Abstract
The amino acid sequences of nicotinic acetylcholine receptors (nAChRs) from diverse species can be compared across extracellular, transmembrane, and intracellular domains. The intracellular domains are most divergent among subtypes, yet relatively consistent among species. The diversity indicates that each nAChR subtype possesses a unique language for communication with its host cell. The conservation across species also suggests that the intracellular domains may play defining functional roles for each subtype. Secondary structure prediction indicates two relatively conserved alpha helices within the intracellular domains of all nAChRs. Among all subtypes, the intracellular domain of α7 nAChR is one of the most-well conserved, and α7 nAChRs have effects in non-neuronal cells independent of generating ion currents, making it likely that the α7 intracellular domain directly mediates signal transduction. There are potential phosphorylation and protein binding sites in the α7 intracellular domain, which are conserved and may be the basis for α7-mediated signal transduction.
Keywords: signal transduction, cys-loop receptors, intracellular domains, protein structure, proteomics, evolution
Ligand-gated ion channel superfamily
Certain key features associated with ligand-gated ion conduction are found in both the bacterial and eukaryotic receptors of the Cys-loop superfamily, suggesting an evolutionary link. While both classes of proteins have in common an extracellular ligand-binding domain, four membrane-spanning helices that include the ion channel, and the signature Cys-loop, which is essential for transducing conformation change between the ligand-binding and the channel domains, there is, however, an additional domain in the eukaryotic proteins that is not present in the bacterial homologs, an intracellular domain between the third and fourth transmembrane domains (TM3 and TM4) (Bocquet et al., 2007, Hilf and Dutzler, 2008). While the presence of an intracellular domain is a consistent feature in all the eukaryotic proteins, it is, in fact, the most variable subdomain in all of these proteins, and an epitope that could be used to uniquely identify one receptor subunit type from all the others. In this paper we will review both the common and diverse features of nicotinic acetylcholine receptor (nAChR) subunit intracellular domains, describing an integrated perspective on vertebrate receptor subunits from a wide range of species including a fish (Danio rerio), an amphibian (Xenopus spp.), a bird (Gallus gallus), a rodent (Rattus rattus), a ruminant (Bos taurus) and a primate (Homo sapiens).
Functional nAChRs form as pentameric complexes of subunits (for review, see (Papke, 2014)). Subunits are classified as either alpha type (α1-α10) or non-alpha (β1-β4, γ, δ, or ε), based on the presence or absence, respectively, of disulfide-linked vicinal cysteines in the extracellular domain. The diversity of nAChR subtypes in vertebrate animals is matched or surpassed by the diversity of nAChR subtypes in insects (Jones et al., 2007) and other invertebrates (Holden-Dye et al., 2013), suggesting that there was broad adaptive radiation of the earliest ancestral proteins. Why are there so many nAChR subtypes, and what different sorts of specializations distinguish one subtype from another? To make these questions more feasible to address, we can focus on just the subtypes in vertebrates, and we see that from fish to primates, there have been retentions of key features that make it possible to observe the phylogenetic continuity of each of the subunits.
Going beyond DNA and predicted protein alignments, intracellular domains may be plotted graphically using Kyte-Doolittle hydrophobicity analyses. Initial predictions about nAChR transmembrane topology depended on hydrophobicity analyses, which are also applicable in our present discussion for the purpose of defining the probable lengths of the intracellular domains as delimited by hydrophobic sequences of TM3 and TM4.
Structural information on nAChR and related proteins has come from several sources, each with intrinsic limitations: the prokaryotic channels (Bocquet et al., 2007, Hilf and Dutzler, 2008), crystal structures of molluscan acetylcholine binding proteins (AChBP) (Parthiban et al., 2009, Sixma and Smit, 2003), and electron micrographic analyses of the receptors that can be purified from the electric organ of Torpedo rays (Unwin et al., 1988, Unwin, 1993, Unwin et al., 2002). Of these sources only the Torpedo receptors have intracellular domains, but these are largely not resolved with this approach. The recently published crystal structure for the mouse 5HT3A receptor (Hassaine et al., 2014) includes helical portions of the intracellular domain, but omits the flexible central loop. With the helices disconnected, it remains uncertain how they would be oriented in the intact protein.
Additionally, the literature contains mutation and deletion studies, identifying various motifs within the intracellular domain required for maturation and locating nAChR. While we begin to get a coherent picture of much of the nAChR proteins from these various sources, we are left with more questions than answers about the evolutionary origins and functional roles for the diverse nAChR intracellular domains. Through an analysis of basic sequence information, we can at least begin to identify some of those questions as a first step in getting answers.
Sources of Perspective
Sequence analyses
An alignment of all human nAChR subunit sequences by Clustal Omega (Sievers et al., 2011) with color-highlighted alpha helices (pink) and beta strands (yellow) predicted by PsiPred (Buchan et al., 2013) is provided in the supplemental data (Figure S1). Included are examples of other pentameric Cys-loop ligand-gated ion channels and an AChBP. Compared with the crystal structure for AChBP (1I9B, (Brejc et al., 2001)), the in silico predictions of helix, coil, and strand locations for the AChBP are in good agreement, greater than 86%. Also, for the extracellular domain, there is generally good alignment among the ligand-gated ion channel sequences and agreement between the structural predictions and the reference crystal structures. As expected, TM1 and TM2 are universally predicted to be helical. However, for many of the nAChR subunits, the predicted TM3 structures were more strand-like than helical, especially toward the cytosolic border.
The perimembrane sections of the intracellular domain align relatively well, and there is predicted helical structure in both of these domains. The middle sections are very highly variable and structurally predicted to be disordered by the PsiPred analysis. These central loop subdomains vary greatly in length, and it is not possible to make a meaningful alignment based on sequence; therefore, they are simply shown center-aligned in the lower portion of Figure S1. Table 1 provides the percent sequence identity for the nAChR intracellular domains of each subunit for the different species studied, and Table S1 (Supplementary Data) provides the percent sequence identity for the nAChR intracellular domains among the human subtypes. The muscle-type α1 intracellular domain is best conserved across species, with 75% sequence identity between human and zebrafish. The short α5 intracellular domain is best conserved among the terrestrial species. As discussed below, the α7 intracellular domain is the best conserved of the ligand-binding neuronal alpha subunits.
Table 1.
Percent sequence identity to human nAChR intracellular domains
| cow | rat | chick | frog | fish | |
|---|---|---|---|---|---|
| alpha1 | 98.04 | 92.16 | 82.35 | 69.61 | 73.53 |
| alpha5 | 84.52 | 89.16 | 86.90 | 82.14 | 66.67 |
| delta | 93.08 | 88.46 | 65.60 | 64.62 | 63.57 |
| alpha7 | 91.67 | 87.50 | 83.33 | 74.31 | 62.24 |
| beta3 | 86.00 | 77.00 | 75.00 | 72.00 | 56.00 |
| alpha3 | 92.20 | 89.36 | 70.21 | 73.05 | 47.86 |
| epsilon | 88.33 | 88.33 | NA | 46.15 | 46.67 |
| alpha6 | 72.31 | 66.67 | 58.46 | 39.34 | 45.38 |
| alpha2 | 70.21 | 66.20 | 51.41 | 49.61 | 42.03 |
| beta2 | 93.85 | 89.23 | 64.17 | 67.20 | 41.80 |
| alpha9 | 80.95 | 80.95 | 46.03 | 41.27 | 40.48 |
| beta1 | 92.25 | 85.27 | 58.91 | 51.59 | 38.89 |
| alpha4 | 72.18 | 68.46 | 52.55 | 49.02 | 36.21 |
| gamma | 86.86 | 81.75 | 46.56 | 45.04 | 35.77 |
| alpha10 | 87.76 | 80.00 | 46.94 | 36.73 | 35.71 |
| beta4 | 74.81 | 67.94 | 53.60 | 47.06 | 35.61 |
The intron-exon borders are also shown in Figure S1. Several of the splice sites in the extracellular domain are well conserved among all the human nAChR, in spite of the fact that the sixteen nAChR genes shown (Supplemental Data) are located on eight different chromosomes (Supplemental Data). Splice sites in other parts of the sequences are more variable, although there appear to be similarities among functional subgroups, e.g. among the muscle receptor subunits and also among the homomeric receptor-forming alpha subunits (α7, α9, and α10). They are notably lacking in the highly variable central section of the intracellular domain of the neuronal nAChR.
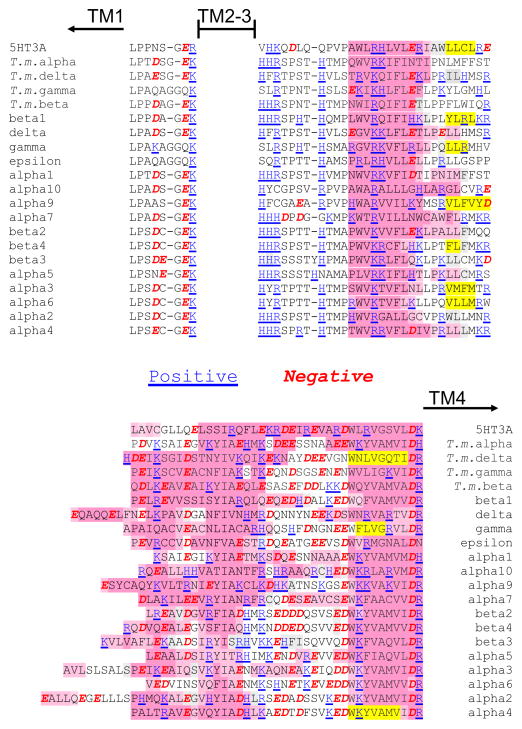
A closer look at the perimembrane portions of the intracellular domains reveals several interesting patterns, including some relatively well conserved charged amino acids (Figure 1). Following TM3, but before the disordered interior domain, there appears to be a loop followed by a predicted helix. In most subunits (all but α9 and α10) there are two or, more often, three positively charged residues in the loop before the predicted helix and additional positive charge within the predicted helix; adjacent positively charged residues, dibasic motifs, have been implicated in trafficking, see below. Negatively charged residues in this section are fewer and more scattered. In contrast, there are both positively and negatively charged residues in the subdomain prior to TM4, and many of these are within a putative helix, accounting for the hypothetical amphipathic character of this domain. It is also interesting to note that the linker between TM1 and TM2, which also is proposed to form a short intracellular loop, contains two conserved negatively charged residues in all of the nAChR subunits except for the muscle γ and ε subunits. Since the expression of these two subunits is developmentally regulated, γ present in embryonic muscle and ε in adult, both forms of the muscle receptor will contain one small intracellular loop without negative charge. This may be significant for the configuration of submembrane ion portals observed (see below).
Figure 1.
Charged residues of the nAChR in the short intracellular loop between TM1 and TM2 and in the segments of the large intracellular domains closest to the transmembrane domains. Note that histidine, which is marked as positively charged, is borderline between polar and charged, while arginine and lysine are always likely to be positively charged and not as polar as histidine. The positively charged residues are colored blue and underlined. The negatively charged residues are in red italics. Secondary structure as predicted by PsiPred (http://bioinf.cs.ucl.ac.uk/psipred/) is indicated with pink for helices and yellow for beta strands (as in Figure S1). Torpedo marmorata subunits are included with human nAChR and 5HT3A subunits.
Hydrophobicity analyses
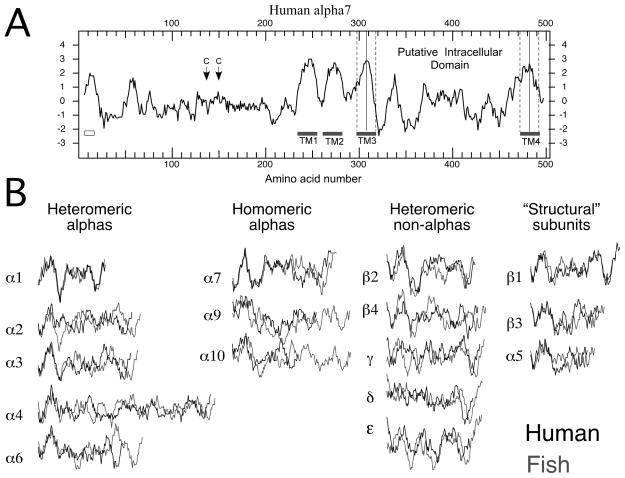
Additional perspective can be gained from Kyte-Doolittle analyses (Kyte and Doolittle, 1982) of hydrophobicity. A representative plot is shown in Figure 2A. The putative intracellular sequence can be defined as the region delimited by TM3 and TM4, each of which are identified as the twenty amino acids centered on the local hydrophobic peak.
Figure 2.
A) Kyte-Doolittle hydrophobicity plot of human α7 from DNA Strider (version 1.4f17 CEA France). Plotted are the average hydrophobicity scores of 13 amino acids in a rolling window. Long stretches of amino acids with positive scores are likely to be lipid associated. The amino terminal signal sequence is indicated by the box under the plot on the left. Position of the two cysteine residues which define the signature Cys-loop are indicated by the arrows. Gray bars indicate the regions associated with the transmembrane domains. The dashed lines indicate how the putative intracellular domains were selected as delimited by 10 amino acids from the hydrophobic peaks of TM3 and TM4. B) The putative intracellular domains of all the nAChR excised from the Kyte-Doolittle hydrophobicity plots for each subunit. The profiles in black are from human sequences, and those in gray are from zebrafish sequences. Vertical alignments are based on the Kyte-Doolittle scores, and the horizontal alignments are based on the putative amino terminals of the respective intracellular domains determined as described for panel A.
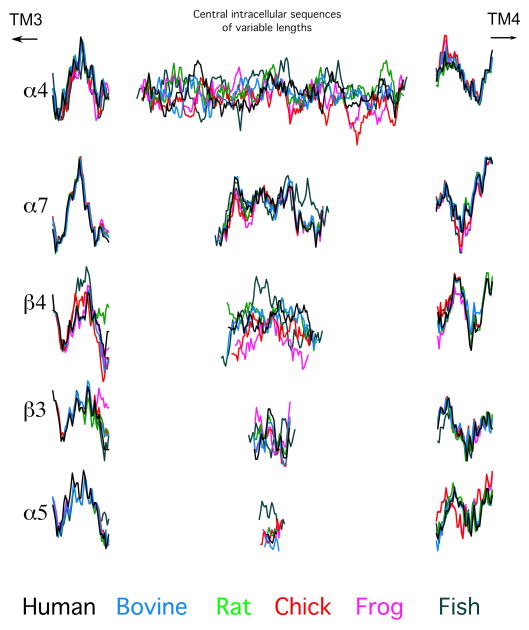
Shown in Figure 2B are the intracellular domain segments excised from all of the human (black) and zebrafish (gray) sequences. The sequences are grouped by subunit function, beginning with those from alpha subunits that form heteromeric complexes that require assembly with non-alpha subunits to create effective agonist binding sites (muscle α1 and neuronal α2, α3, α4, and α6). The second group has the intracellular domains of alpha subunits that will assemble without non-alpha subunits. The third contains the intracellular domains of non-alphas that will co-assemble with subunits of the first group to form agonist-binding sites in heteromeric receptors, and the last group has the intracellular domains of subunits that will only serve as structural subunits. Several features are evident from this visual organization; foremost is the overall diversity in these structures. It is apparent that, over the large phylogenetic distance between zebrafish and humans, there has been more conservation in some subunits, such as α1, than in others, such as β4. Also, the pattern of relative conservation and divergence described by the sequence analysis in the previous section is obvious from this presentation. In this presentation the sequences were aligned at their amino termini, and, while there are clear similarities between the human and fish at the beginning of many of the sequences, they fall progressively more out of register toward the middle sections. It is also apparent that for most of the sequences the same shift in register in the mid section would occur if the sequences were initially aligned at the carboxy termini. As the sequence alignments suggest, these putative intracellular domains should therefore be considered in three segments, with relatively conserved putative helices at either end and a central disordered portion which is highly variable. This is illustrated in Figure 3, where the intracellular sequences of five representative nAChR subunits have been segmented into three sections with the profiles from six diverse species overlaid for comparison.
Figure 3.
Representative intracellular domains from each of the subgroups presented in Figure 1B, of each of the six vertebrate species analyzed (color coded as in the legend at the bottom). Since sequence analysis (Figure S1) indicated that there was relatively high homology in the putative helix-forming subdomains following and preceding TM3 and TM4, respectively, we have split each intracellular domain into three segments, 25 amino acids following TM3, central intracellular sequences of variable length, and 30 amino acids preceding TM4.
Consistent with the sequence analyses, it is apparent from all of the representative sequences in Figure 3 that for each of the subunits depicted the putative helices nearest TM3 are the most well conserved intracellular subdomain. The conservation in the putative amphipathic domain (near TM4) is more variable, with the most hydrophobic part, closest to the membrane, being the most conserved portion. The variations in the middle section of the intracellular domain are particularly interesting. While these domains vary greatly in length, with α5 the shortest and α4 the longest, a feature consistent across species, the hydrophobicity profiles are variously conserved or diverse across the species. Of those illustrated, the profile of α7’s central intracellular domain has been best conserved across species. The profiles of β3 and α5 also appear well conserved, aside from single outliers in the frog and fish sequences, respectively. In contrast, the profiles of α4 and β4 from these six species appear to lack significant conservation of hydrophobicity.
Portals for ion flow
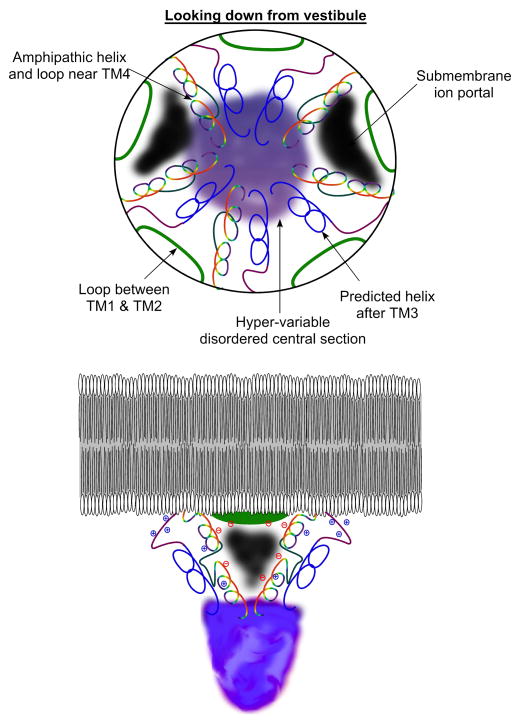
In the reported structure of the Torpedo marmorata receptor, the limited image of the intracellular domains appeared similar for all of the five subunits (two α1s and single β, γ, and δ subunits) (Unwin, 2005). The studies of the Torpedo electron microscopy images suggest that a function for the amphipathic helices near TM4 might be to delimit portals for ions to flow through the walls of the extended intracellular domain (Hales et al., 2006, Miyazawa et al., 1999, Unwin, 2005). Hydrophobic interactions among the helices could be important for the formation of such portals. The short loops of intracellular sequence between first and second transmembrane domains are also likely to contribute to these portals (Papke and Grosman, 2014, Unwin, 2005). It is tempting to speculate about the functions of the highly conserved positively and negatively charged residues in these intracellular areas just below the membrane. The negatively charged residues near the membrane in the putative amphipathic region could assist the cation flow when the channel opens, helping to define the conduction pathway through the portals. Alternatively, it may be that these conserved charges help anchor the position of the attached helices by promoting specific interactions with the phosphatidylcholine head groups of the associated membrane lipids. It is more difficult to ascribe significance to the positively charged residues that predominate in the sequence after the third transmembrane domain. They might also interact with the phosphate groups of the lipid bilayer or contribute to some sort of voltage sensors associated with rectification or as inactivation gates, promoting conformational changes in the nearby helices or the submembrane portals.
An admittedly speculative model is shown in Figure 4. The putative amphipathic alpha helices are positioned as the frames to the submembrane portals (black areas) as suggested by Unwin (Unwin, 2005). As noted above, sequence analyses (Figures 1 and S1) suggest that these subdomains have two sections predicted to be helical (drawn as variegated spirals in Figure 4) separated by highly charged regions (dark grey) that are less likely to be helical. Also contributing to the portals are the loops between TM1 and TM2 (green). The predicted helices near TM3 (blue) are preceded by a strand rich in positive charge (purple), which may orient away from the hydrophobic interior of the protein. These helices may serve a role as backbone structures extending below the portals, providing the outer framework that is then lined by the putative amphipathic helices near TM4, as well as providing attachment points for the variable intracellular segments that have unresolvable structure. Could these helices also serve as pushrods communicating conformational change between the gated transmembrane domains and intracellular sites on the receptor, mediating interactions with other protein partners? To what degree would such conformational coupling be reciprocal so that extracellular ligand binding would affect intracellular signaling and intracellular factors affect ion channel gating?
Figure 4.
Hypothetical model of the Torpedo receptor intracellular domain based on the electron microscopy images (Unwin, 2005) and our sequence analyses (Figure 1). The upper figure is a view looking into the cell from the extracellular ion channel vestibule. The small intracellular loops between TM1 and TM2 are represented by the green segments. The relatively conserved putative helices coming from TM3 are in blue, preceded by the predicted loop indicated in purple. The putative amphipathic helices extending up to TM4 are in two sections shown in variegated color connected by a loop (dark gray). We show the possible location of submembrane portals (black) lined by the amphipathic helices and the TM1-TM2 loop. The lower cartoon provides a view of the model from the side, with a representation of the disordered and variable domains in lavender hues connecting the perimembrane helices and extending into the cytoplasm. The locations of some of the conserved charges (Figure 1) in the short loop between TM1 and TM2, in the loop following TM3, and in the amphipathic helix are indicated.
If these assignments are correct, it should be noted that the functions of the helices near TM3 would be more universal among the subunits since these are the most conserved feature of the intracellular domains. The amphipathic helices proximal to TM4 are more variable, which would suggest that if submembrane portals are a universal feature of the Cys-loop receptors, they may vary significantly in their impact on the conduction properties of receptor subtypes. Support for function of these helices in conduction come from analysis of 5HT3A receptors showing that the residues that limit receptor conductance are in this subdomain (Kelley et al., 2003).
The hypothetical portals will vary significantly with subunit composition and appear to be important determinants of channel conductance (Hales et al., 2006). It is worth considering that, just as voltage-gated channels have separate activation and inactivation “gates”, nAChR may have multiple mechanisms for opening and closing the conduction pathway and that the process of desensitization could involve conformational changes in the submembrane portals. A couple of lines of data support this hypothesis. Firstly, the electron microscopy studies of Torpedo receptors in the putative open and closed states (Miyazawa et al., 2003, Unwin, 1995, Unwin and Fujiyoshi, 2012) have failed to identify a distinct closed conformation associated with desensitization. Secondly, one of the most consistent findings for functional effects of an nAChR phosphorylation state has been on desensitization (Charpantier et al., 2005, Hoffman et al., 1994, Hopfield et al., 1988, Huganir et al., 1986, Nishizaki and Sumikawa, 1998), in some cases associated with especially persistent desensitization (Eilers et al., 1997, Fenster et al., 1999, Paradiso and Brehm, 1998), implicating interference with ion conduction.
Interactions of nAChR subunits dependent on intracellular elements
The very earliest studies of Torpedo nAChR identified the intracellular domains as the sites for cytoskeletal interactions associated with receptor stability and localization (Frail et al., 1988, Maimone and Enigk, 1999, Ramarao and Cohen, 1998). More recently, receptor proteomic analyses have identified numerous possible protein partners for α7 and other nAChR (Jones et al., 2010, Paulo et al., 2009).
One well established role of the large intracellular loop of nAChRs is in regulating receptor maturation (reviewed by Tsetlin, (Tsetlin et al., 2011)). Maturation of nAChRs initiates with insertion of the subunits as they are being translated into the endoplasmic reticulum (ER) membrane and ends with a functional and properly localized receptor at the outer membrane. Folding, assembly, and post-translational modifications of nAChR subunits occurs within the ER; only fully folded and assembled receptors exit the ER and are trafficked to the plasma membrane via the Golgi apparatus. Multiple proteins have been shown to facilitate and regulate this process (Treinin, 2008, Tsetlin et al., 2011). Importantly, and as described below, effects of many of these proteins are mediated by motifs residing within the large intracellular domain.
Mechanisms regulating ER exit or retention/retrieval are important determinants of surface expression and stability of membrane proteins. Receptors retained in the ER are more likely to be degraded than are fully assembled and folded receptors that exited the ER on their way to the plasma membrane. Motifs governing ER exit are found in the intracellular loops of β2, β4, and muscle subunits (Keller et al., 2001, Srinivasan et al., 2011). In human β2 and β4 subunit ER export, LFM motifs reside at the distal ends of the TM3 proximal domains (Figure 1) (Srinivasan et al., 2011). In addition, an ER retention/retrieval (RRQR) motif was found in the non-conserved middle segment of β2 (Srinivasan et al., 2011). Interestingly, increasing the number of ER exit motifs in the fully assembled α4β2 nAChR led to an increase in the number of ER exit sites as seen using SEC24 as a marker. This suggests that increasing the number of ER exit motifs in the fully assembled receptor enhances assembly of ER exit sites leading to more efficient export of nAChRs, increased surface expression and increased stability of nAChRs (Srinivasan et al., 2011). In contrast, a mutation associated with Amyotrophic Lateral Sclerosis (ALS), R349C near the ER exit motif of β4, decreased the number of ER exit sites and surface expression of the receptors (Richards et al., 2011, Sabatelli et al., 2009, Srinivasan et al., 2011). Additionally, nicotine, functioning as a pharmacological chaperone, stabilizes α3β4 receptors having a (α3)2(β4)3 stoichiometry and thus having more ER export motifs. This effect is a likely explanation for the enhanced surface expression and stability of α3β4 receptors following chronic exposure to nicotine (Mazzo et al., 2013). Last, in muscle receptors, assembly was suggested to mask dibasic ER retention/retrieval motifs (COPI binding motifs) in the TM3 proximal helices (a motif found in many nAChR subunits, Figure 1), thus restricting ER exit to fully assembled receptors (Keller et al., 2001). Therefore, motifs regulating ER export in the intracellular loop are key to functional expression of nAChRs.
Heterogeneity of intracellular loop sequences and of the motifs governing maturation within them suggests that differences in surface expression and localization of the specific nAChR subtypes may depend on these sequences. Indeed, substituting the intracellular loop of α7 with that of α3 shifted localization of the chimera from perisynaptic to synaptic (Williams et al., 1998). Moreover, detailed analysis identified motifs within the middle segment of the intracellular loops of α7 and α4 that target receptors containing these motifs to dendrites or axons, respectively (Xu et al., 2006).
The mouse tumor suppressor protein adenomatous polyposis coli (APC) and the adaptor protein 14-3-3 also affect nAChR maturation via the intracellular loop. APC was shown to interact with the TM3 TM4 intracellular loop of β1 and has been shown to regulate clustering of muscle-type receptors (Wang et al., 2003). APC was also shown to affect clustering of α3-containing receptors via the 14-3-3 adaptor protein and a 14-3-3 binding motif (RSSSSES) within the unstructured middle segment of the intracellular domain (Rosenberg et al., 2008). The 14-3-3 adaptor protein also interacts with the α4 subunit via a similar RSLSVQ motif in the middle segment of the intracellular domain, leading to increased surface expression of α4β2 receptors (Jeanclos et al., 2001). APC may also regulate α7 nAChR clustering in a manner that is dependent on Wnt signaling (Farias et al., 2007). Additionally, nAChR may share some interactions in common or in competition with glutamate receptors. The protein interacting with C kinase (PICK1), which can promote clustering of glutamate receptors, has been suggested to interact with the intracellular domain of α7 but with the effects of reducing receptor clustering (Baer et al., 2007).
Many studies have indicated reciprocal cross talk between phosphorylation-dependent signaling and nAChR function, and all known phosphorylation sites in nAChR subunits are in the intracellular loop (Talwar and Lynch, 2014). However, in many cases it is not clear whether the receptor itself is the phosphoprotein. Likewise, when receptor activation has been implicated to regulate kinase activity, it is usually assumed that channel-mediated currents (usually calcium) are required intermediates to phosphorylation events (Chatterjee et al., 2009, Cheng and Yakel, 2014, El Kouhen et al., 2009, Gubbins et al., 2010, Marrero and Bencherif, 2009, Nuutinen et al., 2006, Ren et al., 2005). Although there have been studies that have shown nAChR to be phosphorylated (Charpantier et al., 2005, Guo and Wecker, 2002, Pollock et al., 2009, Pollock et al., 2007) and their phosphorylation status to have effects on turnover, assembly, and subcellular localization (Hopfield et al., 1988, Huganir et al., 1986, Swope et al., 1999) (Yamada et al., 2010), the question remains: to what degree is there direct functional cross talk between phosphorylation sites in the intracellular domain and extracellular and transmembrane sites that regulate channel function? Some of the best data for such coupling come from studies of Torpedo, which have shown that phosphorylation of intracellular sites dynamically regulates nAChR function (Paradiso and Brehm, 1998).
Recently, studies of another member of the Cys-loop superfamily, α3 glycine receptors, have provided good evidence for phosphorylation-dependent conformational coupling between the receptor’s intracellular loop and the extracellular and transmembrane domains that did not rely on ion channel activation (Han et al., 2013). This sort of coupling is consistent with an important functional role for the intracellular domain and potential dual ionotropic and metabotropic roles for Cys-loop receptors, since the inter-domain coupling was not dependent on ion channel activation.
The greatest challenge for understanding the potential functional diversity for nAChRs and other members of the Cys-loop superfamily is identification of potential roles for the diverse, intrinsically disordered central segments of the intracellular domains. From an evolutionary perspective, the diversity in these domains suggests that radiation of the gene family was, to a significant degree, driven by the incorporation of sequence from different sources, which, to date, are unidentified. It is interesting to note that, not only is it likely that these domains came from different unidentified homologs, but selective pressure for conservation of sequence in these domains has varied significantly, as suggested by the variously consistent or diverse hydrophobicity profiles shown in Figure 3. It has been proposed that protein flexibility is of intrinsic importance for molecular recognition enabling protein-protein interactions (Janin and Sternberg, 2013). Apparently disordered proteins may, in fact, be proteins looking for partners. Such interactions may be important for signaling and, as described above, have a role in interactions with proteins regulating receptor maturation.
Specific analyses of alpha7
The α7 nAChR is a particularly attractive candidate for a dual ionotropic-metabotropic receptor since it has been amply demonstrated to play a role in channel-independent signal transduction related to inflammation (de Jonge and Ulloa, 2007). Such modulation of immune cell function has been reported to be most effectively accomplished by ligands that have little or no ion channel efficacy in cells that express α7 nAChR capable of ion channel function (Briggs et al., 2009, Thomsen and Mikkelsen, 2012), suggesting that signal transduction is associated with a conformation of the channel when the ion channels are “desensitized”. Also, as noted previously, the α7 intracellular domain has been generally well conserved. Therefore we look at additional bioinformatic analyses of the α7 intracellular domain that may identify particular candidate sites of interest for further studies.
Numerous putative functional sites are identified by the Eukaryotic Linear Motif resource for Functional Sites in Proteins (ELM) (Dinkel et al., 2014) and ProSite (Sigrist et al., 2013) in the α7 intracellular domain sequences of the six species, as well as in the intracellular domain sequences of the other human nAChR. In fact, the majority of these sites are located in the central flexible portion of the intracellular domains. Some of the sites would be important for receptor assembly, positioning, and disassembly, as well as for potential participation in cell-signaling cascades. A simplified overview, which reports the number of potential sites of various types found in α7 by the ELM analysis is given in Table 2. Some sites are more common in α7 of multiple species than they are in other subunits (see Supplemental Data Table S4 for results with other subunits). Examples include: the Mitogen-activated protein kinase (DOC_MAPK_1) signaling family, which includes extracellular signal-regulated kinases (ERKs) and c-Jun N-terminal kinases (JNKs); a cAMP-dependent protein kinase A (MOD_PKA_1/CAMP-Phospho) phosphorylation site; and a tyrosine-based sorting signal responsible for the interaction with mu subunit of the adaptor protein complex (TRG_ENDOCYTIC_2). Also present in α7 are tyrosine kinase phosphorylation sites and a possible destruction motif (DEG_APCC_DBOX_1). The cAMP-PKA phosphorylation site is in the predicted solvent-accessible region of the intracellular domain, and this has been shown to affect function (Moss et al., 1996), as are tyrosine kinase sites (Charpantier et al., 2005).
Table 2.
Putative association/consensus sites in α7 nAChR
| human | bovine | rat | chick | frog | fish | |
|---|---|---|---|---|---|---|
| ELM analysis | ||||||
| CLV_C14_Caspase3-7 | 1 | |||||
| DEG_APCC_DBOX_1 | 1 | 1 | 1 | 1 | 1 | |
| DEG_Nend_UBRbox_1 | 1 | 1 | ||||
| DEG_Nend_UBRbox_3 | 1 | 1 | 1 | |||
| DEG_SCF_TRCP1_1 | 1 | |||||
| DOC_CKS1_1 | 1 | |||||
| DOC_MAPK_1 | 1 | 1 | 1 | 1 | 1 | |
| DOC_PP2B_2 | ||||||
| DOC_WW_Pin1_4 | 2 | 2 | 2 | 2 | 3 | 2 |
| LIG_14-3-3-2 | 1 | 1 | ||||
| LIG_SH2_STAT5 | 2 | 2 | 2 | 2 | 2 | 2 |
| LIG_SH3_3 | 2 | 1 | 1 | |||
| LIG_TRAF2_1 | 2 | 1 | ||||
| MOD_CK1_1 | 1 | 1 | 1 | 1 | 1 | 5 |
| MOD_CK2_1 | 1 | 1 | 1 | 3 | 1 | 1 |
| MOD_GSK3_1 | 1 | 1 | 1 | 2 | 2 | 4 |
| MOD_NEK2_1 | 1 | |||||
| MOD_NEK2_2 | 1 | 1 | ||||
| MOD_PKA_1 | 1 | 1 | 1 | 1 | 1 | |
| MOD_PKA_2 | 1 | 1 | 1 | 2 | 2 | 1 |
| MOD_PLK | 1 | 1 | 1 | 1 | 1 | |
| MOD_ProDKin_1 | 2 | 2 | 2 | 2 | 3 | 2 |
| TRG_ENDOCYTIC_2 | 1 | 1 | 1 | 1 | 1 | 1 |
| TRG_ER_diArg_1 | 1 | |||||
| TRG_LysEnd_APsAcLL_1 | 1 | 1 | 1 | |||
| TRG_PEX_1 | 1 | 1 | 1 | 1 | 1 | 1 |
CLV_C14_Caspase3-7 (Caspase3 and Caspase7 cleavage site), DEG_APCC_DBOX_1 (destruction motif that binds to the Cdh1 and Cdc20), DEG_Nend_UBRbox_1 (N-terminal motif that initiates protein degradation by binding to the UBR-box of N-recognins), DEG_Nend_UBRbox_3 (N-terminal motif that initiates protein degradation by binding to the UBR-box of N-recognins), DEG_SCF_TRCP1_1 (The DSGxxS phospho-dependent degron binds the F box protein of the SCF-betaTrCP1 complex), DOC_CKS1_1 (Phospho-dependent motif that mediates docking of CDK substrates and regulators to cyclin-CDK-bound Cks1), DOC_MAPK_1 (Mitogen-activated protein kinase), DOC_PP2B_2 (Docking motif in calcineurin substrates that binds at the interface of the catalytic CNA and regulatory CNB subunits), DOC_WW_Pin1_4 (The Class IV WW domain interaction motif is recognised primarily by the Pin1 phosphorylation-dependent prolyl isomerase), LIG_14-3-3-2 (Longer mode 2 interacting phospho-motif for 14-3-3 proteins), LIG_SH2_STAT5 (STAT5 Src Homology 2 (SH2) domain binding motif), LIG_SH3_3 (This is the motif recognized by those SH3 domains with a non-canonical class I recognition specificity), LIG_TRAF2_1 (Major TRAF2-binding consensus motif), MOD_CK1_1 (CK1 phosphorylation site), MOD_CK2_1 (CK2 phosphorylation site), MOD_GSK3_1 (GSK3 phosphorylation recognition site), MOD_NEK2_1 (NEK2 phosphorylation motif), MOD_NEK2_2 (NEK2 phosphorylation motif), MOD_PKA_1 (cAMP-dependent protein kinase A), MOD_PKA_2 (Secondary preference for PKA-type AGC kinase phosphorylation), MOD_PLK (Site phosphorylated by the Polo-like kinase), MOD_ProDKin_1 (Proline-Directed Kinase (e.g. MAPK) phosphorylation site), TRG_ENDOCYTIC_2 (Adaptor Protein complex binding motif), TRG_ER_diArg_1 (di-Arg ER retention motif), TRG_LysEnd_APsAcLL_1 (Sorting signal directing type I transmembrane proteins), TRG_PEX_1 (peroxisomal import receptor Motif)
Additionally, the molecular chaperone RIC-3 (Treinin, 2008), which is necessary for the functional expression of α7 ion channels in some cells (Williams et al., 2005), may require recognition sites in the α7 intracellular domain. RIC-3 is an evolutionarily conserved ER resident chaperone known to affect stability, assembly, trafficking, and surface expression of nAChRs (Alexander et al., 2010, Treinin, 2008, Wang et al., 2009). Effects of RIC-3 are receptor subtype-specific; both positive and negative effects were observed, depending on identity of the co-expressed nAChR and on the experimental system (Halevi et al., 2003, Lansdell et al., 2005). More recently it was shown that RIC-3 affects assembly of α7 but not of α4β2 receptors (Dau et al., 2013). Moreover, structure-function analysis showed that RIC-3 interacts differently with different receptor subunits (Biala et al., 2009, Cohen Ben-Ami et al., 2009). Such specificity suggests that subunit-specific motifs mediate subunit-specific effects of RIC-3. Indeed, a motif within the intracellular loop is likely to specifically mediate the positive effects of RIC-3 on homomeric α7 receptors (Castillo et al., 2006, Gee et al., 2007). Mutation to alanine of any one of five residues (three hydrophobic and two positively charged residues) within a putative amphipathic helix present in the domain preceding TM4 eliminated the positive effects of RIC-3 on α7 expression (Castillo et al., 2006). The last three residues of this motif (RFR residues) are only present in α7 (Figure 1).
Also present in α7 are SH2- and SH3-domain protein binding sites (Table 2), which may provide mechanisms for adapter proteins to bind to α7 (Reebye et al., 2012) after either phosphorylation or a conformation shift of α7 is triggered by ligand binding. This would provide a mechanism for kinases to then bind to the α7 complex for further signaling. This, however, awaits experimental proof as SH2 or SH3 binding site motifs are not always active.
The identification of these numerous sites of potential interaction is consistent with published proteomic analyses (Paulo et al., 2009) and defines the future challenges for identifying the functional partners of the intracellular domain of α7 and other nAChR. It has been speculated that α7 nAChR are directly coupled to G-proteins via sites in their intracellular domain (Kabbani et al., 2013); however, direct evidence for this intriguing hypothesis is still lacking, since it is hard to eliminate channel-dependent calcium signals as intermediates between the nicotinic receptor activation and G-protein signals (Nordman and Kabbani, 2014). As noted above, support for direct α7 modulation of signal transduction that is independent of channel activation comes from studies of cholinergic modulation of inflammation (de Jonge and Ulloa, 2007) and the observation that some of the most effective modulators of α7-mediated anti-inflammatory responses are silent agonists (Chojnacka et al., 2013, Papke et al., 2014), which are unable to produce channel activation but can induce desensitized conformations of the receptor (Briggs et al., 2009, Thomsen and Mikkelsen, 2012).
Conclusions
In summary, multiple potential functions can be ascribed to the intracellular domains of nAChR, and several perspectives suggest guideposts for future investigations and clues to signal transduction mechanisms not presently well understood. The conserved portions have predicted helical structure, have been shown to be required for some receptor maturation and assembly, and have positive and negative charged amino acid residues that may be important for ion channel function and perhaps voltage- or use-dependent changes in channel function. The clustered charged residues in the intracellular loops near the membrane spanning domains are especially interesting as possible regulatory elements that might modulate the function of submembrane portals. It may be worth pursuing differences in these regions for anionic versus cationic channels. The activity (and indeed the existence) of the portals themselves also needs to be further investigated. The flexible central loop section of the intracellular domain is intriguing as to its purpose and origins, as it differs greatly in length and composition among subunits.
Evolution relies on diversity to promote unique functional adaptations, whether on the level of species or molecular subtypes. The intracellular domains of nAChR, highly variable and ostensibly largely disordered, are nonetheless predicted to be accessible for intracellular protein binding, enzymatic activity, and modulatory functions. While the intracellular domains are most variable among subunit types, they are, to varying degrees, relatively conserved across species within subunit type. Noticing patterns of intron-exon borders and chromosome mapping leads one to wonder about evolution and how those domains came to be, since there were none in bacterial homologs. While we can generalize about the extracellular ligand-binding domains and the well-conserved transmembrane domains of all the nAChR subtypes, if we want insights into the functional roles of specific nAChR subtypes, we will have to make efforts to reveal the hidden functions of their intracellular domains.
Supplementary Material
Highlights.
Intracellular domains of Cys-loop receptors are variable and unique to each subunit.
Intracellular subdomains may contribute to ion-conducting submembrane portals.
The α7 intracellular domains contain well conserved sites for protein interactions.
Acknowledgments
The authors thank Drs. Nicole Horenstein, Dietlind Gerloff, and Eliot Spindel for comments and suggestions. This work was supported by National Institute of Health grant RO1-GM57481.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander JK, Sagher D, Krivoshein AV, Criado M, Jefford G, Green WN. Ric-3 promotes alpha7 nicotinic receptor assembly and trafficking through the ER subcompartment of dendrites. J Neurosci. 2010;30:10112–10126. doi: 10.1523/JNEUROSCI.6344-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer K, Burli T, Huh KH, Wiesner A, Erb-Vogtli S, Gockeritz-Dujmovic D, Fuhrer C. PICK1 interacts with alpha7 neuronal nicotinic acetylcholine receptors and controls their clustering. Mol Cell Neurosci. 2007;35:339–355. doi: 10.1016/j.mcn.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biala Y, Liewald JF, Ben-Ami HC, Gottschalk A, Treinin M. The conserved RIC-3 coiled-coil domain mediates receptor-specific interactions with nicotinic acetylcholine receptors. Mol Biol Cell. 2009;20:1419–1427. doi: 10.1091/mbc.E08-08-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocquet N, Prado de Carvalho L, Cartaud J, Neyton J, Le Poupon C, Taly A, Corringer PJ. A prokaryotic proton-gated ion channel from the nicotinic acetylcholine receptor family. Nature. 2007;445:116–119. doi: 10.1038/nature05371. [DOI] [PubMed] [Google Scholar]
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- Briggs CA, Gronlien JH, Curzon P, Timmermann DB, Ween H, Thorin-Hagene K, Gopalakrishnan M. Role of channel activation in cognitive enhancement mediated by alpha7 nicotinic acetylcholine receptors. Br J Pharmacol. 2009;158:1486–1494. doi: 10.1111/j.1476-5381.2009.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo M, Mulet J, Gutierrez LM, Ortiz JA, Castelan F, Gerber S, Criado M. Role of the RIC-3 protein in trafficking of serotonin and nicotinic acetylcholine receptors. J Mol Neurosci. 2006;30:153–156. doi: 10.1385/JMN:30:1:153. [DOI] [PubMed] [Google Scholar]
- Charpantier E, Wiesner A, Huh KH, Ogier R, Hoda JC, Allaman G, Fuhrer C. Alpha7 neuronal nicotinic acetylcholine receptors are negatively regulated by tyrosine phosphorylation and Src-family kinases. J Neurosci. 2005;25:9836–9849. doi: 10.1523/JNEUROSCI.3497-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee PK, Al-Abed Y, Sherry B, Metz CN. Cholinergic agonists regulate JAK2/STAT3 signaling to suppress endothelial cell activation. Am J Physiol Cell Physiol. 2009;297:C1294–1306. doi: 10.1152/ajpcell.00160.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Yakel JL. Presynaptic alpha7 nicotinic acetylcholine receptors enhance hippocampal mossy fiber glutamatergic transmission via PKA activation. J Neurosci. 2014;34:124–133. doi: 10.1523/JNEUROSCI.2973-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chojnacka K, Papke RL, Horenstein NA. Synthesis and evaluation of a conditionally-silent agonist for the alpha7 nicotinic acetylcholine receptor. Bioorg Med Chem Lett. 2013;23:4145–4149. doi: 10.1016/j.bmcl.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Ben-Ami H, Biala Y, Farah H, Elishevitz E, Battat E, Treinin M. Receptor and subunit specific interactions of RIC-3 with nicotinic acetylcholine receptors. Biochemistry. 2009;48:12329–12336. doi: 10.1021/bi901234a. [DOI] [PubMed] [Google Scholar]
- Dau A, Komal P, Truong M, Morris G, Evans G, Nashmi R. RIC-3 differentially modulates alpha4beta2 and alpha7 nicotinic receptor assembly, expression, and nicotine-induced receptor upregulation. BMC neuroscience. 2013;14:47. doi: 10.1186/1471-2202-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge WJ, Ulloa L. The alpha7 nicotinic acetylcholine receptor as a pharmacological target for inflammation. Br J Pharmacol. 2007;151:915–929. doi: 10.1038/sj.bjp.0707264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkel H, Van Roey K, Michael S, Davey NE, Weatheritt RJ, Born D, Gibson TJ. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 2014;42:D259–266. doi: 10.1093/nar/gkt1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers H, Schaeffer E, Bickler P, Forsayeth J. Functional deactivation of the major neuronal nicotinic receptor caused by nicotine and a protein kinase C-dependent mechanism. Mol Pharm. 1997;52:1105–1112. [PubMed] [Google Scholar]
- El Kouhen R, Hu M, Anderson DJ, Li J, Gopalakrishnan M. Pharmacology of alpha7 nicotinic acetylcholine receptor mediated extracellular signal-regulated kinase signalling in PC12 cells. Br J Pharmacol. 2009;156:638–648. doi: 10.1111/j.1476-5381.2008.00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias GG, Valles AS, Colombres M, Godoy JA, Toledo EM, Lukas RJ, Inestrosa NC. Wnt-7a induces presynaptic colocalization of alpha 7-nicotinic acetylcholine receptors and adenomatous polyposis coli in hippocampal neurons. J Neurosci. 2007;27:5313–5325. doi: 10.1523/JNEUROSCI.3934-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster CP, Beckman ML, Parker JC, Sheffield EB, Whitworth TL, Quick MW, Lester RA. Regulation of alpha4beta2 nicotinic receptor desensitization by calcium and protein kinase C. Mol Pharmacol. 1999;55:432–443. [PubMed] [Google Scholar]
- Frail DE, McLaughlin LL, Mudd J, Merlie JP. Identification of the mouse muscle 43,000-dalton acetylcholine receptor-associated protein (RAPsyn) by cDNA cloning. J Biol Chem. 1988;263:15602–15607. [PubMed] [Google Scholar]
- Gee VJ, Kracun S, Cooper ST, Gibb AJ, Millar NS. Identification of domains influencing assembly and ion channel properties in alpha 7 nicotinic receptor and 5-HT3 receptor subunit chimaeras. Br J Pharmacol. 2007;152:501–512. doi: 10.1038/sj.bjp.0707429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbins EJ, Gopalakrishnan M, Li J. Alpha7 nAChR-mediated activation of MAP kinase pathways in PC12 cells. Brain Res. 2010;1328:1–11. doi: 10.1016/j.brainres.2010.02.083. [DOI] [PubMed] [Google Scholar]
- Guo X, Wecker L. Identification of three cAMP-dependent protein kinase (PKA) phosphorylation sites within the major intracellular domain of neuronal nicotinic receptor alpha4 subunits. J Neurochem. 2002;82:439–447. doi: 10.1046/j.1471-4159.2002.01027.x. [DOI] [PubMed] [Google Scholar]
- Hales TG, Dunlop JI, Deeb TZ, Carland JE, Kelley SP, Lambert JJ, Peters JA. Common determinants of single channel conductance within the large cytoplasmic loop of 5-hydroxytryptamine type 3 and alpha4beta2 nicotinic acetylcholine receptors. J Biol Chem. 2006;281:8062–8071. doi: 10.1074/jbc.M513222200. [DOI] [PubMed] [Google Scholar]
- Halevi S, Yassin L, Eshel M, Sala F, Sala S, Criado M, Treinin M. Conservation within the RIC-3 gene family. Effectors of mammalian nicotinic acetylcholine receptor expression. J Biol Chem. 2003;278:34411–34417. doi: 10.1074/jbc.M300170200. [DOI] [PubMed] [Google Scholar]
- Han L, Talwar S, Wang Q, Shan Q, Lynch JW. Phosphorylation of alpha3 glycine receptors induces a conformational change in the glycine-binding site. ACS chemical neuroscience. 2013;4:1361–1370. doi: 10.1021/cn400097j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassaine G, Deluz C, Grasso L, Wyss R, Tol MB, Hovius R, Nury H. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature. 2014;512:276–281. doi: 10.1038/nature13552. [DOI] [PubMed] [Google Scholar]
- Hilf RJ, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- Hoffman PW, Ravindran A, Huganir RL. Role of phosphorylation in desensitization of acetylcholine receptors expressed in Xenopus oocytes. J Neurosci. 1994;14:4185–4195. doi: 10.1523/JNEUROSCI.14-07-04185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden-Dye L, Joyner M, O’Connor V, Walker RJ. Nicotinic acetylcholine receptors: a comparison of the nAChRs of Caenorhabditis elegans and parasitic nematodes. Parasitology international. 2013;62:606–615. doi: 10.1016/j.parint.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Hopfield JF, Tank DW, Greengard P, Huganir RL. Functional modulation of the nicotinic acetylcholine receptor by tyrosine phosphorylation. Nature. 1988;336:677–680. doi: 10.1038/336677a0. [DOI] [PubMed] [Google Scholar]
- Huganir R, Delcour AH, Greengard P, Hess GP. Phosphorylation of the nicotinic acetylcholine receptor regulates its rate of desensitization. Nature. 1986;321:774–776. doi: 10.1038/321774a0. [DOI] [PubMed] [Google Scholar]
- Janin J, Sternberg MJ. Protein flexibility, not disorder, is intrinsic to molecular recognition. F1000 biology reports. 2013;5:2. doi: 10.3410/B5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanclos EM, Lin L, Treuil MW, Rao J, DeCoster MA, Anand R. The chaperone protein 14-3-3eta interacts with the nicotinic acetylcholine receptor alpha 4 subunit. Evidence for a dynamic role in subunit stabilization. J Biol Chem. 2001;276:28281–28290. doi: 10.1074/jbc.M011549200. [DOI] [PubMed] [Google Scholar]
- Jones AK, Brown LA, Sattelle DB. Insect nicotinic acetylcholine receptor gene families: from genetic model organism to vector, pest and beneficial species. Invertebrate neuroscience : IN. 2007;7:67–73. doi: 10.1007/s10158-006-0039-6. [DOI] [PubMed] [Google Scholar]
- Jones AK, Buckingham SD, Sattelle DB. Proteins interacting with nicotinic acetylcholine receptors: expanding functional and therapeutic horizons. Trends Pharmacol Sci. 2010;31:455–462. doi: 10.1016/j.tips.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Kabbani N, Nordman JC, Corgiat BA, Veltri DP, Shehu A, Seymour VA, Adams DJ. Are nicotinic acetylcholine receptors coupled to G proteins? Bioessays. 2013 doi: 10.1002/bies.201300082. [DOI] [PubMed] [Google Scholar]
- Keller SH, Lindstrom J, Ellisman M, Taylor P. Adjacent basic amino acid residues recognized by the COP I complex and ubiquitination govern endoplasmic reticulum to cell surface trafficking of the nicotinic acetylcholine receptor alpha-Subunit. J Biol Chem. 2001;276:18384–18391. doi: 10.1074/jbc.M100691200. [DOI] [PubMed] [Google Scholar]
- Kelley SP, Dunlop JI, Kirkness EF, Lambert JJ, Peters JA. A cytoplasmic region determines single-channel conductance in 5-HT3 receptors. Nature. 2003;424:321–324. doi: 10.1038/nature01788. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lansdell SJ, Gee VJ, Harkness PC, Doward AI, Baker ER, Gibb AJ, Millar NS. RIC-3 enhances functional expression of multiple nicotinic acetylcholine receptor subtypes in mammalian cells. Mol Pharmacol. 2005;68:1431–1438. doi: 10.1124/mol.105.017459. [DOI] [PubMed] [Google Scholar]
- Maimone MM, Enigk RE. The intracellular domain of the nicotinic acetylcholine receptor alpha subunit mediates its coclustering with rapsyn. Mol Cell Neurosci. 1999;14:340–354. doi: 10.1006/mcne.1999.0779. [DOI] [PubMed] [Google Scholar]
- Marrero MB, Bencherif M. Convergence of alpha 7 nicotinic acetylcholine receptor-activated pathways for anti-apoptosis and anti-inflammation: central role for JAK2 activation of STAT3 and NF-kappaB. Brain Res. 2009;1256:1–7. doi: 10.1016/j.brainres.2008.11.053. [DOI] [PubMed] [Google Scholar]
- Mazzo F, Pistillo F, Grazioso G, Clementi F, Borgese N, Gotti C, Colombo SF. Nicotine-modulated subunit stoichiometry affects stability and trafficking of alpha3beta4 nicotinic receptor. J Neurosci. 2013;33:12316–12328. doi: 10.1523/JNEUROSCI.2393-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Stowell M, Unwin N. Nicotinic acetylcholine receptor at 4.6 A resolution: transverse tunnels in the channel wall. J Mol Biol. 1999;288:765–786. doi: 10.1006/jmbi.1999.2721. [DOI] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- Moss SJ, McDonald BJ, Rudhard Y, Schoepfer R. Phosphorylation of the predicted major intracellular domains of the rat and chick neuronal nicotinic acetylcholine receptor alpha 7 subunit by cAMP-dependent protein kinase. Neuropharmacology. 1996;35:1023–1028. doi: 10.1016/s0028-3908(96)00083-4. [DOI] [PubMed] [Google Scholar]
- Nishizaki T, Sumikawa K. Effects of PKC and PKA phosphorylation on desensitization of nicotinic acetylcholine receptors. Brain Res. 1998;812:242–245. doi: 10.1016/s0006-8993(98)00836-1. [DOI] [PubMed] [Google Scholar]
- Nordman JC, Kabbani N. Microtubule dynamics at the growth cone are mediated by alpha7 nicotinic receptor activation of a Galphaq and IP3 receptor pathway. FASEB J. 2014;28:2995–3006. doi: 10.1096/fj.14-251439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuutinen S, Ekokoski E, Lahdensuo E, Tuominen RK. Nicotine-induced upregulation of human neuronal nicotinic alpha7-receptors is potentiated by modulation of cAMP and PKC in SH-EP1-halpha7 cells. Eur J Pharmacol. 2006;544:21–30. doi: 10.1016/j.ejphar.2006.06.038. [DOI] [PubMed] [Google Scholar]
- Papke D, Grosman C. The role of intracellular linkers in gating and desensitization of human pentameric ligand-gated ion channels. J Neurosci. 2014;34:7238–7252. doi: 10.1523/JNEUROSCI.5105-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papke RL. Merging old and new perspectives on nicotinic acetylcholine receptors. Biochem Pharmacol. 2014 doi: 10.1016/j.bcp.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papke RL, Chojnacka K, Horenstein NA. The minimal pharmacophore for silent agonism of alpha7 nAChR. Journal of Pharmacology and Experimental Therapeutics. 2014;350:665–680. doi: 10.1124/jpet.114.215236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradiso K, Brehm P. Long-term desensitization of nicotinic acetylcholine receptors is regulated via protein kinase A-mediated phosphorylation. J Neurosci. 1998;18:9227–9237. doi: 10.1523/JNEUROSCI.18-22-09227.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthiban M, Rajasekaran MB, Ramakumar S, Shanmughavel P. Molecular modeling of human pentameric alpha(7) neuronal nicotinic acetylcholine receptor and its interaction with its agonist and competitive antagonist. J Biomol Struct Dyn. 2009;26:535–547. doi: 10.1080/07391102.2009.10507269. [DOI] [PubMed] [Google Scholar]
- Paulo J, Brucker W, Hawrot E. Proteomic Analysis of an alpha7 Nicotinic Acetylcholine Receptor Interactome. J Proteome Res. 2009;8:1849–1858. doi: 10.1021/pr800731z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock VV, Pastoor T, Katnik C, Cuevas J, Wecker L. Cyclic AMP-dependent protein kinase A and protein kinase C phosphorylate alpha4beta2 nicotinic receptor subunits at distinct stages of receptor formation and maturation. Neuroscience. 2009;158:1311–1325. doi: 10.1016/j.neuroscience.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock VV, Pastoor TE, Wecker L. Cyclic AMP-dependent protein kinase (PKA) phosphorylates Ser362 and 467 and protein kinase C phosphorylates Ser550 within the M3/M4 cytoplasmic domain of human nicotinic receptor alpha4 subunits. J Neurochem. 2007;103:456–466. doi: 10.1111/j.1471-4159.2007.04853.x. [DOI] [PubMed] [Google Scholar]
- Ramarao MK, Cohen JB. Mechanism of nicotinic acetylcholine receptor cluster formation by rapsyn. Proc Natl Acad Sci U S A. 1998;95:4007–4012. doi: 10.1073/pnas.95.7.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reebye V, Frilling A, Hajitou A, Nicholls JP, Habib NA, Mintz PJ. A perspective on non-catalytic Src homology (SH) adaptor signalling proteins. Cellular signalling. 2012;24:388–392. doi: 10.1016/j.cellsig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Ren K, Puig V, Papke RL, Itoh Y, Hughes JA, Meyer EM. Multiple calcium channels and kinases mediate alpha7 nicotinic receptor neuroprotection in PC12 cells. J Neurochem. 2005;94:926–933. doi: 10.1111/j.1471-4159.2005.03223.x. [DOI] [PubMed] [Google Scholar]
- Richards CI, Srinivasan R, Xiao C, Mackey ED, Miwa JM, Lester HA. Trafficking of alpha4* nicotinic receptors revealed by superecliptic phluorin: effects of a beta4 amyotrophic lateral sclerosis-associated mutation and chronic exposure to nicotine. J Biol Chem. 2011;286:31241–31249. doi: 10.1074/jbc.M111.256024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MM, Yang F, Giovanni M, Mohn JL, Temburni MK, Jacob MH. Adenomatous polyposis coli plays a key role, in vivo, in coordinating assembly of the neuronal nicotinic postsynaptic complex. Mol Cell Neurosci. 2008;38:138–152. doi: 10.1016/j.mcn.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatelli M, Eusebi F, Al-Chalabi A, Conte A, Madia F, Luigetti M, Zollino M. Rare missense variants of neuronal nicotinic acetylcholine receptor altering receptor function are associated with sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:3997–4006. doi: 10.1093/hmg/ddp339. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular systems biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Xenarios I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41:D344–347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixma TK, Smit AB. Acetylcholine binding protein (AChBP): A Secreted Glial Protein that Provides a High-Resolution Model for the Extracellular Domain of Pentameric Ligand-Gated Ion Channels. Annu Rev Biophys Biomol Struct. 2003;21:21. doi: 10.1146/annurev.biophys.32.110601.142536. [DOI] [PubMed] [Google Scholar]
- Srinivasan R, Pantoja R, Moss FJ, Mackey ED, Son CD, Miwa J, Lester HA. Nicotine up-regulates alpha4beta2 nicotinic receptors and ER exit sites via stoichiometry-dependent chaperoning. J Gen Physiol. 2011;137:59–79. doi: 10.1085/jgp.201010532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swope SL, Moss SJ, Raymond LA, Huganir RL. Regulation of ligand-gated ion channels by protein phosphorylation. Adv Second Messenger Phosphoprotein Res. 1999;33:49–78. doi: 10.1016/s1040-7952(99)80005-6. [DOI] [PubMed] [Google Scholar]
- Talwar S, Lynch JW. Phosphorylation mediated structural and functional changes in pentameric ligand-gated ion channels: implications for drug discovery. The international journal of biochemistry & cell biology. 2014;53:218–223. doi: 10.1016/j.biocel.2014.05.028. [DOI] [PubMed] [Google Scholar]
- Thomsen MS, Mikkelsen JD. The alpha7 nicotinic acetylcholine receptor ligands methyllycaconitine, NS6740 and GTS-21 reduce lipopolysaccharide-induced TNF-alpha release from microglia. J Neuroimmunol. 2012;251:65–72. doi: 10.1016/j.jneuroim.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Treinin M. RIC-3 and nicotinic acetylcholine receptors: biogenesis, properties, and diversity. Biotechnol J. 2008;3:1539–1547. doi: 10.1002/biot.200800179. [DOI] [PubMed] [Google Scholar]
- Tsetlin V, Kuzmin D, Kasheverov I. Assembly of nicotinic and other Cys-loop receptors. J Neurochem. 2011;116:734–741. doi: 10.1111/j.1471-4159.2010.07060.x. [DOI] [PubMed] [Google Scholar]
- Unwin M, Toyoshima C, Kubalek E. Arrangement of the acetylcholine receptor subunits in the resting and desensitized states determined by cryoelectron microscopy of crystallized Torpedo postsynaptic membranes. J Cell Biol. 1988;107:1123–1138. doi: 10.1083/jcb.107.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin N. The nicotinic acetylcholine receptor at 9A resolution. J Mol Biol. 1993;229:1101–1124. doi: 10.1006/jmbi.1993.1107. [DOI] [PubMed] [Google Scholar]
- Unwin N. Acetylcholine receptor imaged in the open state. Nature. 1995;373:37–43. doi: 10.1038/373037a0. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Unwin N, Fujiyoshi Y. Gating movement of acetylcholine receptor caught by plunge-freezing. J Mol Biol. 2012;422:617–634. doi: 10.1016/j.jmb.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin N, Miyazawa A, Li J, Fujiyoshi Y. Activation of the nicotinic acetylcholine receptor involves a switch in conformation of the alpha subunits. J Mol Biol. 2002;319:1165–1176. doi: 10.1016/S0022-2836(02)00381-9. [DOI] [PubMed] [Google Scholar]
- Wang J, Jing Z, Zhang L, Zhou G, Braun J, Yao Y, Wang ZZ. Regulation of acetylcholine receptor clustering by the tumor suppressor APC. Nat Neurosci. 2003;6:1017–1018. doi: 10.1038/nn1128. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yao Y, Tang XQ, Wang ZZ. Mouse RIC-3, an endoplasmic reticulum chaperone, promotes assembly of the alpha7 acetylcholine receptor through a cytoplasmic coiled-coil domain. J Neurosci. 2009;29:12625–12635. doi: 10.1523/JNEUROSCI.1776-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BM, Temburni MK, Levey MS, Bertrand S, Bertrand D, Jacob MH. The long internal loop of the alpha 3 subunit targets nAChRs to subdomains within individual synapses on neurons in vivo. Nat Neurosci. 1998;1:557–562. doi: 10.1038/2792. [DOI] [PubMed] [Google Scholar]
- Williams ME, Burton B, Urrutia A, Shcherbatko A, Chavez-Noriega LE, Cohen CJ, Aiyar J. Ric-3 promotes functional expression of the nicotinic acetylcholine receptor alpha7 subunit in mammalian cells. J Biol Chem. 2005;280:1257–1263. doi: 10.1074/jbc.M410039200. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhu Y, Heinemann SF. Identification of sequence motifs that target neuronal nicotinic receptors to dendrites and axons. J Neurosci. 2006;26:9780–9793. doi: 10.1523/JNEUROSCI.0840-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Yaguchi T, Kanno T, Mukasa T, Nishizaki T. Auto-positive feedback regulation for nicotinic acetylcholine receptors by protein kinase C activation. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2010;26:247–252. doi: 10.1159/000320524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.