 19 amino acids, contains 6,859 samples (1×19×19×19 = 6,859). The library contains 19 mixtures of OXXX, 19 mixtures of XOXX, 19 mixtures of XXOX and 19 mixtures XXXO and thus allows reasonable number of NMR experiments (19 + 19 + 19 + 19 = 76). Each mixture was prepared at 100 mM stock in DMSO-d6. The influenza RNA promoter was dissolved in buffer composed of 150 mM potassium phosphate, 150 mM NaCl, 0.1 mM MgCl2 at pH 6.4 for screening. A final concentration of 2 mM (10 μl of 100 mM DMSO stock) of each mixture was added to 10 μM of RNA. NMR spectra were collected using 600 MHz Bruker Avance with TCI cryoprobe and analyzed with TOPSPIN2.1 (Bruker Biospin, MA). Both peak intensity of imino proton of U26 and the chemical shift displacement for H1′ ribose peak of adenosine 12 at approximately 5.714 ppm were monitored for compound-RNA binding. However, none of the mixtures caused a significant perturbation of the NMR signal.
Guanidino and amidino compounds 18 to 24 were commercially available (Supplementary Table 1). Compound stocks (in DMSO-d6) were diluted in buffer (10 mM potassium phosphate, 150 mM NaCl, pH 6.0) and added to a solution containing 50 μM influenza RNA promoter at final concentration of 25 μM. We monitored the decrease in peak intensities of imino protons of U26, G13 and G24 for binding. The chemical shift perturbations at the H1′ of adenosine 12 at ~5.714 ppm were also monitored.
ATPlite™ assay for cytotoxicity assessment
Cytotoxicity assays were carried out using the ATPlite™ assay kit (PerkinElmer) following the manufacturer’s protocol. In short, in a white flat bottom 96-well plate, 25,000 MDCK cells were seeded and compounds and control (oseltamivir phosphate) were serial diluted (3x) and added to the well with final concentrations ranging from 250 μM to 38 nM. After a 24-hour incubation period, 10 μl of ATPlite solution was added to each well. The fluorescence reading was recorded using VictorTM X5, 2030 multilabel reader (PerkinElmer) and the CC50 values were plotted using GraphPad PRISM 6.
WSN-Ren luciferase assay to measure viral replication
To assess the ability of selected compounds to inhibit viral replication we adopted a WSN-Ren luciferase assay.(13) The coding sequence of Renilla luciferase was engineered in A/WSN/33 influenza virus in the place of hemagglutanin (HA) and a complementary MDCK cell line expressing HA (MDCK-HA) was used to allow multiple cycles of replication. In a white 96-well plate, 25,000 MDCK-HA cells were seeded and compounds and control (oseltamivir phosphate) were serial diluted (3x) and then added to treat the cells at the final concentration ranging from 250 μM to 38 nM. Renilla luciferase substrate (Omega) was added following manufacturer’s protocol after 4 hours of compound treatment and then incubated overnight. Fluorescence signal was read at 24 hours post infection using VictorTM X5, 2030 multilabel reader (PerkinElmer). The IC50 values were calculated with GraphPad PRISM 6.
Viral replication assay using Real-time PCR to measure viral mRNA level
MDCK cells were plated in 96-well plate at 25,000 cell/well and incubated at 37°C overnight. Cells were treated separately with 50 μM of oseltamivir phosphate, 50 μM of compounds 7, 8 and 10, and DMSO as control and then infected with wild-type influenza virus (A/Puerto Rico/8/1935) at MOI = 0.2. Infected cells were harvested after 24 h post infection with one PBS wash and the cells were lysed with RA1 buffer (RA1 with 1% β–mercaptoethanol) following the Macherey-Nagel manufacture’s protocol in 96 well format for RNA extraction. The purified total RNA was then reverse transcribed using iScript™ cDNA Synthesis Kit from BIO-RAD (Catalog #170-8891). Real time PCR is then used to measure the mRNA production of viral nucleoprotein (A/PR8/34 nucleoprotein) and cellular β-actin in the infected cells for standardization.
Results
We previously suggested that the influenza RNA promoter represents a novel and highly conserved druggable target to combat influenza infections (13) and we identified compound 1 (Figure 1A) as an influenza RNA promoter binding small molecule. Using solution NMR spectroscopy, we reported that the interactions between compound 1 and the viral RNA promoter are mediated by the methoxy groups of compound 1, forming hydrogen bonds with adenine 12 and cytosine 22 of the RNA (Figure 1A), and by electrostatic interactions between the phosphate backbone and the primary amine, likely protonated at physiological pH, in the compound. Moreover, the binding of compound 1 to the viral RNA caused a straightening of the bend close to the AA-U motif in the interloop, which has been suggested to important for RdRp recognition.(10) As a consequence, the compound-bound RNA promoter maintains an A-form helical structure, which presumably is not a good substrate for the RdRp.
We also noticed that in the compound-RNA complex, the piperazine of compound 1 is oriented toward the cavity of the major groove of the RNA helix (Figure 1B). We therefore hypothesized that we might improve the compound binding potency and/or pharmacological properties by modifying the secondary piperazinyl amine.
We assembled a total of 15 analogues of compound 1 (namely compounds 3 to 16, that were synthesized in house, and compound 17 that was purchased from Asinex), all carrying modifications on the secondary amine of the piperazine (Table 1). Compounds 6 to 16 were synthesized using one step coupling using different starting materials (Figure 2). Compound 3 was synthesized using Boc-Gly-Gly-Gly-OH with oxyma pure, DIC, DIEA and DMF at room temperature for 18 hours followed by deprotection of Boc group with 50% TFA in dichloromethane at room temperature for 3 hours (Supplementary Figure 2). Compounds 4 and 5 were synthesized as detailed in Supplementary Figure 2.
Figure 2. Synthesis of 4-amino-6,7-dimethoxy-2-(piperazin-1-yl)quinazoline derivatives (compounds 6 to 16).
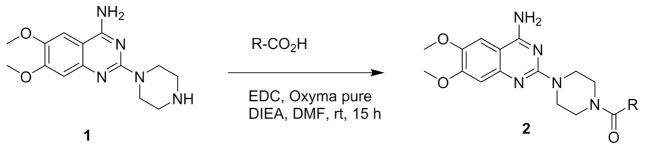
Compounds 6 to 16 were synthesized from commercially available compound 1 using standard coupling conditions (EDC, oxyma pure, DIEA in DMF incubated at rt for 15h) with different starting materials as detailed in Figure 2. 6) R = Et; (7) R = i-Pr; (8) R = Pr; (9) R = 2-Hydroxyethyl; (10) R = Cyclopropyl; (11) R = 1-Methoxyethyl; (12) R = 1-adamantyl; (13) R = tetrahydrofuran; (14) R = Furan; (15) R = thiophene; (16) R = Phenyl.
We first assessed if the compounds were able to bind to the RNA promoter using 1D 1H NMR as we reported previously (10). This primary binding assay confirmed that compounds 3 to 17 were all able to interact with the promoter (Supplementary Figure 1). All compounds seemed to bind to the RNA promoter in a fashion similar to compound 1, causing line broadenings of the imino protons of U26, G13 and G24. In addition, some compounds (3, 4, 5, 7, 9, 11 and 14) also caused line broadening of the imino protons of U27 as compound 1 did (Supplementary Figure 1), again indicating that they likely bind to the RNA in a similar fashion as compound 1.
We then tested compound 3 to 17 for cytotoxicity and ex vivo anti-influenza replication activities. Compound cytotoxicity was assessed using ATPlite™ assay (Perkin Elmer) in MDCK-HA cells. Three compounds were found to be toxic to MDCK-HA cells. Compound 14 showed CC50 value of 78.23 μM, compound 15 showed CC50 value of 66.97 μM and compound 12 was determined to be the most toxic among this set of analogues with CC50 value of 16.4 μM (Table 1). All the remaining compounds did not show significant cytotoxicity when tested up to 250 μM.
We next examined whether the remaining compounds affected influenza viral replication using the WSN-Ren luciferase assay. WSN-Ren luciferase assay was established using a modified influenza virus (A/WSN/33) encoding Renilla luciferase in the place of hemagglutinin (HA) and a complementary MDCK cell line expressing HA (MDCK-HA).(13) We previously reported that compound 1 is active in this assay with an IC50 of 549 μM.(10) By comparing compound-treated to DMSO-treated cells, we found that six compounds (3, 4, 5, 9, 11 and 12) did not possess significant anti-influenza activity in this cell-based assay. The IC50 values were determined for the remaining active compounds. Compounds 6, 7, 8, 10, 13, 14, 15, 16 and 17 displayed improved anti-viral activity compared to compound 1. Compounds 16 and 17 were the least potent yet still had IC50 values of 107.55 ± 49.09 μM and 126.0 ± 24.65 μM respectively, roughly four fold more potent than compound 1 (Table 1). Compounds 6, 7, 8, 10, 13 and 14 displayed IC50 values within a close range (between 25 and 55 μM). Compound 15 had the lowest IC50 value of 16.77 ± 3.99 μM (Table 1), however it was toxic to cells at the upper concentrations tested, which might have skewed the IC50 curve. To more accurately represent the potency of the active compounds, we therefore report the ratio between the IC50 value and CC50 value for each compound in Table 1. The ideal compound would display high inhibitory activity and low cytotoxicity resulting in a low IC50/CC50 ratio. Compounds 7 and 10 were therefore the best performers, displaying IC50/CC50 ratios <0.14, while compounds 6 and 8 also performed well with an IC50/CC50 ratio <0.18 (Table 1).
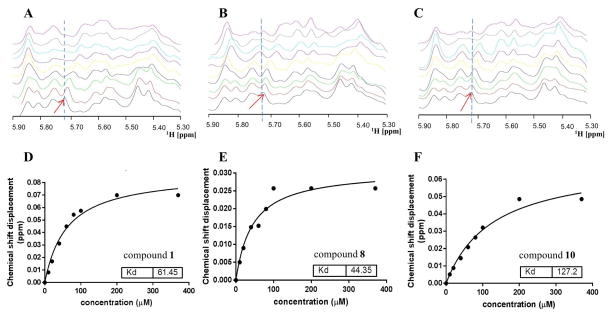
Using careful titrations, Kd values were determined by NMR spectroscopy for compounds 1, 3, 6, 7, 8, 10, 13, 15, 16 and 17. We previously estimated the Kd value for compound 1 monitoring the peak intensity of the imino protons of U26, G13 and G24, resulting in a Kd value of 50.5 ± 9 μM.(10) However, using peak intensity is not an accurate approach since the binding is likely in the intermediate exchange with respect to the time scale related to the chemical shift perturbations at the imino protons. Therefore, we monitored chemical shift perturbations in the ribose region upon compound binding, as a fast exchange regime is observed within the RNA ribose protons upon complex formation. From that, we calculated Kd values using the chemical shift titration of the ribose peak at 5.714 ppm (corresponding to the H1′ of A12) for compound 1 that led to a Kd value of 61.45 μM (Figure 3A and 3D), close to the previously published value (Table 1). Except for compound 8, which displayed a Kd value of 44.35 μM (Figure 3B and 3E), all the other tested compounds had Kd values above 100 μM. As mentioned above, in a Renilla luciferase reporter assay (13, 14), the most potent compounds are 7, 8 and 10 with Kd values of 158.5 μM, 44.4 μM, and 127.2 μM, respectively for binding to the RNA promoter (Figure 3B–F) and IC50 values of 33.9 μM, 44.2 μM, and 34.2 μM, respectively (Table 1). Even though compounds 7 and 10 had Kd values slightly weaker (> 100 μM) compared to compound 1 (61.45 μM), their IC50 values (33.9 μM and 34.2 μM) were much improved from 549 μM of compound 1. Compound 8 possesses both an improved Kd value at 44.4 μM and an improved IC50 value of 44.2 μM compared to compound 1 (Table 1).
Figure 3. NMR based Kd determination for compounds 1, 8 and 10.
(A), (B) and (C) reports 1D 1H NMR spectra in the ribose region of the influenza RNA promoter (50 μM) titrated with 0, 10, 20, 40, 60, 80, 100, 200 and 370 μM of compounds 1, 8 and 10, respectively. Chemical shift perturbations at 5.714 ppm (red arrow, corresponding to the H1′ of adenosine 12) were monitored and the displacement of distance (δ ppm) used to calculate the Kd values in GraphPad PRISM 6 shown in (D), (E) and (F) for compounds 1, 8 and 10, respectively.
In order to further confirm the anti-influenza activity of compounds 7, 8 and 10 in an orthogonal cell-based assay, we measured the nucleoprotein mRNA levels of cells infected with influenza A virus in the presence or absence of compounds. In the WSN-Ren assay conditions, MDCK-HA cells were treated with 50 μM of compounds 7, 8 or 10, using oseltamivir phosphate and compound 1 at the same concentrations as controls (Figure 4A). Compounds 7 and 10 at 50 μM caused a significant inhibition of viral mRNA production, similar to that of oseltamivir, with p-values of 0.003 and 0.0052 respectively in unpaired t-test against DMSO control (Figure 4A). Compound 8 was slightly less active but still significant in reducing viral RNA levels compared to DMSO (p-value = 0.05) (Figure 4A). Compound 10 also demonstrated significant dose-dependent inhibitory activity (one-way ANOVA test P value of 0.0111) in a parallel system where wild-type MDCK cells and wild-type influenza A/PR8/35 were used to measure viral nucleoprotein mRNA production levels (Figure 4B).
Figure 4. Viral replication assay measured by RT-PCR.
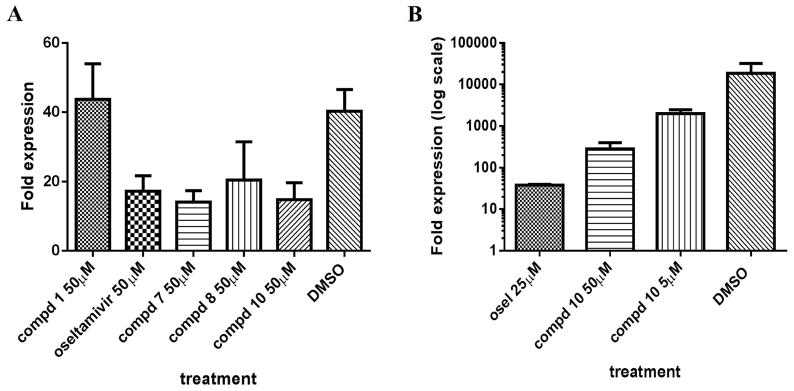
(A) MDCK-HA cells were treated with control or compounds at 50 μM and then were infected with WSN-Ren luciferase virus in the same condition as the WSN-Ren luciferase assay. The mRNA level of WSN nucleoprotein was measured and standardized against beta-actin. The fold expression indicated that compounds 7, 8, and 10 at 50 μM showed similar inhibitory effect as oseltamivir phosphate at the same concentration while compound 1 did not showed significant inhibitory activity at 50 μM. The P value is < 0.0001 analyzed by one-way ANOVA test.
(B) MDCK cells were treated with controls (oseltamivir phosphate or DMSO) or compound and then infected with wild-type influenza A/PR/8/35 virus at MOI= 0.2. Influenza A/PR8 Nucleoprotein mRNA was measured and standardize against beta-actin mRNA level. The fold expressed is shown in log10 scale. Oseltamivir phosphate at 25 μM and compound 10 at 50 μM and 5 μM showed inhibitory effects as the mRNA levels were reduced compared to DMSO treated cells. The P value was analyzed with one-way ANOVA analysis to be P = 0.0111.
Discussion and Conclusions
The need for new anti-influenza therapy is pressing as drug resistant strains of the virus emerge.(15) We have previously reported that the highly conserved RNA promoter sequence on the 3′ and 5′ ends of each viral genomic segment is a novel druggable candidate.(7, 9) Here, we aimed first to identify the amino acid motif binding directly to the RNA promoter employing the HTS by NMR technique.(16) Influenza RNA promoter is bound by a hetero-trimeric RNA dependent RNA polymerase (RdRp), where PB1 subunit makes direct contact with the RNA promoter.(17) Nevertheless, it was only vaguely reported that both the C and N termini of PB1 are required for RNA binding with very little information on where the interaction occurs or any sequence or structural requirements for the interaction.(18) We screened a combinatorial peptide library via 1D proton NMR using our recently developed HTS by NMR method.(16) A tetra-peptide library was assembled in positional scanning mixtures, where one position was systematically fixed as one natural amino acid (except for cysteine) while the other three positions carry all the possible combinations of natural amino acid (except for cysteine). The arrangement of combinatorial library would allow the identification of peptide binding sequence and the interaction location of a specific target. We screened this library by 1D proton NMR monitoring both imino and ribose regions (imino protons of U26 and G13 and the H1′ of A12 at ~5.72 ppm, respectively).(10) However, no clear hits emerged, indicating that there might not be a linear epitope that appreciably binds to the influenza RNA promoter. This result further corroborates the hypothesis that viral RdRp might recognize the overall shape of the RNA helical structure that would function as a ligand itself for the binding.(19)
Taking advantage of the highly sensitive NMR-based binding assay, we assembled a small library of compounds all containing basic residues such as guanidine or amidino, given that such functional groups are often favored in nucleic acid binding compounds (Supplementary Table 1).(20) We proceeded to test these compounds in 1D proton NMR in the same fashion as described for compounds 1 to 17 and found that compounds 20 and 23 caused significant line broadening at the imino protons of G13 and chemical shift perturbations in the ribose peak at 5.714 ppm, indicating binding to influenza RNA promoter (Supplementary Table 1). However, other guanidine or amidino containing compounds did not show appreciable binding to the RNA promoter indicating that the binding of compounds 20 and 23 may be specific and not merely driven by electrostatic interactions. Unfortunately, both compounds were found to be cytotoxic (CC50 values of 6.8 μM and 20.2 μM, respectively), and therefore unsuitable for further evaluation and optimization.
These results further substantiated our interest in compound 1 as a starting point for the development of novel RNA promoter binding antagonists. We reported that compound 1, albeit binding the RNA promoter with a Kd = 61.45 μM, was not particularly effective in inhibiting viral replication, exhibiting only a modest IC50 = 549 μM using a luciferase-based assay.(10) Accordingly, when tested in an orthogonal viral replication assay measuring nucleoprotein mRNA level using qPCR, compound 1 was not effective at 50 μM (Figure 4A). Based on the NMR structure of compound 1 in complex with the RNA promoter, we observed that the interactions were mediated primarily by the methoxy groups of compound 1, and its primary amine (Figure 1A), suggesting that the secondary amine on the piperazine ring could be used to improve either the binding affinity and/or the pharmacological properties of the compound. Compounds 3, 6, 7, 8, 10 and 17 had Kd values that are similar to compound 1 (Table 1). However, even though the biding affinity of the compound analogues might not have improved significantly compared to compound 1, most of the compounds exerted dramatically improved inhibitory activities in ex vivo assays. For all the compounds that demonstrated inhibitory activities (compounds 6, 7, 8, 10, 13, 14, 15, 16 and 17), the IC50 values were roughly ten-fold better compared to IC50 of 549 μM of compound 1 (Table 1). For example compound 8 had IC50 value average at 44.18 μM and had an improved affinity with a Kd = 44.35 μM (Supplementary Figure 3). Moreover, when using the IC50/CC50 ratio to evaluate the potency of this compound series, compounds 7, 8 and 10 were identified to be the inhibitors with the largest cellular therapeutic window (IC50/CC50 ratio) in this series. Even though compounds 3, 4, 5, 9, 11 and 12 did not inhibit viral replication in cellular assay, they all bound to the influenza RNA promoter (Table 1). Similar to compound 1, these molecules might not exhibit cellular anti-influenza activity, perhaps due to premature degradation, limited cell permeability, binding to other targets, etc. In this regard, recent work clearly suggest that direct inhibition of RdRp targeting PB1-PA interactions with small molecules can be a viable target for inhibition of viral replication in both influenza A and B viruses. (21–23) While we cannot rule out that our compounds may also directly affect RdRp in a similar fashion, our molecules do not share any structural features that resemble the reported chemical inhibitors of PB1-PA interactions. (21–23) Nonetheless, this point needs to be experimentally verified in follow-up studies.
In conclusion, our preliminary attempts to identify novel pharmacological tools binding to the RNA promoter of influenza A virus culminated in the identification of small molecules that possess inhibitory activity against influenza viral replication in both luciferase based cellular assay and RT-PCR, with compounds 7, 8 and 10 in particular displaying IC50/CC50 ratios < 0.18 and activity in the tens of μM in both binding and cell-based assays. These studies further validated the influenza RNA promoter as possible novel druggable target against influenza A viral infections.
Supplementary Material
Supplementary Figure 1. NMR spectra of influenza RNA promoter at the imino region in presence of various test compounds.
(A) Compounds 3, 4 and 13 at the indicated concentrations were added to 50 μM of influenza RNA promoter. The peak intensity reduction of imino protons of U26 and G13 indicated compound binding to the RNA promoter.
(B) Compounds 8, 11, 12, 14 and 15 at 30 μM were added to 30 μM of influenza RNA promoter. The peak intensity reduction of imino protons of U26 and G13 indicated compound binding to RNA promoter.
(C) Compounds 5, 6, 7, 9, 10 and 16 at 30 μM were added to 30 μM of influenza RNA promoter. The peak intensity reduction of imino protons of U26 and G13 indicated compound binding to RNA promoter.
Supplementary Figure 2. Synthesis of compounds 3, 4 and 5.
Compound 4 was prepared by coupling of compound 1 with 5-Boc-aminopeptanoic acid (5-Boc-aminopentanoic acid, oxyma pure, DIC, DIEA, DMF, rt, 18 h) followed by removal of Boc group in the presence of TFA (50% TFA in dichloromethane, rt, 3 h). Similarly, compound 3 was synthesized using Boc-Gly-Gly-Gly-OH (Boc-Gly-Gly-Gly-OH, oxyma pure, DIC, DIEA, DMF, rt, 18 h and deprotection of Boc group with 50% TFA in dichloromethane at rt for 3 h. Compound 1 was coupled with Methanesulfonyl chloride in the presence of base at rt for 6 h to give compound 5.
Supplementary Figure 3. WSN-Ren luciferase assay was used to determine IC50 values.
WSN-Ren luciferase assay was used for assessing viral replication using luciferase production and IC50 values were determined. MDCK-HA cells were seeded at 25,000 cells/well in a white, flat-bottom 96 well plate and allowed to grow overnight at 37°C. Each compound was 3x serially diluted and added to cells to reach final concentrations of 250 μM to 38 nM. WSN-Ren luciferase virus was then used to infect the cells and the Renilla luciferase substrate was added at 4 hour after infection. The luminescence was read at 24 h post infection and was calculated to be percent luminescence standardizing against DMSO treated cells. Supplementary Figure 3A demonstrated that compound 3 did not inhibit the viral replication and had an IC50 value > 250 μM. Figure 3B showed a representative graph for compound 8 had while Figure 3C showed a representative graph for compound 10.
Supplementary Table 1: Chemical structure and binding affinities for compounds 18 to 24 to the influenza A virus RNA promoter.
Compounds were tested as described in the methods section. NB indications no appreciable binding is detected under the chosen experimental conditions. + indicates significant line broadening at the imino protons of U26 and G13 upon complexation under the experimental conditions (see methods).
Acknowledgments
We thank Mr. Stephen Soonthornvacharin for helping with the viral replication assay. Financial support is from grants NIH grant AI098091 (MP) and NIH 9RO1 GM 110569-06 (GV).
Footnotes
Conflict of interest
The authors declare no financial/commercial conflicts of interest.
References
- 1.Sharma M, Li C, Busath DD, Zhou HX, Cross TA. Drug sensitivity, drug-resistant mutations, and structures of three conductance domains of viral porins. Biochimica et biophysica acta. 2011;1808:538–46. doi: 10.1016/j.bbamem.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Clercq E. Antiviral agents active against influenza A viruses. Nature reviews Drug discovery. 2006;5:1015–25. doi: 10.1038/nrd2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoneda M, Okayama A, Kitahori Y. Oseltamivir-resistant seasonal A(H1N1) and A(H1N1)pdm09 influenza viruses from the 2007/2008 to 2012/2013 season in Nara Prefecture, Japan. Japanese journal of infectious diseases. 2014;67:385–8. doi: 10.7883/yoken.67.385. [DOI] [PubMed] [Google Scholar]
- 4.Boivin G. Detection and management of antiviral resistance for influenza viruses. Influenza and other respiratory viruses. 2013;7(Suppl 3):18–23. doi: 10.1111/irv.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boltz DA, Aldridge JR, Jr, Webster RG, Govorkova EA. Drugs in development for influenza. Drugs. 2010;70:1349–62. doi: 10.2165/11537960-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi F, Xie Y, Shi L, Xu W. Viral RNA polymerase: a promising antiviral target for influenza A virus. Current medicinal chemistry. 2013;20:3923–34. doi: 10.2174/09298673113209990208. [DOI] [PubMed] [Google Scholar]
- 7.Bae SH, Cheong HK, Lee JH, Cheong C, Kainosho M, Choi BS. Structural features of an influenza virus promoter and their implications for viral RNA synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10602–7. doi: 10.1073/pnas.191268798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fodor E. The RNA polymerase of influenza a virus: mechanisms of viral transcription and replication. Acta virologica. 2013;57:113–22. doi: 10.4149/av_2013_02_113. [DOI] [PubMed] [Google Scholar]
- 9.Park CJ, Bae SH, Lee MK, Varani G, Choi BS. Solution structure of the influenza A virus cRNA promoter: implications for differential recognition of viral promoter structures by RNA-dependent RNA polymerase. Nucleic acids research. 2003;31:2824–32. doi: 10.1093/nar/gkg387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MK, Bottini A, Kim M, Bardaro MF, Jr, Zhang Z, Pellecchia M, et al. A novel small-molecule binds to the influenza A virus RNA promoter and inhibits viral replication. Chemical communications. 2014;50:368–70. doi: 10.1039/c3cc46973e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT, Cuervo JH. Generation and use of synthetic peptide combinatorial libraries for basic research and drug discovery. Nature. 1991;354:84–6. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- 12.Dooley CT, Houghten RA. The use of positional scanning synthetic peptide combinatorial libraries for the rapid determination of opioid receptor ligands. Life sciences. 1993;52:1509–17. doi: 10.1016/0024-3205(93)90113-h. [DOI] [PubMed] [Google Scholar]
- 13.Bottini A, De SK, Baaten BJ, Wu B, Barile E, Soonthornvacharin S, et al. Identification of small molecules that interfere with H1N1 influenza A viral replication. Chem Med Chem. 2012;7:2227–35. doi: 10.1002/cmdc.201200453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, et al. Human host factors required for influenza virus replication. Nature. 2010;463:813–7. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurt AC. The epidemiology and spread of drug resistant human influenza viruses. Current opinion in virology. 2014;8C:22–9. doi: 10.1016/j.coviro.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Wu B, Zhang Z, Noberini R, Barile E, Giulianotti M, Pinilla C, et al. HTS by NMR of combinatorial libraries: a fragment-based approach to ligand discovery. Chemistry & biology. 2013;20:19–33. doi: 10.1016/j.chembiol.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biswas SK, Nayak DP. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. Journal of virology. 1994;68:1819–26. doi: 10.1128/jvi.68.3.1819-1826.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez S, Ortin J. Distinct regions of influenza virus PB1 polymerase subunit recognize vRNA and cRNA templates. The EMBO journal. 1999;18:3767–75. doi: 10.1093/emboj/18.13.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Varani G. Protein families and RNA recognition. The FEBS journal. 2005;272:2088–97. doi: 10.1111/j.1742-4658.2005.04650.x. [DOI] [PubMed] [Google Scholar]
- 20.Parkesh R, Childs-Disney JL, Nakamori M, Kumar A, Wang E, Wang T, et al. Design of a bioactive small molecule that targets the myotonic dystrophy type 1 RNA via an RNA motif-ligand database and chemical similarity searching. Journal of the American Chemical Society. 2012;134:4731–42. doi: 10.1021/ja210088v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muratore G, Goracci L, Mercorelli B, Foeglein A, Digard P, Cruciani G, et al. Small molecule inhibitors of influenza A and B viruses that act by disrupting subunit interactions of the viral polymerase. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6247–52. doi: 10.1073/pnas.1119817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lepri S, Nannetti G, Muratore G, Cruciani G, Ruzziconi R, Mercorelli B, et al. Optimization of small-molecule inhibitors of influenza virus polymerase: from thiophene-3-carboxamide to polyamido scaffolds. Journal of medicinal chemistry. 2014;57:4337–50. doi: 10.1021/jm500300r. [DOI] [PubMed] [Google Scholar]
- 23.Pagano M, Castagnolo D, Bernardini M, Fallacara AL, Laurenzana I, Deodato D, et al. The fight against the influenza A virus H1N1: synthesis, molecular modeling, and biological evaluation of benzofurazan derivatives as viral RNA polymerase inhibitors. Chem Med Chem. 2014;9:129–50. doi: 10.1002/cmdc.201300378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. NMR spectra of influenza RNA promoter at the imino region in presence of various test compounds.
(A) Compounds 3, 4 and 13 at the indicated concentrations were added to 50 μM of influenza RNA promoter. The peak intensity reduction of imino protons of U26 and G13 indicated compound binding to the RNA promoter.
(B) Compounds 8, 11, 12, 14 and 15 at 30 μM were added to 30 μM of influenza RNA promoter. The peak intensity reduction of imino protons of U26 and G13 indicated compound binding to RNA promoter.
(C) Compounds 5, 6, 7, 9, 10 and 16 at 30 μM were added to 30 μM of influenza RNA promoter. The peak intensity reduction of imino protons of U26 and G13 indicated compound binding to RNA promoter.
Supplementary Figure 2. Synthesis of compounds 3, 4 and 5.
Compound 4 was prepared by coupling of compound 1 with 5-Boc-aminopeptanoic acid (5-Boc-aminopentanoic acid, oxyma pure, DIC, DIEA, DMF, rt, 18 h) followed by removal of Boc group in the presence of TFA (50% TFA in dichloromethane, rt, 3 h). Similarly, compound 3 was synthesized using Boc-Gly-Gly-Gly-OH (Boc-Gly-Gly-Gly-OH, oxyma pure, DIC, DIEA, DMF, rt, 18 h and deprotection of Boc group with 50% TFA in dichloromethane at rt for 3 h. Compound 1 was coupled with Methanesulfonyl chloride in the presence of base at rt for 6 h to give compound 5.
Supplementary Figure 3. WSN-Ren luciferase assay was used to determine IC50 values.
WSN-Ren luciferase assay was used for assessing viral replication using luciferase production and IC50 values were determined. MDCK-HA cells were seeded at 25,000 cells/well in a white, flat-bottom 96 well plate and allowed to grow overnight at 37°C. Each compound was 3x serially diluted and added to cells to reach final concentrations of 250 μM to 38 nM. WSN-Ren luciferase virus was then used to infect the cells and the Renilla luciferase substrate was added at 4 hour after infection. The luminescence was read at 24 h post infection and was calculated to be percent luminescence standardizing against DMSO treated cells. Supplementary Figure 3A demonstrated that compound 3 did not inhibit the viral replication and had an IC50 value > 250 μM. Figure 3B showed a representative graph for compound 8 had while Figure 3C showed a representative graph for compound 10.
Supplementary Table 1: Chemical structure and binding affinities for compounds 18 to 24 to the influenza A virus RNA promoter.
Compounds were tested as described in the methods section. NB indications no appreciable binding is detected under the chosen experimental conditions. + indicates significant line broadening at the imino protons of U26 and G13 upon complexation under the experimental conditions (see methods).