Abstract
Bacterial blight and bacterial leaf streak are serious, economically damaging, diseases of rice caused by the bacteria Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola. Bacillus amyloliquefaciens FZB42 was shown to possess biocontrol activity against these Xanthomonas strains by producing the antibiotic compounds difficidin and bacilysin. Analyses using fluorescence, scanning electron and transmission electron microscopy revealed difficidin and bacilysin caused changes in the cell wall and structure of Xanthomonas. Biological control experiments on rice plants demonstrated the ability of difficidin and bacilysin to suppress disease. Difficidin and bacilysin caused downregulated expression of genes involved in Xanthomonas virulence, cell division, and protein and cell wall synthesis. Taken together, our results highlight the potential of B. amyloliquefaciens FZB42 as a biocontrol agent against bacterial diseases of rice, and the utility of difficidin and bacilysin as antimicrobial compounds.
The Gram-negative bacterial genus Xanthomonas can infect at least 350 different plants, resulting in significant economic losses in agriculture worldwide1,2,3. Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola are important rice pathogens, which cause bacterial rice blight and bacterial leaf streak of rice, respectively2. Traditional management practices, especially copper chemicals, increase the cost of production, leave residuals on crops and soil, and develop resistance in populations of target pathogens3,4. Therefore, there is a pressing need to develop cost-effective and convenient strategies that minimize environmental impact. Biological control agents, for example plant growth-promoting bacteria Pseudomonas and Bacillus5, have received a great deal of attention on account of being environmentally-friendly and versatile in their mode of action.
Bacillus spp. are attractive for use in farming systems because of their ability to form heat- and desiccation-resistant endospores which can survive the preparation of bacterial formulations6. Bacillus amyloliquefaciens FZB42 is the type strain for a group of plant-associated Bacillus spp. classified as B. amyloliquefaciens subsp. plantarum. Bacillus amyloliquefaciens FZB42 has the impressive ability to stimulate plant growth and to suppress plant pathogenic organisms, which distinguishes it from the related model organism B. subtilis 168, and has been commercially applied to a broad range of host plants7,8. The genome of strain FZB42 was sequenced and it harbors an array of giant gene clusters that produce several secondary metabolites with antimicrobial activity9. Its antifungal activity is attributed mainly to the nonribosomally synthesized cyclic lipopeptides bacillomycin D and fengycin8, its antibacterial activity is mainly due to non-ribosomal synthesis of polyketides10, its nematicidal activity is due to the ribosomally synthesized peptide antibiotic plantazolicin11, whilst its algicidal activity arises from the nonribosomal dipeptide bacilysin12.
In a previous study, we demonstrated that rice plants treated with B. amyloliquefaciens FZB42 suspensions showed significant improvement in resistance to X. oryzae pv. oryzae over untreated plants13. The aim of the present study was to identify the antibacterial substance(s) present in the culture suspensions of FZB42 and to gain insight into the underlying mechanisms responsible for the antagonistic effect against Xanthomonas spp. The results demonstrate that difficidin and bacilysin from B. amyloliquefaciens FZB42 have antibacterial activity against X. oryzae pv. oryzae and X. oryzae pv. oryzicola, and that cytotoxic effects cause apparent changes in the bacterial plasma membrane and structure.
Results
Difficidin and bacilysin have antibacterial activities against X. oryzae pv. oryzae and X. oryzae pv. oryzicola
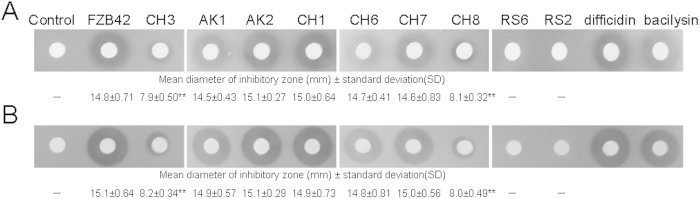
To identify the active substances produced by B. amyloliquefaciens FZB42, we initially used a mutant strain devoid of non-ribosomal synthesis of lipopeptides and polyketides (strain CH3). In agar diffusion assays, strain CH3 resulted in a small zone of inhibition against Xanthomonas, and that zone was very significantly different (P < 0.01) from wild-type B. amyloliquefaciens FZB42, indicating that one or more lipopeptides and/or polyketides and/or other metabolites synthesized through the sfp-dependent pathway had the ability to suppress growth of Xanthomonas (Fig. 1).
Figure 1. Detection of antagonistic action against Xanthomonas oryzae pv. oryzae.
(A) and Xanthomonas oryzae pv. oryzicola (B) by paper-disc agar diffusion assay. Bactericidal activity was tested as described in Methods. Control (Landy medium), FZB42 (wild type, producer of lipopeptides, polyketides and bacilysin), CH3 (Δsfp::Emr, deficient in lipopeptide and polyketide synthesis), AK1 (ΔbmyA::Emr, deficient in bacillomycin D synthesis), AK2 (ΔfenA::Cmr, deficient in fengycin synthesis), CH1 (ΔsrfA::Emr, deficient in surfactin synthesis), CH6 (Δbae::Cmr, no synthesis of bacillaene), CH7 (Δmln::Cmr, no synthesis of macrolactin), CH8 (Δdfn::Emr, no synthesis of difficidin), RS6 (Δsfp::Emr Δbac::Cmr, no lipopeptides, polyketides or bacilysin), and RS2 (Δbac::Cmr Δdfn::Emr, deficient in bacilysin and difficidin). The diameters of inhibition zones (mm) included the paper disk diameter (5 mm). Data are expressed as means ± standard deviation (SD). − indicates no inhibitory activity. **indicates an extremely significant difference compared with FZB42 (P < 0.01).
To identify the anti-Xanthomonas substances, single mutants of B. amyloliquefaciens deficient in production of surfactin (CH1), bacillomycin D (AK1), fengycin (AK2), bacillaene (CH6), macrolactin (CH7) difficidin (CH8) and a double mutant (RS6), blocked in synthesis of lipopeptides and polyketides and production of bacilysin, were tested. Strains complemented for those genes were also examined. Agar diffusion tests illustrate that the inhibitory effect exerted by CH8 was clearly reduced relative to the wild-type (P < 0.01) and RS6 yielded no inhibition zone, whilst the corresponding complemented strains resulted in similar bactericidal effects to the wild-type, suggesting that difficidin and bacilysin act as antagonists of X. oryzae pv. oryzae and X. oryzae pv. oryzicola (Fig. 1, Fig. S2). This conclusion was corroborated by the absence of an antagonistic effect of strain RS2, which is devoid of difficidin and bacilysin, and efficient suppression of Xanthomonas by difficidin and bacilysin purified from FZB42 culture filtrates (Fig. 1).
Effect of difficidin and bacilysin on viability of Xanthomonas spp. cells
We characterized Xanthomonas spp. cell development in the absence and presence of difficidin and bacilysin using phase contrast/fluorescence microscopy in combination with LIVE/DEAD BacLight bacterial viability staining (Figs S3 and S4; Table 1). Incubation of Xanthomonas spp. cell suspensions with the probes did not result in an increase in dead cells (red fluorescence) and, accordingly, the vast majority (95.58%, 96.09%) of the cell population remained alive (green fluorescence). In contrast, after 12 h of exposure to 10 μg/ml difficidin or bacilysin, the proportions of red fluorescent cells increased to 35.81%–40.48%. A large number of dead cells (86.67%–91.17%) were observed in the presence of 50 μg/ml difficidin or bacilysin as a consequence of the extensive accumulation of the red probe into the bacterial cells in response to membrane damage.
Table 1. Quantification of the viability of Xanthomonas cells after exposure to difficidin and bacilysin.
| Treatment |
Xanthomonas oryzae pv. oryzae |
Xanthomonas oryzae pv. oryzicola |
|||
|---|---|---|---|---|---|
| live cells (%) | dead cells (%) | live cells (%) | dead cells (%) | ||
| untreated | 96.09 | 3.91 | 95.58 | 4.42 | |
| difficidin | 10 μg/ml | 59.52 | 40.48 | 62.85 | 37.15 |
| 50 μg/ml | 8.83 | 91.17 | 10.28 | 89.72 | |
| bacilysin | 10 μg/ml | 63.65 | 36.35 | 64.19 | 35.81 |
| 50 μg/ml | 11.25 | 88.75 | 13.33 | 86.67 | |
The number of total, live (green) and dead (red) cells was counted in ten different microscopic fields. The values indicate the percentages of live and dead cells in the suspensions.
Morphological and ultrastructural changes of Xanthomonas cells in the presence of difficidin and bacilysin
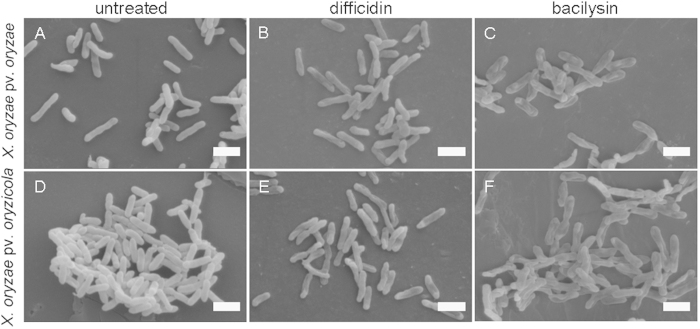
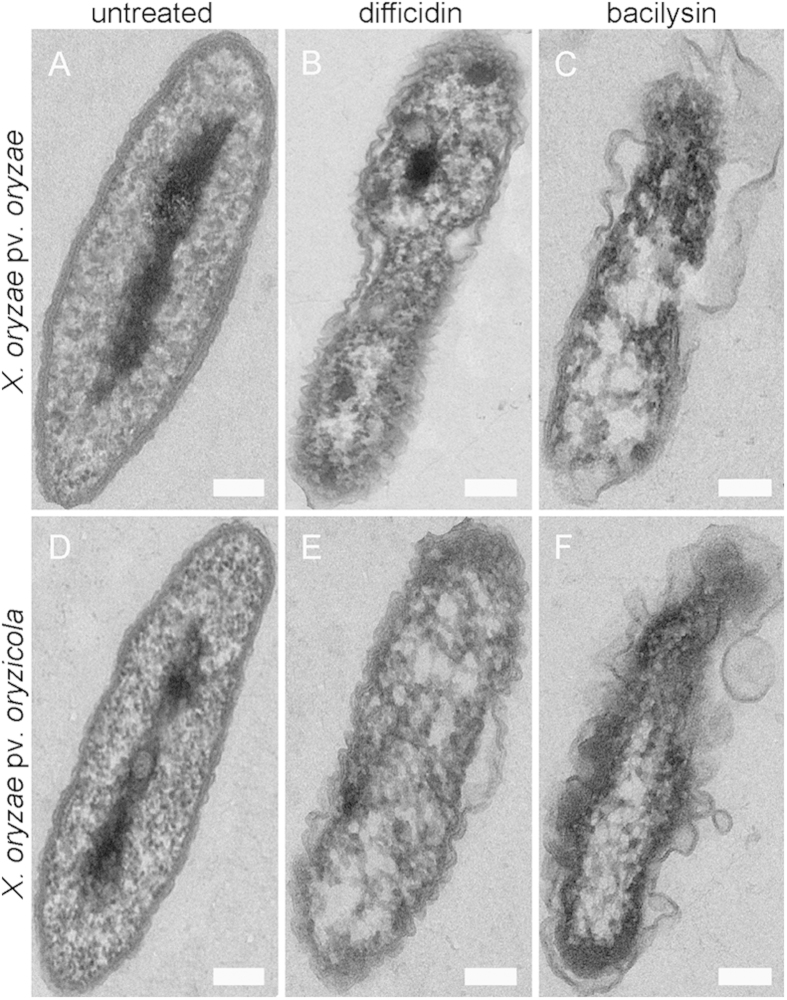
Visualization of the cellular damage caused to X. oryzae pv. oryzae and X. oryzae pv. oryzicola at the ultrastructural level by difficidin and bacilysin was undertaken by SEM and TEM analyses (Figs 2 and 3). In the SEM study, untreated control cells appeared intact, plump and typically rod-shaped with a smooth exterior (Fig. 2A,D). Upon exposure to difficidin or bacilysin, cell walls became loose and porous, distorted from their normal shape or even ruptured (Fig. 2B to C and Fig. 2E to F). By TEM, untreated cells showed a very distinct cell wall and a uniformly distributed electro-dense cytoplasm (Fig. 3A,D). After 12 h of treatment with difficidin or bacilysin, the lysis of the bacterial cell or a partial vesiculation of the membrane were clearly visible and resulted in plasmolysis and efflux of intracellular components. There were no evident electro-dense and basic structures (Fig. 3B to C and Fig. 3E to F). The damage due to bacilysin was more severe than that caused by difficidin.
Figure 2. Morphological changes of Xanthomonas cells after exposure to 50 μg/ml difficidin or bacilysin for 12 h determined by SEM.
(A) untreated X. oryzae pv. oryzae; (B) X. oryzae pv. oryzae treated with difficidin; (C) X. oryzae pv. oryzae treated with bacilysin; (D) untreated X oryzae pv. oryzicola; (E) X. oryzae pv. oryzicola treated with difficidin; (F) X. oryzae pv. oryzicola treated with bacilysin. Bars: 1 μm.
Figure 3. Ultrastructural effects of 50 μg/ml difficidin or bacilysin on Xanthomonas cells after 12 h determined by TEM.
(A) an untreated X. oryzae pv. oryzae cell; (B) X. oryzae pv. oryzae treated with difficidin; (C) X. oryzae pv. oryzae treated with bacilysin; (D) an untreated X. oryzae pv. oryzicola cell; (E) X. oryzae pv. oryzicola treated with difficidin; (F) X. oryzae pv. oryzicola treated with bacilysin. Bars: 0.2 μm.
Biological control of rice diseases caused by Xanthomonas spp
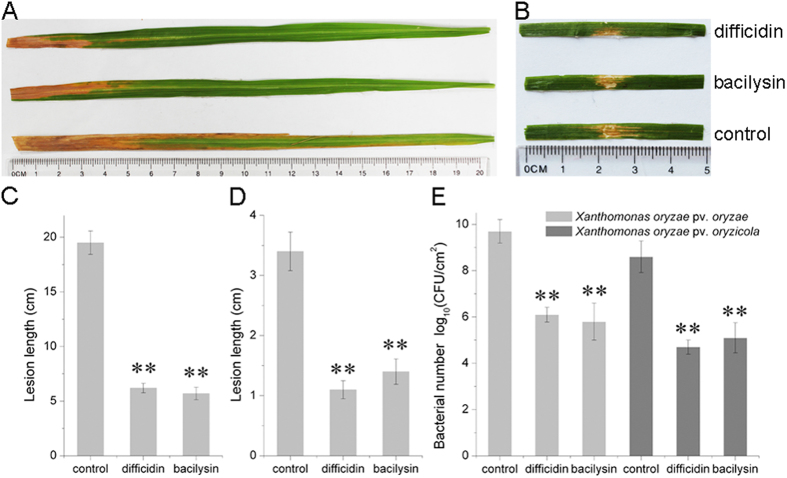
To investigate the role of difficidin and bacilysin in resistance to bacterial leaf blight and bacterial leaf streak of rice, pathogenicity assays were performed. Figure 4 shows that rice plants (cultivar 9311) treated with difficidin and bacilysin exhibited a significant reduction in Xanthomonas virulence relative to controls. The lengths of lesions and the disease severities caused by the pathogens decreased remarkably. The protective rates for difficidin and bacilysin were 58.82%–72.31%. Simultaneously, the biocontrol efficacy of mutants against rice diseases caused by Xanthomonas spp. was also investigated. As previously observed in the in vitro assays, the mutants RS2 and RS6 impaired in the production of difficidin and bacilysin completely lost the ability to control bacterial leaf blight and bacterial leaf streak of rice, and leaves developed symptoms similar to those observed in untreated leaves (Fig. S5). The biocontrol abilities of the difficidin mutants CH3 and CH8 were slightly decreased compared with the wild-type strain (Fig. S5).
Figure 4. Pathogenicity test of Xanthomonas oryzae pv. oryzae and Xanthomonas oryzae pv. oryzicola strains on rice.
(A) Representative result of lesion length symptom tests on the leaves of adult susceptible rice (cultivar 9311, two-month old) after treatment with 50 μg/ml difficidin and bacilysin, respectively. (B) Representative result of water-soaking lesion length tests on rice seedling leaves (cultivar 9311, two-week old) after infiltration with 50 μg/ml difficidin and bacilysin, respectively. (C) Calculated lesion lengths on the leaves of susceptible adult rice. (D) Calculated water-soaking lesion lengths on the leaves of rice seedlings. (E) The number of Xanthomonas cells in adult-susceptible rice leaves and rice seedling leaves after difficidin and bacilysin treatments. Data are expressed as means ± standard deviation (SD); **indicates an extremely significant difference compared with controls (P < 0.01).
The population densities of X. oryzae pv. oryzae and X. oryzae pv. oryzicola on rice leaves were evaluated to associate the observed biocontrol activity with the antagonistic effect of difficidin and bacilysin on the number of phytopathogenic bacteria. A significant decrease of population levels, more than three orders of magnitude, was observed in comparison with controls when difficidin and bacilysin were applied before inoculation of rice with Xanthomonas spp. (Fig. 4E).
Determination of Xanthomonas gene expression after exposure to difficidin and bacilysin
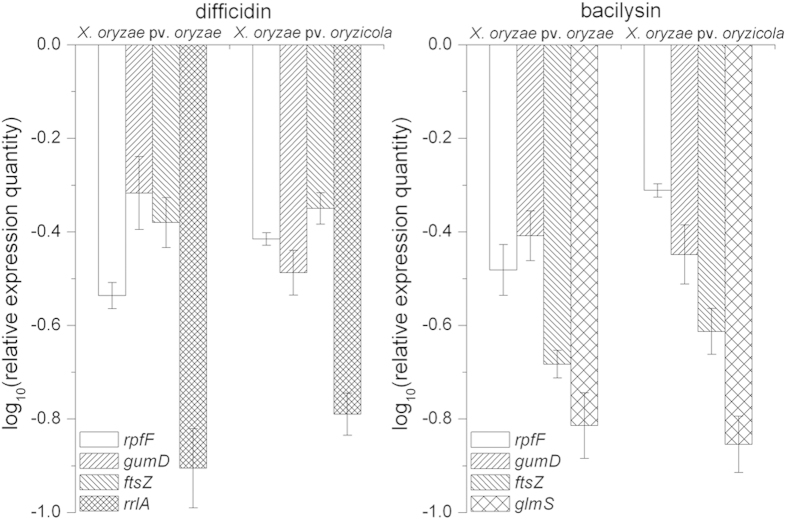
Five genes (rpfF, gumD, ftsZ, rrlA and glmS) were chosen to explore the effects of difficidin and bacilysin on Xanthomonas gene expression. The rpfF gene is involved in production of a diffusible signal factor (DSF)1 and the gumD gene is a gene in the gum operon responsible for extracellular polysaccharide (EPS) biosynthesis2, which are both required for virulence. The ftsZ gene product is involved in cell division4. rrlA, a 23S rRNA gene, has been reported to be a binding site for macrolide antibiotics14,15. The glmS gene encodes glucosamine-6-phosphate synthase, which is important for the biosynthesis of peptidoglycan, a component of the bacterial cell wall16. Figure 5 shows that the transcriptional expression of rpfF, gumD and ftsZ were slightly downregulated, while the levels of rrlA and glmS decreased significantly, on treatment of X. oryzae pv. oryzae and X. oryzae pv. oryzicola with difficidin or bacilysin.
Figure 5. Quantitative real-time PCR analysis of expression of five genes (rpfF, gumD, ftsZ, glmS, rrlA) in Xanthomonas cells in response to difficidin and bacilysin treatment.
Values were normalized to the levels of 16S rRNA, an internal reference gene. The y-axis represents mean expression values ± SD relative to the control. The experiment was independently repeated five times.
Discussion
Numerous studies have demonstrated that biological control is an interesting and efficient strategy that might be applied in the management of plant diseases caused by the genus Xanthomonas. Zeriouh et al. (2011)3 reported that the iturin-like lipopeptides are essential components in the biological control arsenal of B. subtilis against cucurbit pathogenic bacteria X. campestris pv. cucurbitae. Silva et al. (2013)4 showed that alkyl gallates display a potent antibacterial activity against X. citri subsp. citri, the causal agent of Asiatic citrus canker. Wang et al. (2012)17 indicated that chitosan markedly inhibits the growth of pathogenic Xanthomonas isolated from Euphorbia pulcherrima. In the present study, we showed that difficidin and bacilysin from B. amyloliquefaciens FZB42 have antibacterial activities against X. oryzae pv. oryzae and X. oryzae pv. oryzicola.
Difficidin was first detected in fermentation broth of B. subtilis ATCC 39320 and characterized as a highly unsaturated 22-membered macrocylic polyene lactone phosphate ester18. The dipeptide bacilysin, consisting of a non-proteinogenic L-anticapsin and an N-terminal L-alanine, was first isolated by Foster and Woodruff from the soil bacterium B. subtilis19. Genome analysis of B. amyloliquefaciens FZB42 revealed gene clusters encoding the biosynthesis genes for difficidin and bacilysin, and these two substances were found in culture broths by HPLC9,10. The antibacterial activities of difficidin and bacilysin from Bacillus spp. have generally been described against medically important bacteria19,20; only a few reports have addressed their effects on plant pathogenic bacteria21. The results presented in this work show conclusively the crucial role of difficidin and bacilysin in the antagonistic effect against two important rice pathogens and their protective capability against rice diseases.
Study of a large number of macrolides revealed they could inhibit protein synthesis by binding to the large ribosomal subunit22,23. Canu et al. (2002)14 demonstrated that domains V and II of 23S rRNA (rrl gene) and proteins L22 and L4 are binding sites for macrolides. Difficidin, as one kind of macrolide, has been reported to rapidly inhibit protein synthesis and possibly also damage cell membranes24. Antimicrobial activity of bacilysin depends on the anticapsin moiety, which is released by peptidases. Intracellular anticapsin then blocks glucosamine synthetase, and, hence, bacterial peptidoglycan or fungal mannoprotein biosynthesis, resulting in protoplasting and lysis12,19,25. In this report, we found difficidin and bacilysin affected the cell wall, as evidenced by fluorescence and ultramicroscopic observations. The rrlA gene, the potential binding site of difficidin, and the glmS gene, the target for bacilysin, were downregulated significantly, as illustrated by qRT-PCR. This result was further confirmed by the reduction of rrlA expression detected using a transcriptional lacZ reporter, and of the glucosamine synthase activity encoded by glmS (Fig. S6). Moreover, the transcript levels of virulence genes rpfF and gumD decreased, which coincided with a decline in disease severities. A similar phenomenon was observed previously, in that bacilysin caused apparent changes in the algal cell wall and cell organelle membranes12.
In summary, our results support the view that difficidin and bacilysin are the main X. oryzae pv. oryzae and X. oryzae pv. oryzicola suppressing compounds in the culture filtrate of B. amyloliquefaciens FZB42. These strains are the causative agents of the important rice diseases bacterial blight and bacterial leaf streak, respectively. Since difficidin and bacilysin have not been previously used in agricultural management, this finding provides a potential option to use them or their producer strain FZB42 as an alternative to chemical bactericides to control rice diseases.
Methods
Bacterial strains and growth conditions
The bacterial strains used in this study are described in Table S1. Bacillus spp. were cultivated routinely on Luria broth (LB) medium solidified with 1.5% agar and fermented in Landy medium21,26. Nutrient agar (NA) medium3,27 was used to culture X. oryzae pv. oryzae and X. oryzae pv. oryzicola. When required, antibiotics were added to the following final concentrations: ampicillin 100 μg/ml, chloramphenicol 5 μg/ml, rifampicin 100 μg/ml and erythromycin 10 μg/ml.
In vitro evaluation of antibacterial activity
The antibacterial activity of B. amyloliquefaciens cell-free culture filtrates was roughly analyzed as previously described25. The B. amyloliquefaciens strains were grown on Landy medium at 30°C with agitation for 38 h. After centrifugation at 12,000 × g for 10 min, the supernatants were filtered through 0.22 μm Millipore membranes. Five microliters of culture filtrate obtained after centrifugation and filtration were applied to a paper disk (5 mm diameter) placed on NA agar inoculated with X. oryzae pv. oryzae or X. oryzae pv. oryzicola. Landy medium was used instead of culture supernatant as the control. The plates were incubated at 28 °C for 48 h and the inhibition zones (mm) included the paper disk diameter.
Purification of difficidin and bacilysin
To purify difficidin, culture filtrates from B. amyloliquefaciens FZB42 grown in Landy medium were absorbed onto an amberlite XAD16 column which was washed with distilled water and eluted with 100% methanol. The eluate was lyophilized and dissolved in methanol containing 10% distilled water. High-performance liquid chromatography-electrospray ionization (HPLC-ESI) of difficidin was performed essentially as previously described10,21. The retention time of difficidin was 8.574 min as detected by absorbance at 280 nm and the expected molecular mass of 544 Da. Eluate at the corresponding retention time was collected and lyophilized to obtain pure difficidin (Fig. S1). Pure bacilysin was produced as in Wu et al. (2014)12.
LIVE/DEAD BacLight bacterial viability staining
The viability assay was performed using the LIVE/DEAD BacLight bacterial viability staining kit L7012 (Invitrogen, Molecular Probes, USA) as previously described3. The kit consists of two colored fluorescence stains: a green-fluorescent SYTO 9 stain and a red-fluorescent propidium iodide (PI) stain. When used in an appropriate mixture, live bacteria with intact membranes fluoresce green, while bacteria with damaged membranes fluoresce red. Xanthomonas cells treated with difficidin or bacilysin (10 μg/ml, 50 μg/ml) for 12 h were centrifuged at 1000 × g for 10 min, and resuspended in 10 mM sodium phosphate buffer (pH 7.4) at a concentration of 107–108 cells/ml. Then, 10 μl of the molecular probes, prepared as recommended by the manufacturer, were added, and the cell suspensions were incubated for 15 min at 25 °C in the dark. The samples were analyzed by an Olympus BX43 microscope using cellSens Standard Software (Tokyo, Japan).
SEM and TEM studies
Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) analysis were used to determine the effects of difficidin and bacilysin on Xanthomonas cells at the ultrastructural level. X. oryzae pv. oryzae and X. oryzae pv. oryzicola treated with 50 μg/ml difficidin or bacilysin were centrifuged and prefixed with 2.5% glutaraldehyde. Fixed cells were rinsed three times for 10 minutes with 100 mM phosphate buffer, postfixed for 3 h in 1% osmium tetroxide, and dehydrated through an ethanol gradient. For SEM analysis, samples were coated with gold and analyzed on a Hitachi S-3000N scanning electron microscope (Hitachi, Japan). For TEM analysis, samples were embedded in Epon 812, sectioned with an ultramicrotome and examined under a Hitachi H-600 transmission electron microscope.
Pathogenicity test in rice plants
Pathogenicity assays were conducted in a glasshouse at 25−28 °C as previously described2,13. In brief, X. oryzae pv. oryzae and X. oryzae pv. oryzicola strains were cultivated in NA broth at 28 °C with appropriate antibiotics. Two days before inoculation with the bacterial pathogen, rice (cultivar 9311) leaves were sprayed with 50 μg/ml difficidin, bacilysin, or water as the control. For observation of lesion length due to X. oryzae pv. oryzae, two-month-old rice plants were inoculated with a suspension of 108 CFU (colony forming unit)/ml of strain PXO99A by the leaf-clipping method. For observation of water-soaking due to X. oryzae pv. oryzicola, a suspension (108 CFU/ml) of strain RS105 was infiltrated into the leaves of two-week old rice seedlings by needleless syringe. The disease symptoms were recorded after 15 days of incubation and the protective rate was calculated by using the following equation: protective rate (%) = (1 − T/C) × 100, where T (treatment) and C (control) are lesion lengths with and without treatment, respectively. Population levels of X. oryzae pv. oryzae and X. oryzae pv. oryzicola in leaf tissue were estimated by serial dilutions and colony counts on plates of selective medium after 2 days of incubation at 28 °C.
Quantitative real time-PCR analysis
For the determination of gene expression, Xanthomonas spp. were exposed to 50 μg/ml difficidin or bacilysin for 2 h, respectively. Total RNA was extracted using a Bacterial RNA Kit (Omega Bio-Tek, USA) according to the manufacturer’s instructions. First-strand cDNA was synthesized using Reverse Transcriptase (TaKaRa Bio Inc, Dalian, China) with random hexamer primers and the resulting cDNA was used as the template for subsequent PCR amplification. qRT-PCR was performed with SYBR Premix Ex Taq (TaKaRa Bio Inc, Dalian, China) using a 7500 Fast Real-Time PCR Detection System. Gene 16S rRNA was used as the internal reference for normalization. Primers for these genes are listed in Table S2.
Statistical analysis
Each experiment was independently repeated at least three times. Data were analyzed using analysis of variance, followed by a Fisher least significant difference test; the statistics software SPSS v16.0 (SPSS Inc., Chicago, USA) was employed26,27.
Additional Information
How to cite this article: Wu, L. et al. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci. Rep. 5, 12975; doi: 10.1038/srep12975 (2015).
Supplementary Material
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (31100056 and 31471811), Agro-Scientific Research in the Public Interest (20130315), the Special Fund for the Fundamental Research Funds for the Central Universities (KYZ201404), the Doctoral Fund of the Ministry of Education of China (20100097120011), and the National High-Tech R&D Program of China (2012AA101504).
Footnotes
The authors declare no competing financial interests.
Author Contributions L.M.W., H.J.W. and X.W.G. conceived and designed the experiments. L.M.W. performed most of the experiments. L.N.C. and X.F.Y. performed the quantitative real time-PCR and pathogenicity test in rice plants, respectively. R.B. supplied B. amyloliquefaciens strains. L.M.W. analyzed the experimental data and wrote the manuscript.
References
- Chatterjee S. & Sonti R. V. rpfF mutants of Xanthomonas oryzae pv. oryzae are deficient for virulence and growth under low iron conditions. Mol. Plant Microbe. Interact. 15, 463–471 (2002). [DOI] [PubMed] [Google Scholar]
- Qian G. et al. Proteomic analysis reveals novel extracellular virulence-associated proteins and functions regulated by the diffusible signal factor (DSF) in Xanthomonas oryzae pv. oryzicola. J. Proteome Res. 12, 3327–3341 (2013). [DOI] [PubMed] [Google Scholar]
- Zeriouh H. et al. The iturin-like lipopeptides are essential components in the biological control arsenal of Bacillus subtilis against bacterial diseases of cucurbits. Mol Plant Microbe Interact. 24, 1540–1552 (2011). [DOI] [PubMed] [Google Scholar]
- Silva I. C. et al. Antibacterial activity of alkyl gallates against Xanthomonas citri subsp. citri. J Bacteriol. 195, 85–94 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira A. G. et al. Evaluation of the antibiotic activity of extracellular compounds produced by the Pseudomonas strain against the Xanthomonas citri pv. citri 306 strain. Bio Control. 56, 125–131 (2011). [Google Scholar]
- Chowdhury S. P. et al. Effects of Bacillus amyloliquefaciens FZB42 on lettuce growth and Health under Pathogen Pressure and Its Impact on the Rhizosphere Bacterial Community. PLoS One. 8, e68818, 10.1371/journal.pone.0068818 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B. et al. Efficient colonization of plant roots by the plant growth promoting bacterium Bacillus amyloliquefaciens FZB42, engineered to express green fluorescent protein. J Biotechnol. 151, 303–311 (2011). [DOI] [PubMed] [Google Scholar]
- Koumoutsi A. et al. Structural and Functional Characterization of Gene Clusters Directing Nonribosomal Synthesis of Bioactive Cyclic Lipopeptides in Bacillus amyloliquefaciens Strain FZB42. J Bacteriol. 186, 1084–1096 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. H. et al. Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J Biotechnol. 140, 27–37 (2009). [DOI] [PubMed] [Google Scholar]
- Chen X. H. et al. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J Bacteriol. 188, 4024–4036 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. et al. The highly modified microcin peptide plantazolicin is associated with nematicidal activity of Bacillus amyloliquefaciens FZB42. Appl Microbiol Biotechnol. 97, 10081–10090 (2013). [DOI] [PubMed] [Google Scholar]
- Wu L. et al. Bacilysin from Bacillus amyloliquefaciens FZB42 has specific bactericidal activity against harmful algal bloom specie. Appl Environ Microbiol. 80, 7512–7520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. Comparative proteomic analysis of rice seedlings in response to inoculation with Bacillus cereus. Lett Appl Microbiol. 56, 208–215 (2013). [DOI] [PubMed] [Google Scholar]
- Canu A. et al. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 46, 125–131 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory S. T., Cate J. H. & Dahlberg A. E. Spontaneous erythromycin resistance mutation in a 23S rRNA gene, rrlA, of the extreme thermophile Thermus thermophilus IB-21. J Bacteriol. 183, 4382–4385 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciechowski M., Milewski S., Mazerski J. & Borowski E. Glucosamine-6-phosphate synthase, a novel target for antifungal agents. Molecular modelling studies in drug design. Acta Biochim. Pol. 52, 647–653 (2005). [PubMed] [Google Scholar]
- Wang Y. et al. Action of chitosan against Xanthomonas pathogenic bacteria isolated from Euphorbia pulcherrima. Molecules 17, 7028–7041 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K. E. et al. Difficidin and oxydifficidin: novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis. II. Isolation and physico-chemical characterization. J Antibiot. 40, 1682–1691 (1987). [DOI] [PubMed] [Google Scholar]
- Kenig M. & Abraham E. P. Antimicrobial activities and antagonists of bacilysin and anticapsin. J Gen Microbiol. 94, 37–45 (1976). [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B. et al. Difficidin and oxydifficidin: novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis. I. Production, taxonomy and antibacterial activity. J Antibiot. 40, 1677–1681 (1987). [DOI] [PubMed] [Google Scholar]
- Chen X. H. et al. Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J Biotechnol. 140, 38–44 (2009). [DOI] [PubMed] [Google Scholar]
- Wilson D. N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 12, 35–48 (2014). [DOI] [PubMed] [Google Scholar]
- Krokidis M. G. et al. Insights into the mode of action of novel fluoroketolides, potent inhibitors of bacterial protein synthesis. Antimicrob Agents Chemother. 58, 472–480 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweerink M. M. & Edison A. Difficidin and oxydifficidin: novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis. III. Mode of action of difficidin. J Antibiot. 40, 1692–1697 (1987). [DOI] [PubMed] [Google Scholar]
- Wu L. et al. Bacilysin overproduction in Bacillus amyloliquefaciens FZB42 markerless derivative strains FZBREP and FZBSPA enhances antibacterial activity. Appl Microbiol Biotechnol. 99, 4255–4263 (2015). [DOI] [PubMed] [Google Scholar]
- Chen X. H., Koumoutsi A., Scholz R. & Borriss R. More than anticipated - production of antibiotics and other secondary metabolites by Bacillus amyloliquefaciens FZB42. J Mol Microbiol Biotechnol. 16, 14–24 (2009). [DOI] [PubMed] [Google Scholar]
- Salzberg S. L. et al. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC genomics 9, 204, 10.1186/1471-2164-9-204 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.