Abstract
Flower development is one of the major developmental processes that governs seed setting in angiosperms. However, little is known about the molecular mechanisms underlying flower development in legumes. Employing RNA-seq for various stages of flower development and few vegetative tissues in chickpea, we identified differentially expressed genes in flower tissues/stages in comparison to vegetative tissues, which are related to various biological processes and molecular functions during flower development. Here, we provide details of experimental methods, RNA-seq data (available at Gene Expression Omnibus database under GSE42679) and analysis pipeline published by Singh and colleagues in the Plant Biotechnology Journal (Singh et al., 2013), along with additional analysis for discovery of genes involved in shoot apical meristem (SAM) development. Our data provide a resource for exploring the complex molecular mechanisms underlying SAM and flower development and identification of gene targets for functional and applied genomics in legumes.
Keywords: Shoot apical meristem, Flower development, Chickpea, RNA-seq, Differential expression
| Specifications | |
|---|---|
| Organism/cell line/tissue | Cicer arietinum L. genotype ICC 4958 |
| Sequencer or array type | Illumina Genome Analyzer IIx |
| Data format | Raw data: FASTQ files, analyzed data: txt files |
| Experimental factors | Tissues/organs |
| Experimental features | RNA-seq dataset for gene expression profiling in shoot apical meristem and flower development in chickpea |
| Sample source location | New Delhi, India |
Direct link to deposited data
Deposited data can be found here: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42679.
Materials and methods
Sample collection and RNA isolation
Chickpea (Cicer arietinum L. genotype ICC 4958) seeds were grown in culture room and field for collection of vegetative and reproductive tissues as described [1]. Three vegetative tissues [germinating seedling (GS), young leaves (YL) and shoot apical meristem (SAM)], and four stages each of flower bud (FB1–FB4) and flower (FL1–FL4) were harvested in at least three biological replicates (Fig. 1). Harvested tissues were snap frozen into liquid nitrogen and stored at − 80 °C till RNA isolation. Total RNA was extracted from each tissue using TRI reagent (Sigma Life Science) according to manufacturer's instructions after grinding tissues in pre-chilled (in liquid nitrogen) mortar and pestle. Total RNA concentration and purity were determined using Nanodrop 1000 Spectrophotometer (Thermo Fisher Scientific) and microfludics-based RNA nano chip on 2100 Bioanalyzer (Agilent Technologies).
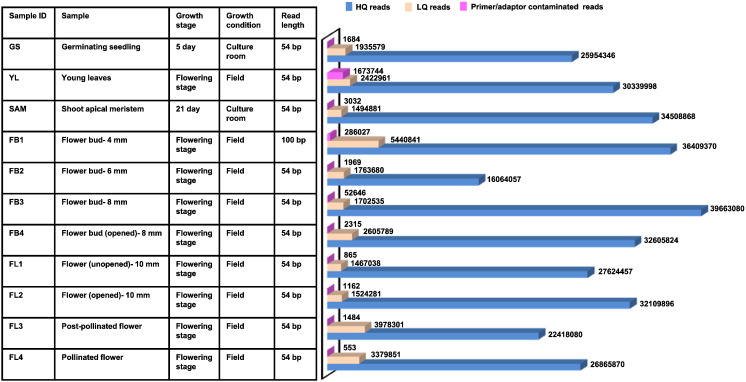
Fig. 1.
Details of samples used for RNA-seq and summary of data after filtering low-quality (LQ) and primer/adaptor contaminated reads using NGS QC ToolKit. These data are taken from Singh et al. [5].
Generation of RNA-seq data
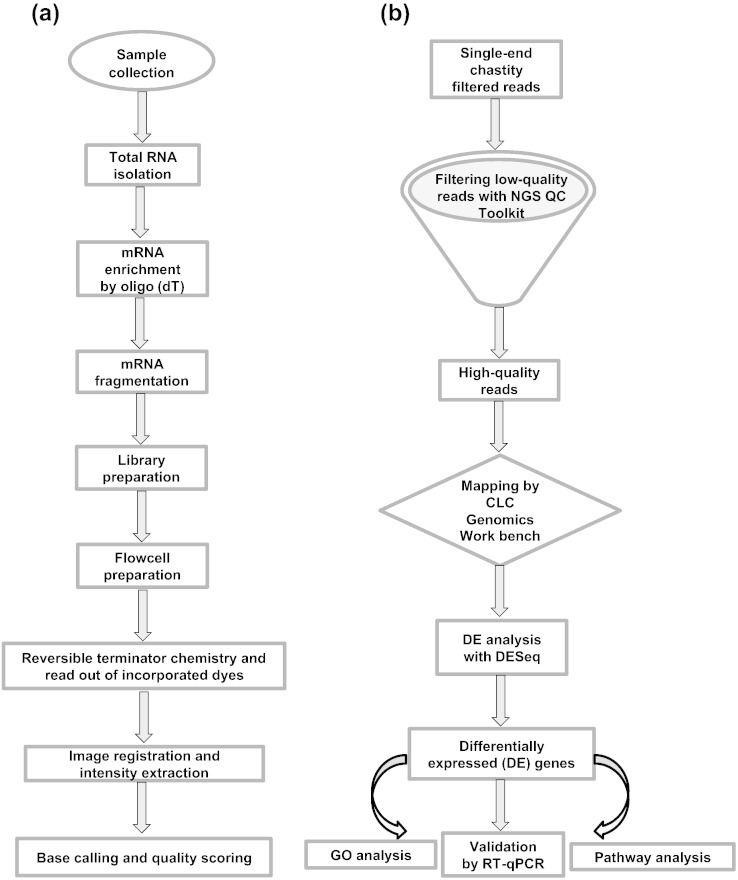
To understand the global expression profiles during flower development in chickpea, we performed RNA-seq analysis. Library for each sample was constructed using mRNA-Seq Sample Prep kit and single-end sequencing was performed following standard protocol of Illumina so as to generate 54 nucleotide long reads using Genome Analyzer IIx (Illumina Inc., San Diego, CA). High quality chastity-filtered reads were obtained in FASTQ file format after filtering the raw data through standard Illumina pipeline. Whole RNA-seq data were submitted to the Gene Expression Omnibus (series accession number GSE42679). An outline of strategy adopted for sample preparation and sequencing is provided in Fig. 2a.
Fig. 2.
Workflow for RNA-seq experiments conducted in the study. (a) General strategy for sample preparation and sequencing. Total RNA was isolated from collected chickpea samples, cDNA libraries were prepared and single-end sequencing was performed followed by image analysis to obtain sequence data. (b) Differential gene expression analysis pipeline. Low-quality (LQ) reads were removed using NGS QC ToolKit (v2.2.3) and high-quality reads were mapped on chickpea transcriptome using CLC Genomics Workbench. Differentially expressed genes in different tissues were identified using DESeq, followed by various analyses thereof.
Data processing and differential expression quantification
The strategy used for data processing and differential gene expression analysis is represented in Fig. 2b. The single-end chastity-filtered reads obtained in FASTQ format were further subjected to stringent sequence quality controls, using NGS QC Toolkit (v2.2.3) at default parameters [2]. This resulted into removal of 4–15% of low-quality sequences (Fig. 1). The high-quality reads were then aligned to the chickpea transcriptome (v1.0) available at Chickpea Transcriptome Database (v2.0; http://www.nipgr.res.in/ctdb.html) [3] with a tolerance of up to two mismatches using CLC Genomics Workbench (v4.7.2). The first round of alignment resulted into mapping of 66–88% of reads for individual samples. For the sequences that did not map to transcriptome, a second round of alignment was performed, in which we trimmed 14 bases from the 3′-end of unmapped reads after optimization of trimming one base recursively. This procedure enabled us to recover reads showing some misalignments caused by spurious or low-quality bases present at the read ends. After this step, total mapping percentage of reads increased to > 90% for each sample. Read count obtained after mapping was normalized to ‘reads per kilobase of exon model per million mapped reads (RPKM) for each sample. The genes having ≥ 1 RPKM at least in one sample were considered to be expressed. In total, 33,584 genes were found expressed in chickpea tissues analyzed, which represented 96.6% of the chickpea transcriptome.
Differential gene expression analysis was performed between tissues considering total reads mapped to chickpea transcripts using Bioconductor package DESeq (v1.5.24) [4]. DESeq uses a negative binomial distribution to model total read counts for biological and technical variation by a generalized Poisson distribution model. For differential gene expression analysis, data were normalized across the samples with size factors and variance. To estimate dispersion between samples without replicates, method = “blind” together with sharingMode = “fit-only” was used. Genes with P-value ≤ 0.05 and fold change ≥ 2 were regarded as differentially expressed. This represented a 100% linear fold change i.e. log22 = 1 or 100%. The expression of 1572 genes was significantly different in floral tissues as compared to vegetative tissues [5]. Among these, 1304 genes were up-regulated and 269 genes were down-regulated in floral tissues. These data have been integrated into the Chickpea Transcriptome Database (v2.0; http://www.nipgr.res.in/ctdb.html).
Differentially expressed transcripts in shoot apical meristem
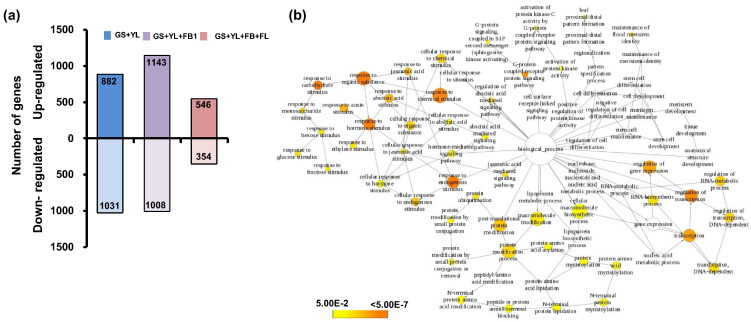
Using similar strategy mentioned above, gene expression in SAM was compared with three combinations of other tissues to identify differentially expressed transcripts. In the first combination, differentially expressed genes in SAM were identified as compared to vegetative tissues (GS + YL) only, leading to identification of 882 up- and 1031 down-regulated genes (Fig. 3a). In the second combination, SAM was compared with vegetative tissues and first stage of flower bud development (FB1), which resulted into 1143 up- and 1008 down-regulated genes in SAM (Fig. 3a). Further, in the third combination, SAM was compared with all the tissues (GS + YL + FB1–4 + FL1–4) and 546 exclusively up- and 354 exclusively down-regulated genes in SAM were identified (Fig. 3a).
Fig. 3.
Differential gene expression analysis in shoot apical meristem (SAM) (a) The number of up- (upper side) and down-regulated (lower side) genes in SAM as compared to vegetative tissues (GS + YL), vegetative (GS + YL) and flower bud (FB1), and all other tissues (GS + YL + FB + FL) are shown. (b) Gene ontology enrichment analysis of genes up-regulated in SAM as compared to vegetative (GS + YL) and flower bud (FB1). Analysis was performed using BiNGO and the biological process terms showing significant enrichment are shown. Node size is proportional to the number of transcripts in each category and colors according to the significance level (white—no significant difference; color scale, yellow—P-value = 0.05, orange—P-value < 0.0000005).
We performed gene ontology (GO) analysis using the BiNGO (v3.0.2) tool with P-value cut-off of ≤ 0.05 after applying Benjamini Hochberg correction to reveal functional categories enriched in the up-regulated genes in SAM. We found genes involved in various processes of SAM development, including G-protein coupled receptor protein signaling pathway, RNA biosynthetic process, regulation of RNA metabolic process, cell differentiation, stem cell differentiation and maintenance, leaf proximal distal/pattern formation, abscisic acid mediated signaling pathways, anatomical structure development, cell differentiation, responses to organic substances, responses to hormone stimulus, response to auxin stimulus and protein ubiquitination etc. enriched in SAM (Fig. 3b). The biological process GO terms found enriched in our study are in very good agreement with previous reports. For example, G-protein coupled receptor protein regulates cell proliferation, physiological responses of abscisic acid and gibberellins during SAM development [6], [7], [8], [9], [10]. Auxin signaling has been shown to promote leaf differentiation in SAM [11]. Furthermore, processes like abscisic acid mediated signaling pathways, anatomical structure development, responses to organic substances and responses to hormone stimulus have found in up-regulated genes in shoot apex of Arabidopsis by movement of WUSCHEL protein [12], [13]. In addition, we found many differentially expressed genes involved in regulation of processes responsible for floral initiation, such as abscisic acid mediated signaling pathway, activation of protein kinases, regulation of transcription, nucleobase, nucleoside and nucleic acid metabolic processes and maintenance of floral meristem identity (Fig. 3b) [14]. These genes could be used to modulate flowering time in chickpea and may prove to be of agronomic importance.
Discussion
We describe here a RNA-seq dataset, which catalogs the transcriptome of SAM and flower development in chickpea. This dataset revealed differentially expressed genes during SAM and flower development, involved in regulation of their various processes [5; this study]. This dataset is of high quality and can help improve our understanding of molecular mechanisms regulating SAM and flower development in legumes and can be mined and integrated with results of other experiments to explore regulatory networks, specific pathways or genes active during development of SAM and flower in chickpea.
Conflict of interest
Authors declare no conflict of interest.
Acknowledgments
This work was funded by the Department of Biotechnology, Government of India, under the Next Generation Challenge Programme on Chickpea Genomics (grant number BT/PR12919/AGR/02/676/2009 from 2009 to 2014). VKS acknowledges the award of research fellowship from the Department of Biotechnology, Government of India.
References
- 1.Garg R., Sahoo A., Tyagi A.K., Jain M. Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.) Biochem. Biophys. Res. Commun. 2010;396:283–288. doi: 10.1016/j.bbrc.2010.04.079. [DOI] [PubMed] [Google Scholar]
- 2.Patel R.K., Jain M. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garg R., Patel R.K., Jhanwar S., Priya P., Bhattacharjee A., Yadav G., Bhatia S., Chattopadhyay D., Tyagi A.K., Jain M. Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant Physiol. 2011;156:1661–1678. doi: 10.1104/pp.111.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh V.K., Garg R., Jain M. A global view of transcriptome dynamics during flower development in chickpea by deep sequencing. Plant Biotechnol. J. 2013;11:691–701. doi: 10.1111/pbi.12059. [DOI] [PubMed] [Google Scholar]
- 6.Bommert P., Je B., Goldshmidt A., Jackson D. The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature. 2013;502:555–558. doi: 10.1038/nature12583. [DOI] [PubMed] [Google Scholar]
- 7.Chen J.G., Pandey S., Huang J., Alonso J.M., Ecker J.R., Assmann S.M., Jones A.M. GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol. 2004;135:907–915. doi: 10.1104/pp.104.038992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandey S., Chen J.G., Jones A.M., Assmann S.M. G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol. 2006;141:243–256. doi: 10.1104/pp.106.079038. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Ullah H., Chen J.G., Young J.C., Im K.H., Sussman M.R., Jones A.M. Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science. 2001;292:2066–2069. doi: 10.1126/science.1059040. [DOI] [PubMed] [Google Scholar]
- 10.Wong C.E., Singh M.B., Bhalla P.L. The dynamics of soybean leaf and shoot apical meristem transcriptome undergoing floral initiation process. PLoS ONE. 2013;8:e65319. doi: 10.1371/journal.pone.0065319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vernoux T., Besnard F., Traas J. Auxin at the shoot apical meristem. Cold Spring Harb. Perspect. Biol. 2010;2:a001487. doi: 10.1101/cshperspect.a001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yadav R.K., Perales M., Gruel J., Ohno C., Heisler M., Girke T., Jönsson H., Reddy G.V. Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 2013;9:654. doi: 10.1038/msb.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav R.K., Perales M., Gruel J., Girke T., Jönsson H., Reddy G.V. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011;25:2025–2030. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong C.E., Singh M.B., Bhalla P.L. Molecular processes underlying the floral transition in the soybean shoot apical meristem. Plant J. 2009;57:832–845. doi: 10.1111/j.1365-313X.2008.03730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]