An analysis of Pseudomonas syringae AvrB-induced stomatal invasion uncovers an unexpected role of plasma membrane H+-ATPase in jasmonate signaling.
Abstract
Stomata are natural openings through which many pathogenic bacteria enter plants. Successful bacterial pathogens have evolved various virulence factors to promote stomatal opening. Here, we show that the Pseudomonas syringae type III effector protein AvrB induces stomatal opening and enhances bacterial virulence in a manner dependent on RPM1-INTERACTING4 (RIN4), which promotes stomatal opening by positively regulating the Arabidopsis plasma membrane H+-ATPase (AHA1), which is presumed to directly regulate guard cell turgor pressure. In support of a role of AHA1 in AvrB-induced stomatal opening, AvrB enhances ATPase activity in plants. Unexpectedly, AHA1 promotes the interaction between the jasmonate (JA) receptor CORONATINE INSENSITIVE1 (COI1) and JASMONATE ZIM-DOMAIN (JAZ) proteins and enhances JA signaling. JA signaling is required for optimum stomatal infection in AHA1-active plants. Similarly, AvrB also induces the COI1-JAZ9 interaction and the degradation of multiple JAZ proteins. AvrB-induced stomatal opening and virulence require the canonical JA signaling pathway, which involves the COI1 and NAC transcription factors. The findings thus point to a previously unknown pathway exploited by P. syringae that acts upstream of COI1 to regulate JA signaling and stomatal opening.
INTRODUCTION
Stomatal pores, delineated by a pair of guard cells, are important structures in the epidermis of terrestrial plants that primarily function in gas exchange and transpiration. As a natural opening, stomata are exploited by many bacterial pathogens as a gateway for invasion. The stomatal aperture is regulated by the volume of guard cells and is subject to regulation by the circadian clock, CO2 concentration, light, temperature, humidity, and drought to coordinate photosynthesis activities and control water status (Kim et al., 2010). Plants actively close stomata to restrict attempted bacterial entry, and some bacterial pathogens are capable of opening stomata to gain access to the interior of plant tissues (Melotto et al., 2006).
Coronatine (COR), a bacterial factor known to open stomata, is a small molecule produced by a number of Pseudomonas syringae isolates (Mittal and Davis, 1995; Melotto et al., 2006). COR-induced stomatal opening plays a major role in bacterial entry into the leaf tissue when spray-inoculated (Melotto et al., 2006). The ability of P. syringae to open stomata is not limited to isolates containing COR; different P. syringae strains use a variety of strategies to open stomata. Syringolin A, which is produced by some P. syringae pv syringae strains, acts as a proteasome inhibitor to open stomata and counteract stomatal innate immunity in bean (Phaseolus vulgaris) and Arabidopsis thaliana plants (Schellenberg et al., 2010). Furthermore, some the P. syringae effector proteins HopZ1a and HopX1 have recently been shown to assist bacterial entry through stomata (Jiang et al., 2013; Gimenez-Ibanez et al., 2014).
Accumulating evidence points to the jasmonate (JA) signaling pathway as a target actively manipulated by P. syringae for virulence. COR is a structural and functional analog of the active form of JA, JA-Ile (Katsir et al., 2008). Both COR and JA-Ile are perceived by their receptor COI1, an F-box protein that recruits transcription repressor JASMONATE ZIM-DOMAIN (JAZ) proteins to the SCFCOI1 E3 ligase complex for degradation (Sheard et al., 2010). The removal of JAZ proteins enables the transcription of several transcription activators and triggers JA responses. Arabidopsis coi1 mutants do not respond to COR (Xie et al., 1998; Kloek et al., 2001) and do not support COR-induced stomatal opening and virulence (Mittal and Davis, 1995; Melotto et al., 2006). Similarly, HopZ1a acetylates and promotes the degradation of JAZ proteins, whereas HopX1 is a cysteine protease that directly degrades JAZ proteins, resulting in the activation of JA signaling and stomatal opening (Jiang et al., 2013; Gimenez-Ibanez et al., 2014). Recent findings indicate that JA signaling regulates stomatal opening through conserved NAM-ATAF-CUC2 (NAC) transcription factors, including ARABIDOPSIS NACs (ANAC019, ANAC055, and ANAC072) and tomato (Solanum lycopersicum) JA2-LIKE (JA2L) (Zheng et al., 2012; Du et al., 2014).
The P. syringae effector protein AvrB triggers immunity in plants carrying the cytoplasmic immune receptor RPM1 (Grant et al., 1995). The immune activation requires an RPM1-interacting protein called RIN4, which is a plasma membrane (PM)-associated protein (Mackey et al., 2002). RIN4 also interacts with AvrB and RIPK, a receptor-like cytoplasmic kinase (Liu et al., 2011), which leads to a specific phosphorylation of RIN4 at Thr-166 that is essential for RPM1 activation (Mackey et al., 2002; Chung et al., 2011; Liu et al., 2011). The AvrB-RIN4 interaction has been hypothesized to promote bacterial virulence in plants lacking RPM1 (Dangl and Jones, 2001; Kim et al., 2005), but direct evidence is lacking. Nonetheless, we have previously shown that AvrB can substitute COR to induce JA response gene expression in a manner dependent on COI1 and RIN4 (He et al., 2004; Shang et al., 2006; Cui et al., 2010). Whether the elevated JA signaling contributes to bacterial virulence remains unknown.
It was recently shown that RIN4 directly interacts with and enhances the activity of the Arabidopsis PM H+-ATPase (AHA1), thereby promoting stomatal opening (Liu et al., 2009). Activation of AHA1 alters plasma membrane potential, which is expected to drive the influx of K+ ion and solutes, resulting in the increased turgor pressure necessary for stomatal opening (Dietrich et al., 2001; Kim et al., 2010), raising the possibility that AvrB regulates stomatal opening through the RIN4-AHA1 pathway. However, AHA1 is not known to influence JA signaling. Furthermore, it is puzzling how a plasma membrane-localized effector (Nimchuk et al., 2000) modulates JA signaling in plants. Here, we show that transgenic expression of AvrB enhances PM H+-ATPase activity, and bacterially delivered AvrB induces stomatal opening and enhances stomatal infection in a RIN4-dependent manner, demonstrating that the RIN4-AHA1 pathway is exploited by AvrB for virulence. Surprisingly, ost2-2D, a gain-of-function mutant of AHA1, displayed elevated expression of JA response genes, including ANAC019, ANAC055, and ANAC072, suggesting that AHA1 regulates stomatal opening through JA signaling. Consistent with this notion, the ost2-2D coi1 double mutant displayed reduced disease susceptibility to P. syringae compared with ost2-2D. Furthermore, the COI1-JAZ9 interaction in Arabidopsis was enhanced upon the expression of AvrB or in the ost2-2D background. RIN4, AHA1, and AvrB all induced the degradation of multiple JAZ proteins when transiently expressed in Nicotiana benthamiana. AvrB-induced stomatal opening in tomato requires COI1 and JA2L, demonstrating that this stomatal virulence is executed by the canonical JA signaling pathway. Together, the data uncover a previously unknown mechanism in the regulation of JA-mediated stomatal opening that is actively exploited by the P. syringae effector AvrB.
RESULTS
AvrB Complements the cor- Mutation in P. syringae to Induce Stomatal Opening
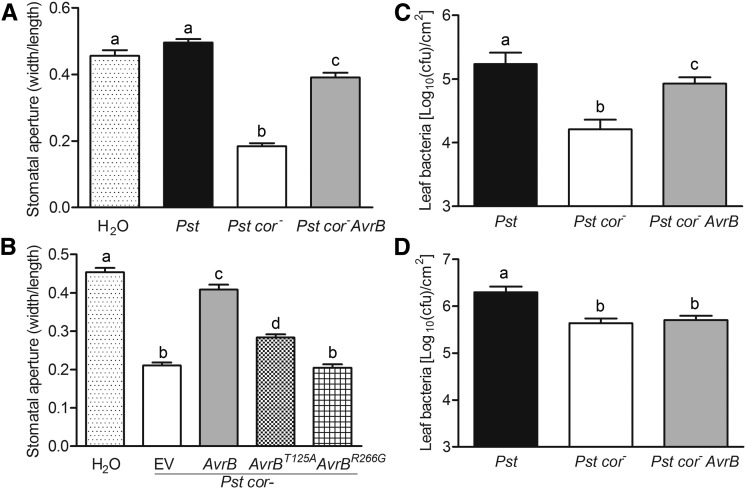
We previously showed that AvrB can substitute COR to induce JA response genes (He et al., 2004). Because a major function of COR is to induce stomatal opening, this raises the possibility that AvrB may similarly promote stomatal opening. To test this, we exposed epidermal strips of rpm1 plants with a COR-deficient strain Pst cor- carrying avrB. In this experiment, the wild-type strain Pst was used as a positive control and Pst cor- containing an empty vector served as a negative control. Consistent with previous reports (Melotto et al., 2006), epidermal strips treated with Pst cor- showed tightly closed stomata 4 h after treatment, whereas those treated with Pst or water displayed open stomata (Figure 1A; Supplemental Figure 1). Epidermal strips treated with Pst cor- avrB consistently showed larger stomatal apertures than those treated with Pst cor- (Figure 1A; Supplemental Figure 1), indicating that the bacterially delivered AvrB largely compensated for the COR deficiency in stomatal opening.
Figure 1.
avrB Complements the Pst cor- Mutation in Stomatal Opening.
(A) AvrB reopens closed stomata in the absence of COR. rpm1 leaves were exposed to water, Pst, Pst cor-, or Pst cor- avrB, and the width and length of stomata were measured after a 4-h treatment. The width-to-length ratio is shown in the graph. Three independent experiments were performed with similar results.
(B) Strains carrying avrB mutants that were previously known to be nonfunctional were impaired in their ability to reopen closed stomata.
(C) AvrB promotes bacterial growth of Pst cor- in rpm1 when plants were spray-inoculated.
(D) avrB does not enhance Pst cor- growth in plants when inoculated by syringe infiltration.
Error bars indicate se. Different letters denote significant differences at P ≤ 0.01; Student’s t test, n ≥ 30 ([A] and [B]) or 7 ([C] and [D]); three biological repeats.
The avrBT125A and avrBR266G mutants are nonfunctional in triggering RPM1 resistance and JA signaling (Ong and Innes, 2006; Cui et al., 2010). We examined if these mutants were affected in their ability to open stomata of rpm1 epidermal strips when delivered by Pst cor-. In contrast to strips inoculated with Pst cor- avrB, which showed open stomata, those inoculated with Pst cor- carrying avrBT125A or avrBR266G showed closed stomata and were indistinguishable from those inoculated with Pst cor- (Figure 1B), indicating that these mutants were unable to open stomata. We next examined if the avrB-enhanced stomatal opening led to increased virulence during spray inoculation. Indeed, Pst cor- avrB grew to a level ∼3-fold higher compared with Pst cor- when spray-inoculated onto rpm1 plants, although lower than that of Pst (Figure 1C). This partial complementation of Pst cor- growth is consistent with the slightly smaller stomatal aperture induced by Pst cor- avrB compared with Pst (Figure 1A; Supplemental Figure 1). However, avrB did not enhance the multiplication of Pst cor- bacteria when inoculated by syringe infiltration (Figure 1D). Together, these results indicate that avrB complements the cor- mutation and contributes to stomatal virulence.
COI1 Is Required for avrB-Induced Stomatal Virulence
Because COR reopens stomata in a manner dependent on COI1 (Melotto et al., 2006), we examined whether avrB-induced stomatal opening and virulence were also dependent on COI1. We first generated an rpm1 coi1 double mutant by crossing rpm1 and coi1. The Pst cor- avrB-induced stomatal reopening 4 h after inoculation was abolished in rpm1 coi1 epidermal strips, but not in rpm1 (Figure 2A; Supplemental Figure 2). Three days after spray inoculation onto rpm1 plants, Pst cor- avrB grew to a higher level than did Pst cor- (Figure 2B). However, the bacterial counts of these two strains were the same in rpm1 coi1 3 d postinoculation (Figure 2B), confirming that the stomatal virulence of avrB requires COI1.
Figure 2.
COI1 Is Required for avrB-Induced Stomatal Virulence.
(A) Pst cor- avrB failed to open stomata in rpm1 coi1. Epidermal peels from coi1 and coi1 rpm1 plants were incubated in water, Pst cor-, or Pst cor- avrB for 4 h, and stomatal apertures were measured.
(B) The avrB-induced virulence was compromised in rpm1 coi1. Bacterial counts in plants were measured 3 d after spray inoculation. Error bars indicate se. Different letters denote significant difference at P ≤ 0.05 (Student’s t test, n ≥ 7, three biological repeats).
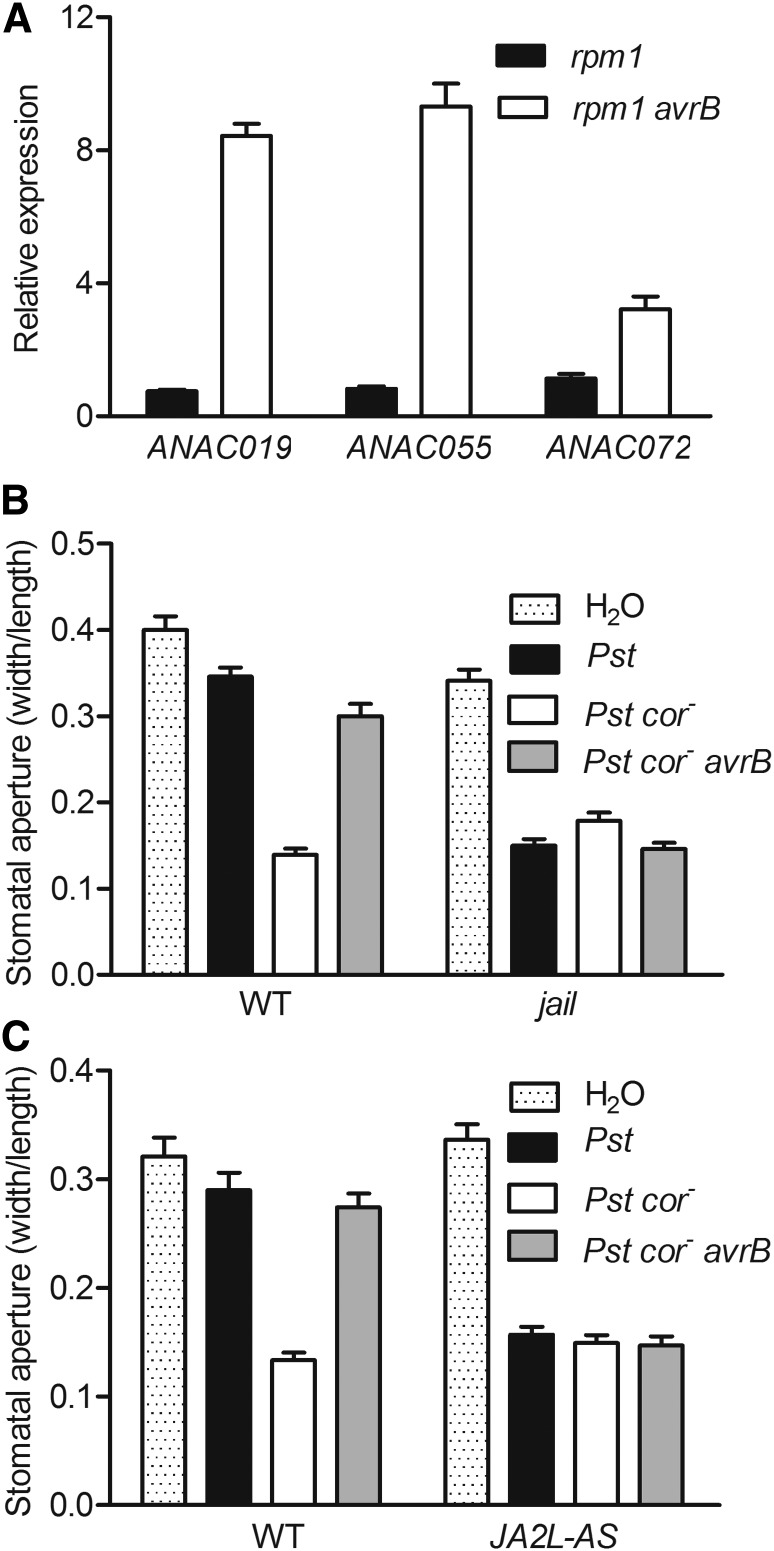
The NAC Transcription Factor JA2L Is Required for AvrB-Induced Stomatal Virulence
The NAC transcription factors ANAC019, ANAC055, and ANAC072 are transcriptionally activated downstream of COI1 and are required for COR-induced stomatal virulence (Zheng et al., 2012). Similarly, the tomato homolog JA2L also acts to mediate COR-induced stomatal virulence (Du et al., 2014). We tested whether AvrB is capable of inducing the expression of ANAC019, ANAC055, and ANAC072 in Arabidopsis. Estradiol-induced expression of the AvrB transgene in the rpm1 background led to strong expression of all three genes tested (Figure 3A). Because the available anac019 anac055 anac072 triple mutant contains RPM1, which may complicate the virulence assays, we sought to determine if avrB induced stomatal virulence in tomato plants, which do not recognize avrB. Figure 3B shows that Pst cor- avrB induced stomatal opening in wild-type tomato plants, but not in jai1 plants, which carry a loss-of-function mutation in the tomato ortholog of COI1 (Li et al., 2004), indicating that JA signaling is essential for avrB-induced stomatal opening in both tomato and Arabidopsis plants. We then tested if avrB was able to induce stomatal opening in tomato plants expressing an antisense version of the JA2L cDNA. The JA2L antisense plants (JA2L-AS) do not respond to COR and show reduced susceptibility to Pst (Du et al., 2014). Epidermal peels of wild-type tomato plants (M82) treated with Pst cor- avrB displayed larger stomatal apertures than those treated with Pst cor- (Figure 3C), indicating that avrB also promotes stomatal opening in tomato plants. By contrast, epidermal peels of JA2L-AS plants showed similar stomatal apertures in response to Pst cor- and Pst cor- avrB, indicating that JA2L is required for the avrB-induced stomatal opening in tomato plants (Figure 3C).
Figure 3.
AvrB Acts through COI1 and NAC Transcription Factors to Induce Stomatal Virulence.
(A) AvrB induces the expression of NAC genes in Arabidopsis. Nontransgenic and a stable AvrB-FLAG transgenic Arabidopsis line in the rpm1 background were induced with estradiol, and NAC transcripts were examined 12 h later. ACTIN8 was used as the internal standard.
(B) and (C) Pst cor- avrB opens stomata in tomato plants in a manner dependent on JAI1 (B) and JA2L (C). Fully expanded healthy leaves from 5-week-old plants were immersed in water or bacterial suspension for 4 h, and stomatal apertures were measured. Tomato cv M82 was used as the wild type.
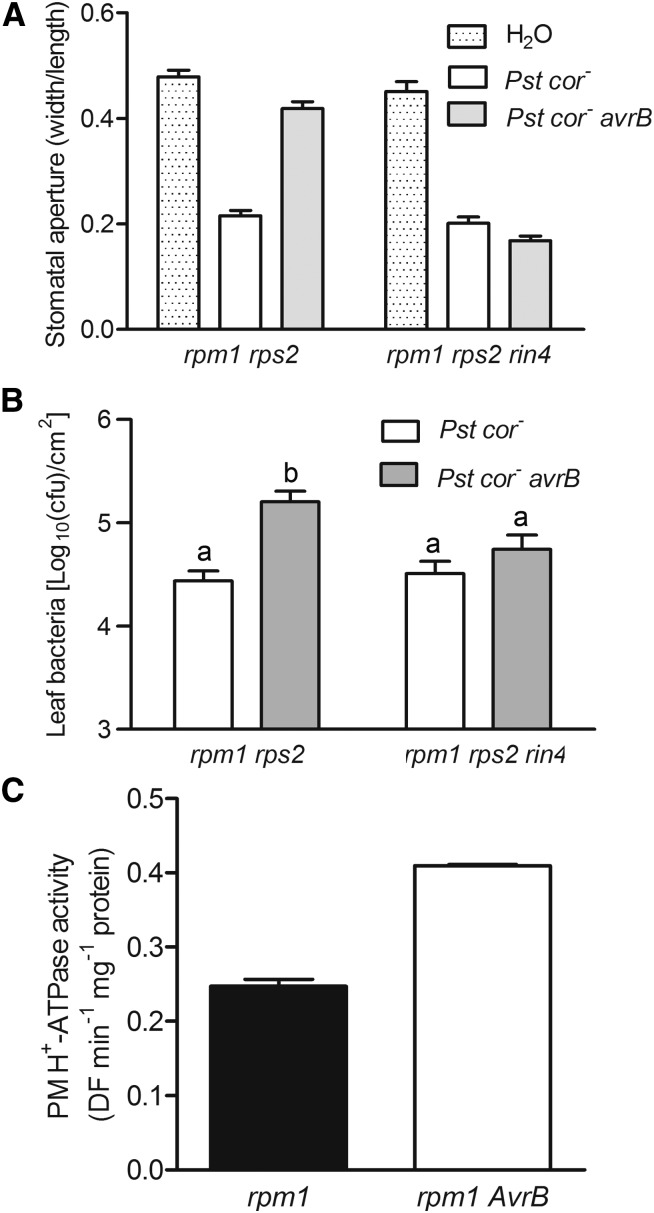
RIN4 Is Required for avrB- and COR-Induced Stomatal Opening
We previously showed that AvrB can complement COR deficiency and induce JA response genes in plants in a RIN4-dependent manner (He et al., 2004; Cui et al., 2010). We sought to test if RIN4 was similarly required for avrB-induced stomatal virulence. We first examined the stomatal apertures in the rpm1 rps2 double mutant and rpm1 rps2 rin4 triple mutant exposed to bacteria. An rpm1 mutant background was used to avoid the AvrB-triggered immune activation through RPM1 (Grant et al., 1995), whereas an rps2 mutant background was used to avoid constitutive activation of the immune receptor RPS2 in the absence of RIN4 (Mackey et al., 2003). Pst cor- avrB reopened stomata of rpm1 rps2 plants after a 4-h treatment, but not those of rpm1 rps2 rin4 plants (Figure 4A; Supplemental Figure 3), indicating that RIN4 is indeed required for avrB-induced stomatal opening. To test if the rin4 mutation compromised avrB-induced stomatal virulence, we spray-inoculated rpm1 rps2 and rpm1 rps2 rin4 plants with Pst cor- or Pst cor- avrB. While Pst cor- avrB grew to a significantly higher level compared with Pst cor- in rpm1 rps2 plants, the two strains grew to similar levels on the rpm1 rps2 rin4 mutant plants (Figure 4B), indicating that RIN4 is required for avrB-induced virulence. Taken together, these results indicate that RIN4 is required for avrB-induced stomatal virulence.
Figure 4.
The RIN4-AHA1 Pathway Is Required for avrB-Induced Stomatal Opening.
(A) Pst cor- avrB does not reopen stomata in rpm1 rps2 rin4 plants. Epidermal peels of the indicated genotypes were incubated with water, Pst cor-, or Pst cor- avrB for 4 h, and stomatal apertures were measured.
(B) avrB-induced stomatal virulence is RIN4 dependent. Plants of the indicated genotypes were spray inoculated with the indicated bacterial strains, and bacterial counts in plants were determined 3 d later. Error bars indicate se. Different letters denote significant differences at P ≤ 0.01 (Student’s t test, n = 11, three biological repeats).
(C) AvrB positively regulates PM H+-ATPase activity. Plasma membrane vesicles were isolated from rpm1 and rpm1 AvrB. PM H+-ATPase activity was initiated by the addition of 3 mM ATP, and 10 μM carbonyl cyanide m-chlorophenylhydrazone was added to dissipate the ΔpH. One representative experiment of three replicates is shown in the figure.
RIN4 is required for Pst-induced stomatal opening and disease susceptibility (Liu et al., 2009), suggesting that RIN4 is also required for COR-induced stomatal opening. To confirm this, we exposed epidermal peels to flg22 and COR to determine the role of RIN4 in COR-induced stomatal opening. As indicated in Supplemental Figure 4, flg22 treatment induced stomatal closure in both rpm1 rps2 and rpm1 rps2 rin4. Simultaneous exposure to flg22 and COR led to stomatal opening in rpm1 rps2 epidermis, but not in rpm1 rps2 rin4, indicating that RIN4 is required for COR-induced stomatal opening.
AvrB Induces PM H+-ATPase Activity
RIN4 directly interacts with AHA1 and enhances the activity of the PM H+-ATPase, thereby promoting stomatal opening (Liu et al., 2009). The requirement of RIN4 in AvrB-induced stomatal opening prompted us to test if AvrB modulated PM H+-ATPase activity in Arabidopsis. We isolated plasma membrane-enriched vesicles from AvrB-expressing plants by aqueous two-phase partitioning and measured H+-transport activities. The PM H+-ATPase activity in rpm1AvrB transgenic plants was significantly higher than that in rpm1 plants (Figure 4C), supporting the notion that AvrB is capable of modulating AHA1 activity.
JA Signaling Is Required for Enhanced Disease Susceptibility in ost2-2D Plants
The activation of PM H+-ATPase induces hyperpolarization of the plasma membrane and inward K+ channels, which are thought to cause stomatal opening directly through increased turgor pressure in the guard cell (Dietrichet al., 2001; Kim et al., 2010). However, this contradicts our observation that a canonical JA signaling pathway is required for AvrB-induced stomatal opening. We therefore tested whether the AHA1 PM H+-ATPase can activate JA signaling by examining the expression of JA response genes NAC019, NAC055, NAC072, and VSP2 in ost2-2D. The NAC genes and VSP2 were all upregulated strongly in ost2-2D (Figure 5A; Supplemental Figure 5), indicating that JA signaling is constitutively activated in this mutant. To investigate if AHA1 activates JA signaling in a COI1-dependent manner, we generated the coi1 ost2-2D double mutant by crossing ost2-2D to coi1. Transcript levels of the three NAC genes in coi1 ost2-2D were all greatly reduced compared with ost2-2D, although slightly higher than that in the wild type and coi1 (Figure 5A). Furthermore, VSP2 expression in coi1 ost2-2D was decreased to a level similar to that in coi1 (Supplemental Figure 5), indicating that constitutive JA signaling in the ost2-2D mutant was largely COI1 dependent.
Figure 5.
AHA1 Regulates Stomatal Opening through the JA Signaling Pathway.
(A) NAC transcripts were constitutively elevated in ost2-2D in a COI1-dependent manner. Four-week-old plants of the indicated genotypes were analyzed for NAC gene expression. Error bars indicate se. Different letters denote significant differences at P ≤ 0.01 (Student’s t test, n = 3, three biological repeats).
(B) ost2-2D exhibits enhanced disease susceptibility to Pst in a COI1-dependent manner. Plants of the indicated genotypes were spray inoculated with Pst bacteria. Bacterial counts in the leaf were determined 3 d postinoculation. Error bars indicate se. Different letters denote significant differences at P ≤ 0.05 (Student’s t test, n ≥ 7, three biological repeats).
We next compared wild-type, coi1, ost2-2D, and coi1 ost2-2D plants for stomatal resistance to Pst. Consistent with a previous report (Liu et al., 2009), Pst grew to a level ∼5-fold higher in ost2-2D compared with wild-type plants when spray-inoculated (Figure 5B). However, Pst grew to a similar level in the wild type and ost2-2D when syringe-infiltrated into leaves (Supplemental Figure 6), confirming that the enhanced disease susceptibility in ost2-2D was caused by a defect in stomatal defense. However, coi1 ost2-2D plants showed a significantly lower Pst counts than did ost2-2D plants following spray inoculation (Figure 5B). The bacterial counts in coi1 ost2-2D were similar to that in wild-type plants, but higher than that in coi1 plants (Figure 5B), indicating that the enhanced disease susceptibility of the ost2-2D mutant was at least partly dependent on COI1.
AvrB, RIN4, and AHA1 Promote JAZ Degradation
The aforementioned results support the notion that AvrB, RIN4, and AHA1 regulate stomatal opening by modulating JA signaling. The requirement of COI1 for these activities suggests that these proteins act upstream of COI1. We therefore asked if AvrB could induce JAZ protein degradation when transiently expressed in N. benthamiana plants. The abundance of all six tested JAZ-HA proteins was greatly reduced in the presence of AvrB-FLAG (Figure 6A), suggesting that AvrB induces the degradation of multiple JAZ proteins. The expression of AvrBR266G and AvrBT125A mutants, which are unable to induce JA signaling (Cui et al., 2010), failed to induce JAZ9-HA degradation (Figure 6B), indicating that the AvrB-induced JAZ protein degradation correlated with its ability to activate JA signaling.
Figure 6.
AvrB, RIN4, and AHA1 Promote JAZ Degradation.
(A) AvrB induces the degradation of JAZ proteins. JAZ-HA and AvrB-FLAG were transiently coexpressed in N. benthamiana, and protein levels were analyzed by immunoblots 24 h later. The same protein gel was stained with Ponceau to show equal loading. This experiment was repeated twice with similar results.
(B) Transient expression of the AvrBT125A and AvrBR266G mutants does not induce JAZ9 degradation in N. benthamiana.
(C) and (D) RIN4 (C) and AHA1 (D) induce the degradation of JAZ proteins. JAZ-HA proteins were transiently coexpressed with RIN4 or AHA1-HA in N. benthamiana and the accumulation of JAZ proteins was determined by immunoblot analysis.
We similarly tested the impact of AHA1 and RIN4 on JAZ degradation in N. benthamiana plants. Again, all three tested JAZ-HA proteins accumulated to much lower levels when RIN4 (Figure 6C) and AHA1-HA (Figure 6D) were overexpressed, indicating that both RIN4 and AHA1 promote the degradation of these JAZ proteins.
AvrB and AHA1 Induce COI1-JAZ Interaction
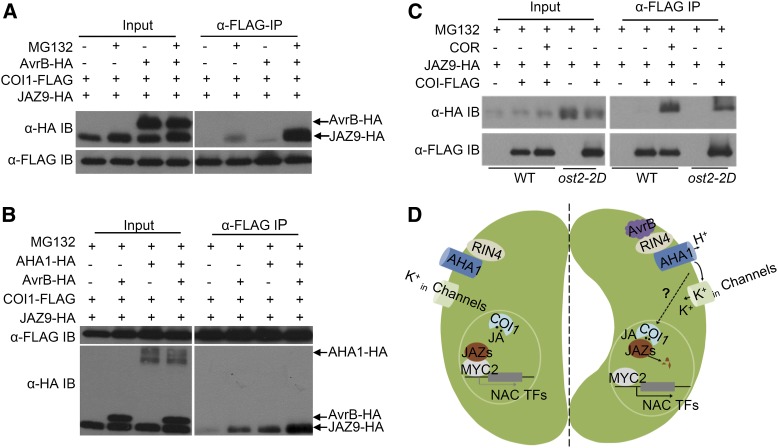
We next determined if AvrB interacted with COI1 or JAZ proteins. We coexpressed AvrB-FLAG with COI1-HA or JAZ9-HA in rpm1 protoplasts and performed coimmunoprecipitation (co-IP) assays. In this experiment, a COR-induced COI1-JAZ9 interaction was used as a positive control. While a COR-induced interaction between COI1-FLAG and JAZ9-HA was clearly detected, AvrB-FLAG did not interact with either COI1-HA or JAZ9-HA (Supplemental Figure 7), suggesting that AvrB does not directly target COI1 and JAZ proteins. We then examined if AvrB induced COI1-JAZ9 interaction in rpm1 protoplasts. In the absence of AvrB, a weak COI1-JAZ9 interaction was observed only in the presence of MG132, an inhibitor of proteasome-dependent protein degradation (Figure 7A). This interaction was greatly enhanced by AvrB-HA, indicative of an AvrB-induced COI1-JAZ9 interaction.
Figure 7.
AvrB and AHA1 Induce COI1-JAZ Interaction.
(A) Transient expression of AvrB promotes COI1-JAZ9 interaction in rpm1 protoplasts in the absence of COR or JA-Ile.
(B) AHA1-HA was transiently expressed in protoplasts, and the interaction between COI1 and JAZ9 was analyzed by co-IP assay.
(C) COI1 and JAZ9 interact in ost2-2D protoplasts in the absence of COR. Protoplasts of the indicated genotypes were treated with or without COR, and a co-IP assay was performed.
(D) Model for AvrB-induced stomatal opening. AvrB interacts with RIN4, which then activates AHA1, resulting in an alteration of plasma membrane potential. This may generate an unknown signal that potentiates the CO1-JAZ interaction to trigger JA signaling, subsequently inducing stomatal opening.
We further tested the impact of AHA1 on COI1-JAZ9 interaction in Arabidopsis rpm1 protoplasts. Similar to AvrB, AHA1-HA overexpression led to an interaction between COI1-FLAG and JAZ9-HA in the presence of MG132 (Figure 7B). This interaction was further enhanced when AHA1-HA was coexpressed with AvrB-HA, indicating that AHA1 and AvrB synergistically induce the COI1-JAZ9 interaction. We further coexpressed JAZ9-HA with COI1-FLAG in wild-type and ost2-2D protoplasts and determined the COI1-JAZ9 interaction by co-IP. In wild-type protoplasts, a COI1-JAZ9 interaction was only detected in the presence of COR (Figure 7C). By contrast, a strong, constitutive COI1-JAZ9 interaction was observed in ost2-2D in the absence of COR exposure, indicating that increased AHA1 activity enhances the COI1-JAZ9 interaction. A co-IP assay indicated that AHA1 did not interact with JAZ9 in protoplasts (Supplemental Figure 8), suggesting that the effect of ost2-2D on the COI1-JAZ9 interaction was indirect. Together these results support the notion that both AvrB and AHA1 enhance JA signaling by promoting the COI1-JAZ interaction.
DISCUSSION
In this study, we report that AvrB induces stomatal opening to confer stomatal virulence to P. syringae through a canonical JA signaling pathway defined by COI1 and NAC transcription factors. We provide evidence that AvrB enhances JA signaling through the RIN4-AHA1 pathway, which promotes JAZ degradation by indirectly inducing the COI1-JAZ interaction, uncovering a previously unknown mechanism that acts upstream of COI1 in the regulation of JA signaling.
AvrB is known to confer apoplastic virulence in soybean (Glycine max) plants lacking the cognate immune receptor RPG1 through an unknown mechanism (Ashfield et al., 1995). A role for AvrB in virulence has not been reported in Arabidopsis. The data presented here indicate that AvrB can substitute for COR function and confers stomatal virulence in Arabidopsis plants lacking the cognate immune receptor RPM1. Importantly, RIN4 is required for the AvrB virulence function, indicating that RIN4 is a virulence target. Because RIN4 is guarded by RPM1 (Mackey et al., 2002), the findings are consistent with the guard model in the evolution of RPM1 resistance (Dangl and Jones, 2001).
AvrB-mediated stomatal virulence required COI1 in Arabidopsis and JAI1 and JA2L in tomato plants, demonstrating that AvrB-mediated virulence is strictly dependent on the JA signaling pathway. This is consistent with our previous findings that bacterially delivered AvrB can substitute for COR function to enhance JA response gene expression (He et al., 2004; Cui et al., 2010).
RIN4 interacts with AHA1 and enhances the PM H+-ATPase activity (Liu et al., 2009). The activation of PM H+-ATPase induces hyperpolarization of the plasma membrane and inward K+ channels, which are thought to increase turgor pressure and stomatal opening (Dietrich et al., 2001; Kim et al., 2010). In this study, we show that the AvrB-mediated stomatal virulence function required RIN4 and that transgenic expression of AvrB enhanced the PM H+-ATPase activity. The data collectively support that AvrB induces stomatal opening through the RIN4-AHA1 pathway.
At first glance, the apparent requirement of JA signaling for AvrB-induced stomatal opening is at odds with the current understanding of AHA1, which is not known to play a role in JA signaling. Our data show that the ost2-2D mutant displayed a strong, constitutive JA response gene expression in Arabidopsis. The constitutive JA signaling phenotype is unlikely to be an effect of chronic activation of AHA1 in the ost2-2D mutant because transient expression of wild-type AHA1 also induced JAZ protein degradation and COI1-JAZ9 interaction. The constitutive JA response gene expression and increased disease susceptibility in ost2-2D were greatly reduced in the ost2-2D coi1 double mutant, indicating that the elevated JA signaling in ost2-2D is intimately linked to the control of stomatal movement. This conclusion is consistent with our previous findings that RIN4 is required for AvrB-induced JA signaling (Cui et al., 2010). Furthermore, transient expression of AvrB, RIN4, and AHA1 induced the degradation of a similar set of JAZ proteins. Together, the data indicate that AvrB regulates JA signaling through RIN4 and AHA1.
AvrB, RIN4, and AHA1 are localized to the plasma membrane (Harper et al., 1990; Nimchuk et al., 2000; Mackey et al., 2002), whereas COI1 is the receptor for JA-Ile and COR and regulates JA response gene expression in the nucleus. Consistent with this, AvrB and AHA1 did not interact with JAZ9. AHA1 and AvrB induced COI1-JAZ9 interaction in a synergistic manner when transiently expressed in Arabidopsis protoplasts, indicating that AvrB and AHA1 indirectly promote COI1-JAZ9 interaction. In addition, the gain-of-function mutant ost2-2D exhibits constitutive COI1-JAZ9 interaction in the absence of exogenous JA. Although the functional redundancy of AHA1 and AHA2 and lethality of the loss-of-function aha1 aha2 double mutant (Arango et al., 2003; Haruta et al., 2010) prevented us from testing if the PM H+-ATPases are required for JA signaling and AvrB-induced stomatal opening, it is highly likely that RIN4 and AHA1 act upstream of the COI1-JAZ interaction to regulate JA signaling and stomatal opening. Consistent with this possibility, the constitutive expression of JA response genes and enhanced disease susceptibility in ost2-2D is largely abolished by the coi1 mutation.
The biochemical mechanism by which the AHA1 activity induces COI1-JAZ interaction is not clear. A recent report shows that the ion channels GLR3.3 and GLR3.6 regulate the distal wound-stimulated expression of JA response genes (Mousavi et al., 2013), indicating that membrane potential plays an important role in JA signaling. The activation of PM H+-ATPase may also induce JA signaling through an alteration of membrane potentials. AvrB and AHA1 may enhance COI1-JAZ interaction by increasing JA biosynthesis. However, this is unlikely because transgenic expression of AvrB in Arabidopsis does not alter JA contents (Cui et al., 2010). An alternative possibility is that the activation of PM H+-ATPase induces a posttranslational modification of JAZ proteins. For example, degradation of JAZ by the bacterial effector HopZ1a is mediated by acetylation of the JAZ proteins (Jiang et al., 2013). A third, more likely, possibility is that the activation of PM H+-ATPase induces the generation of an unknown signal, which potentiates the COI1-JAZ interaction. Indeed, the inositol pyrophosphate InsP5 has been shown to potentiate the assembly of the COI1-JAZ complex in vitro (Sheard et al., 2010). A recent report showed that VIH2, a PPIP5K regulating the biosynthesis of InsP8, plays an important role in JA signaling and suggests that InsP8 is a critical cofactor in COI1-JAZ complex formation (Laha et al., 2015). This model may explain the inability of COR to open stomata in the rin4 mutant, which may be defective in the generation of such a cofactor. Future analysis of inositol pyrophosphates may shed light on the biochemical link between AHA1 and JA signaling.
In summary, existing evidence and the data presented here support the existence of a pathway consisting of RIN4 and AHA1 that positively regulates JA signaling by promoting the COI1-JAZ interaction (Figure 7D). AvrB induces AHA1 activity likely by inducing RIN4 phosphorylation, thus hijacking the JA signaling pathway to enhance stomatal virulence. In the absence of AvrB, the basal activity of this pathway may contribute to COR-induced JA signaling and stomatal opening. The findings underscore a previously unknown regulation of the COI1-JAZ interaction by the AHA1.
METHODS
Plant Materials
In this study, Arabidopsis thaliana plants include rpm1 rps2 (Kim et al., 2005), rpm1 (rps3-1) (Bisgrove et al., 1994), rpm1 rps2 and rpm1 rps2 rin4 (Kim et al., 2005), coi1 (coi1-1) (Xie et al., 1998), and ost2-2D (Merlot et al., 2007). The plants were grown at 23°C with a 10-h-light/14-h-dark photoperiod.
To generate double mutants, coi1 was crossed with rpm1 or ost2-2D. The F1 progeny of the crosses were propagated, and double mutants were identified in the F2 population. To generate the AvrB transgenic plants, the pER8-AvrB-3×FLAG construct under the control of the estradiol-inducing promoter (Shang et al., 2006) was introduced into rpm1 plants.
Tomato (Solanum lycopersicum) plants used in this study included cvM82, jai1 (Li et al., 2004), and JA2L-AS (Du et al., 2014). The tomato plants were grown in growth chambers and maintained at 16 h light at 25°C and 8 h darkness at 18°C.
Bacterial Strains and Bacterial Growth Assay
Bacterial strains used in this study include Pst and the coronatine-deficient mutants Pst cor-, Pst cor- avrB, Pst cor- avrBT125A, and Pst cor-avrBR266G (Cui et al., 2010). For spray inoculation, plants were sprayed with Pst and mutant derivatives at 5 × 108 colony-forming units (cfu)/mL with 0.017% Silwet L-77, and the bacterial population in the leaf was determined after 3 d. All experiments were repeated at least three times.
For syringe infiltration, 4-week-old plants were inoculated with Pst and mutant derivatives at a concentration of 1 × 105 cfu/mL, and the bacterial population in plants was measured after 3 d. All experiments were repeated at least three times.
Stomatal Aperture Measurements
Four-week-old Arabidopsis plants were kept under light for 2 h to ensure that most stomata were open prior to treatment. Epidermal strips were peeled from the rosette leaves and floated on a suspension of 5 × 108 cfu/mL Pst in water or a solution containing flg22 or flg22 and COR in MES buffer (10 mM MES, pH 6.15, 10 mM KCl, and 50 μM CaCl2) for 4 h before being examined under the 40× objective of a microscope (Axio Imager.A2; Zeiss). At least 30 stomata were recorded for each sample.
For stomatal aperture measurements in tomato, 5-week-old plants were kept under light for at least 3 h to ensure that most stomata were open before bacterial inoculation. Fully expanded healthy leaves were immersed in water or 5 × 108 cfu/mL of Pst. Four hours after treatment, the epidermis was peeled off and immediately examined under a 40× objective lens using a microscope (Axio Imager.A2; Zeiss), and at least 30 stomata were recorded for each sample.
Protein Analysis
For the JAZ protein degradation assay in Nicotiana benthamiana, AvrB-FLAG, RIN4, and AHA1-HA were coexpressed individually with JAZ-HA using Agrobacterium tumefaciens-mediated transient expression as previously described (Jiang et al., 2013). Proteins were extracted 24 h after infiltration of Agrobacterium, and the abundance of JAZ proteins was analyzed by immunoblot.
Coimmunoprecipitation Assay
The protoplasts were transfected with the indicated plasmids, incubated for 12 h at 23°C, and then treated with water or 1 μM COR for 1 h. Total protein was extracted with an extraction buffer containing 50 mM HEPES, pH 7.5, 150 mM KCl, 1 mM EDTA, 0.3% Trition X-100, 1 mM DTT, and 1× proteinase inhibitor cocktail (Roche). For anti-FLAG IP, total protein was incubated with 50 μL agarose-conjugated anti-FLAG antibody (Sigma-Aldrich) for 4 h and then washed six times with washing buffer (50 mM HEPES, pH 7.5, 150 mM KCl, 1 mM EDTA, 0.3% Triton X-100, and 1 mM DTT). The protein was eluted with 0.5 mg/mL 3× FLAG peptide for 1 h and then separated by SDS-PAGE and detected by anti-HA and anti-FLAG immunoblot.
PM H+-ATPase Activity Assays
To analyze PM H+-ATPase activity in transgenic Arabidopsis lines expressing the AvrB gene, 4-week-old rpm1 and rpm1 AvrB plants were induced with estradiol, and 15 to ∼20 g in each treatment was harvested 24 h later. Plasma membrane vesicles were isolated using aqueous two-phase partitioning and the H+-ATPase activity assay was performed as described previously (Yang et al., 2010).
RT-PCR
To analyze the abundance of ANAC019, ANAC055, and ANAC072 transcripts in transgenic Arabidopsis lines expressing the AvrB gene, 7-d-old seedlings grown on Murashige and Skoog agar media were induced with estradiol for 12 h. Then, 15 to 20 plants were harvested for RNA isolation for each treatment. To analyze the expression of NAC genes and other JA marker genes in ost2-2D, leaves of 4-week-old plants were used.
RNA was extracted using Trizol reagent (Invitrogen) followed by DNase digestion (Promega) to remove genomic DNA contamination and reverse transcribed using the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen). Quantitative RT-PCR was performed using Power SYBR green chemistry (Takara). Primer sequences used include ANAC019F 5′-GCATCTCGTCGCTCAG-3′ and ANAC019R 5′-CTCGACTTCCTCCTCCG-3′; ANAC055F 5′-GCGCTGCCTCATAGTC-3′ and ANAC055R 5′-CGAGGAATCCCCTCAGT-3′; ANAC072F 5′-TGGGTGTTGTGTCGAAT-3′ and ANAC072R 5′-ATCGTAACCACCGTAACT-3′ (Zheng et al., 2012); VSP2F 5′-CTTTGACCTAGATGATACCCTCCTCTCTAG-3′ and VSP2R 5′-CCAACGGTCACTGAGTATGATGGGTTCAAT-3′.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative and GenBank data libraries under the following accession numbers: AT3G25070, RIN4; AT2G18960, AHA1; AT1G52890, ANAC019; AT3G15500, ANAC055; AT4G27410, ANAC072; AT5G24770, VSP2; AT2G39940, COI1; AT3G17860, JAZ3; AT1G48500, JAZ4; AT1G72450, JAZ6; AT2G34600, JAZ7; AT1G70700, JAZ9; AT3G43440, JAZ11; AT3G07040, RPM1; AT4G26090, RPS2; and M21965, avrB.
Supplemental Data
Supplemental Figure 1. The P. syringae Effector Protein AvrB Complements the cor- Mutation in Stomatal Opening.
Supplemental Figure 2. COI1 Is Required for avrB-Induced Stomatal Virulence.
Supplemental Figure 3. RIN4 Is Required for avrB-Induced Stomatal Opening.
Supplemental Figure 4. COR Reopens Stomata in a RIN4-Dependent Manner.
Supplemental Figure 5. VSP2 Transcripts Were Upregulated in ost2-2D in a COI1-Dependent Manner.
Supplemental Figure 6. ost2-2D Lines Did Not Display Enhanced Susceptibility to Syringe Inoculation with Pst.
Supplemental Figure 7. AvrB Does Not Interact with COI1 or JAZ9 in Protoplasts.
Supplemental Figure 8. AHA1 Does Not Interact with JAZ9.
Supplementary Material
Acknowledgments
We thank Jeffrey Leung for ost2-2D seeds and Gabriel Schaaf for helpful discussion. J.-M.Z. and Z.Z. were funded by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB11020200), by the Chinese Ministry of Science and Technology (2011CB100702 and 2015CB910200), and by the Chinese Natural Science Foundation (31320103909 and 31230007). C.L. was funded by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB11030200).
AUTHOR CONTRIBUTIONS
Z.Z., C.L., Y.G., and J.-M.Z. designed the experiments. Z.Z. and X.Z. generated Arabidopsis transgenic plants. Y.W., Y.Y., and Z.Z. performed proton ATPase assays. Z.Z. and M.D. performed stomatal measurements in tomato. Z.Z. generated the constructs and performed the rest of the experiments. Z.Z. and J.-M.Z. analyzed the data. Z.Z. and J.-M.Z. wrote the article.
Glossary
- COR
coronatine
- JA
jasmonate
- PM
plasma membrane
- co-IP
coimmunoprecipitation
- cfu
colony-forming units
Footnotes
Articles can be viewed online without a subscription.
References
- Arango M., Gévaudant F., Oufattole M., Boutry M. (2003). The plasma membrane proton pump ATPase: the significance of gene subfamilies. Planta 216: 355–365. [DOI] [PubMed] [Google Scholar]
- Ashfield T., Keen N.T., Buzzell R.I., Innes R.W. (1995). Soybean resistance genes specific for different Pseudomonas syringae avirulence genes are allelic, or closely linked, at the RPG1 locus. Genetics 141: 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove S.R., Simonich M.T., Smith N.M., Sattler A., Innes R.W. (1994). A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6: 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E.H., da Cunha L., Wu A.J., Gao Z., Cherkis K., Afzal A.J., Mackey D., Dangl J.L. (2011). Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Wang Y., Xue L., Chu J., Yan C., Fu J., Chen M., Innes R.W., Zhou J.M. (2010). Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 7: 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Dietrich P., Sanders D., Hedrich R. (2001). The role of ion channels in light-dependent stomatal opening. J. Exp. Bot. 52: 1959–1967. [DOI] [PubMed] [Google Scholar]
- Du M., et al. (2014). Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 26: 3167–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Boter M., Fernández-Barbero G., Chini A., Rathjen J.P., Solano R. (2014). The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 12: e1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M.R., Godiard L., Straube E., Ashfield T., Lewald J., Sattler A., Innes R.W., Dangl J.L. (1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846. [DOI] [PubMed] [Google Scholar]
- Harper J.F., Manney L., DeWitt N.D., Yoo M.H., Sussman M.R. (1990). The Arabidopsis thaliana plasma membrane H+-ATPase multigene family. Genomic sequence and expression of a third isoform. J. Biol. Chem. 265: 13601–13608. [PubMed] [Google Scholar]
- Haruta M., Burch H.L., Nelson R.B., Barrett-Wilt G., Kline K.G., Mohsin S.B., Young J.C., Otegui M.S., Sussman M.R. (2010). Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 285: 17918–17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P., Chintamanani S., Chen Z., Zhu L., Kunkel B.N., Alfano J.R., Tang X., Zhou J.M. (2004). Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J. 37: 589–602. [DOI] [PubMed] [Google Scholar]
- Jiang S., Yao J., Ma K.W., Zhou H., Song J., He S.Y., Ma W. (2013). Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 9: e1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.S., Desveaux D., Singer A.U., Patel P., Sondek J., Dangl J.L. (2005). The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc. Natl. Acad. Sci. USA 102: 6496–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Böhmer M., Hu H., Nishimura N., Schroeder J.I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61: 561–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloek A.P., Verbsky M.L., Sharma S.B., Schoelz J.E., Vogel J., Klessig D.F., Kunkel B.N. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26: 509–522. [DOI] [PubMed] [Google Scholar]
- Laha D., et al. (2015). VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and jasmonate-dependent defenses in Arabidopsis. Plant Cell 27: 1082–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao Y., McCaig B.C., Wingerd B.A., Wang J., Whalon M.E., Pichersky E., Howe G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Elmore J.M., Fuglsang A.T., Palmgren M.G., Staskawicz B.J., Coaker G. (2009). RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Elmore J.M., Lin Z.J., Coaker G. (2011). A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey D., Belkhadir Y., Alonso J.M., Ecker J.R., Dangl J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey D., Holt B.F. III, Wiig A., Dangl J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754. [DOI] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980. [DOI] [PubMed] [Google Scholar]
- Merlot S., Leonhardt N., Fenzi F., Valon C., Costa M., Piette L., Vavasseur A., Genty B., Boivin K., Müller A., Giraudat J., Leung J. (2007). Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 26: 3216–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal S., Davis K.R. (1995). Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol. Plant Microbe Interact. 8: 165–171. [DOI] [PubMed] [Google Scholar]
- Mousavi S.A.R., Chauvin A., Pascaud F., Kellenberger S., Farmer E.E. (2013). GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500: 422–426. [DOI] [PubMed] [Google Scholar]
- Nimchuk Z., Marois E., Kjemtrup S., Leister R.T., Katagiri F., Dangl J.L. (2000). Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 101: 353–363. [DOI] [PubMed] [Google Scholar]
- Ong L.E., Innes R.W. (2006). AvrB mutants lose both virulence and avirulence activities on soybean and Arabidopsis. Mol. Microbiol. 60: 951–962. [DOI] [PubMed] [Google Scholar]
- Schellenberg B., Ramel C., Dudler R. (2010). Pseudomonas syringae virulence factor syringolin A counteracts stomatal immunity by proteasome inhibition. Mol. Plant Microbe Interact. 23: 1287–1293. [DOI] [PubMed] [Google Scholar]
- Shang Y., Li X., Cui H., He P., Thilmony R., Chintamanani S., Zwiesler-Vollick J., Gopalan S., Tang X., Zhou J.M. (2006). RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA 103: 19200–19205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yang Y., Qin Y., Xie C., Zhao F., Zhao J., Liu D., Chen S., Fuglsang A.T., Palmgren M.G., Schumaker K.S., Deng X.W., Guo Y. (2010). The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 22: 1313–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X.Y., Spivey N.W., Zeng W., Liu P.P., Fu Z.Q., Klessig D.F., He S.Y., Dong X. (2012). Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.