Summary
Circadian clocks regulate membrane excitability in master pacemaker neurons to control daily rhythms of sleep and wake. Here we find that two distinctly timed electrical drives collaborate to impose rhythmicity on Drosophila clock neurons. In the morning, a voltage-independent sodium conductance via the NA/NALCN ion channel depolarizes these neurons. This current is driven by the rhythmic expression of NCA localization factor-1, linking the molecular clock to ion channel function. In the evening, basal potassium currents peak to silence clock neurons. Remarkably, daily antiphase cycles of sodium and potassium currents also drive mouse clock neuron rhythms. Thus, we reveal an evolutionarily ancient strategy for the neural mechanisms that govern daily sleep and wake.
Introduction
Circadian clocks have evolved to align organismal biochemistry, physiology and behavior to daily environmental oscillations. At the core of these clocks in all multicellular organisms are conserved transcriptional feedback loops (Allada and Chung, 2010; Hardin, 2011). In Drosophila, the bHLH-PAS transcription factor heterodimer CLOCK (CLK) and CYCLE (CYC) directly binds E-boxes (CACGTG) in target promoters of the clock genes, period (per) and timeless (tim), and activates their transcription. PER and TIM proteins feed back to repress CLK/CYC activity. The temporal separation of transcriptional activation and repression and/or mRNA and protein oscillations, in some cases by many hours (Lee et al., 1998), results in robust daily oscillations of per, tim, and other rhythmic transcripts. These molecular clocks in turn, control a broad range of cellular and physiological responses likely via the rhythmic transcription of clock output genes.
While molecular clocks are expressed in a variety of cell types, those in specific circadian clock neurons in the brain exhibit special properties. These so-called “master” circadian pacemakers, such as the mammalian suprachiasmatic nucleus (SCN) and the Drosophila lateral and dorsal neurons, drive robust 24-hour rhythms of sleep and wake behavior (Helfrich-Forster, 2005; Mohawk and Takahashi, 2011). Unlike generic clock cells, these clock neurons are interconnected via neural networks and as a result, produce coherent and sustained free running molecular and behavioral rhythmicity under constant conditions (Flourakis and Allada, 2015; Guo et al., 2014; Peng et al., 2003; Seluzicki et al., 2014; Shafer et al., 2002; Yang and Sehgal, 2001; Yao and Shafer, 2014). Although the anatomical features of brain pacemaker networks are highly divergent between mammals and invertebrates such as Drosophila, their ability to control sleep and wake cycles uniformly depends on daily rhythms of membrane excitability (Cao and Nitabach, 2008; Colwell, 2011; de Jeu et al., 1998; Kuhlman and McMahon, 2004; Sheeba et al., 2008). However, the mechanistic links between the molecular clock and the machinery controlling cellular excitability are not well understood.
Using patch-clamp analysis of the Drosophila DN1p, we show for the first time that circadian clock control of membrane excitability operates via resting sodium leak conductance through the NARROW ABDOMEN (NA) channel, providing timed depolarizing drive to circadian pacemaker neurons. We demonstrate that the sodium leak rhythm depends on rhythmic expression of NCA localization factor −1, linking the molecular clock and membrane excitability. We reveal that both flies and mice, separated by hundreds of millions of years in evolution, utilize antiphase oscillations of sodium and potassium conductances to drive clock control of membrane potential. Thus the conservation of clock mechanisms between invertebrates and vertebrates extends from core timing mechanisms to the control of membrane excitability in the master clock neurons governing sleep and wake.
Results
Rhythmic resting potassium and sodium leak currents collaborate to drive clock-controlled excitability of the Drosophila circadian neurons
To elucidate the mechanistic basis of daily changes in membrane excitability in Drosophila clock neurons, we performed whole-cell patch-clamp electrophysiology on the posterior dorsal neurons 1 (DN1p) on explanted brains (Flourakis and Allada, 2015; Seluzicki et al., 2014). DN1p neurons harbor molecular circadian clocks, and, under 12 hours light- 12 hours dark (LD) conditions, they contribute to increases in locomotor activity in advance of lights-on (i.e., morning anticipation) and lights-off (i.e., evening anticipation) (Zhang et al., 2010a; Zhang et al., 2010b). In addition to their established function in circadian behavior, the DN1p are an attractive target for patch clamp analysis as we can selectively label and identify DN1p neurons using the Clk4.1M-GAL4 driver in combination with UAS-CD8-GFP (Zhang et al., 2010a; Zhang et al., 2010b) (Fig. 1A). Furthermore, the DN1p neurons are easily accessible by electrode as they are located near the brain surface (Flourakis and Allada, 2015; Seluzicki et al., 2014).
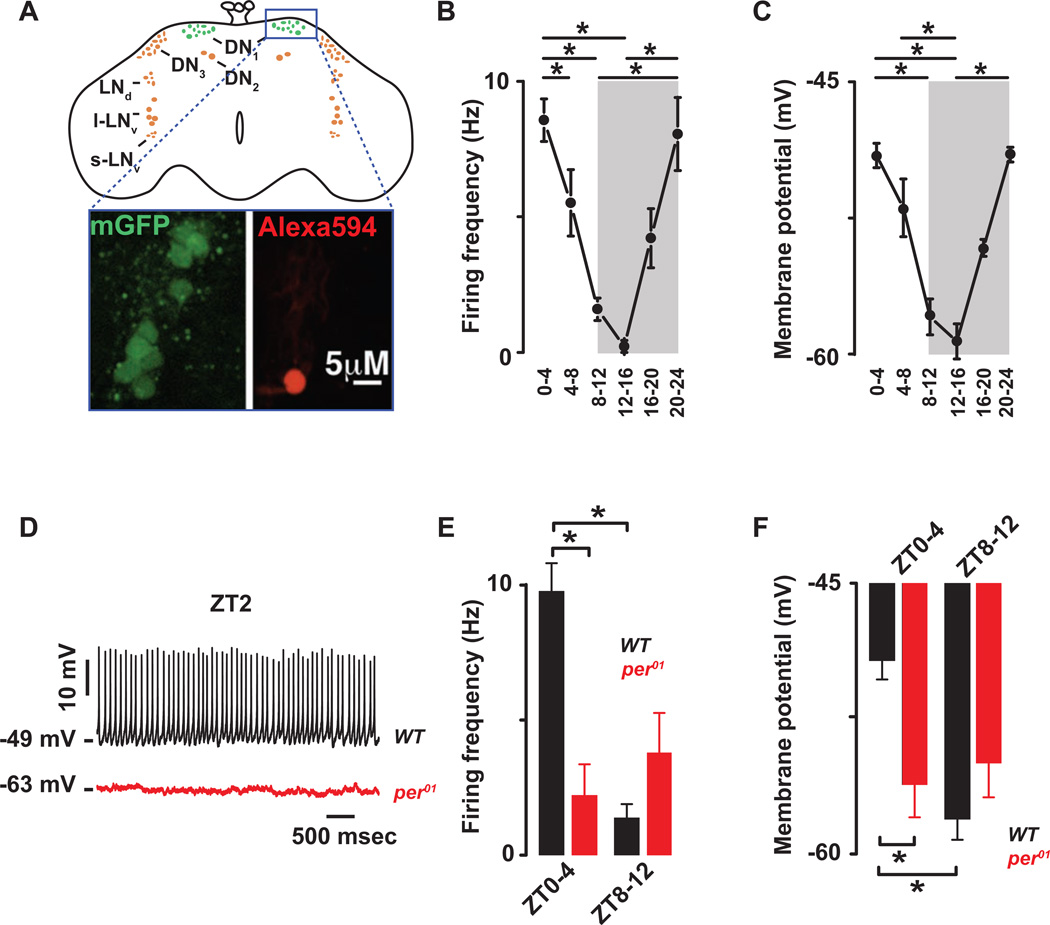
Figure 1. The cellular excitability of the Drosophila DN1p circadian pacemaker neurons is clock controlled.
(A) Schematic and image of the Drosophila brain indicating the location of the DN1ps and other clock neurons. Representative images of the GFP-expressing DN1ps in the intact Drosophila brain are shown below. The DN1ps were labeled by using the Clk4.1M–G4 driving the expression of U-CD8-GFP. Whole-cell access to GFP labeled neurons was confirmed following diffusion of Alexa Fluor 594 biocytin included in intracellular recording solution. All recorded WT neurons are plotted against time of day (in 4 hours bins) to show daily rhythms of firing frequency (B) and membrane potential (C). Grey areas represent the dark phase of the LD cycle. Asterisks indicate statistical significance (p<0.05) from a one-way ANOVA, Tukey’s post-hoc test. (D) Representative current clamp recordings at Zeitgeber Time-2 (ZT2) showing that the per01 DN1ps neurons (red) are hyperpolarized and silent compared to WT DN1p neurons (black). Histogram showing the decrease in firing frequency (E) and membrane potential (F) and lack of daily rhythm in per01 (red, 2.2±1.1Hz, −56±2 mV, n=15 at ZT0-4 and 3.9±1.5Hz, −55±1.9 mV, n=10 at ZT8–12, p>0.41) when compared to WT (black) DN1p neurons. Results are expressed as mean±SEM. Asterisks indicate statistical significance (p<0.05) from t-test performed in WT at ZT0-4 vs ZT8–12. See also Figures. S.1–2 and Tables S.1–2.
Using whole-cell patch clamp analysis, a large daily variation in the firing frequency was detected (Fig. 1B, p<0.05 and Fig. S.1A). The wild type (WT) DN1ps fire at ∼10Hz in the morning (Zeitgeber Time, ZT0-4) and are nearly silent in the evening (ZT8–12) (Table S.1A). The firing frequency in cell-attached configuration was comparable to that observed in whole-cell mode (Fig S.1B–D) suggesting that dialysis did not alter measurements of firing rates. The membrane potential also exhibited a temporal pattern: more depolarized in the morning than in the evening (Fig 1C, p<0.05 and Table S.1B). The neurons show daily rhythmic cellular excitability: more responsive to depolarizing currents in the morning than in the evening (Fig S.1E and Table S.2A). The input resistance had no significant diurnal rhythm (Fig.S.1F, Table S.1C). The rhythms in firing frequency and membrane potential were not evident in the arrhythmic core clock mutant per01, indicating that the canonical clock controls daily changes in intrinsic membrane properties. Compared to WT, the per01 neurons are hyperpolarized (Fig. 1D), and show no rhythm in firing frequency (Fig. 1E, p=0.41), membrane potential (Fig. 1F, p=0.66) or cellular excitability (Fig S.2A, p>0.41). The per01 neurons also require more depolarizing current to fire at the same rates as WT (Fig. S.2B and table S.2B). Importantly, the high amplitude daily rhythm in firing frequency observed in WT neurons exceed those previously described in another set of Drosophila circadian neurons (LNvs) and more closely approximate those described in mammalian SCN clock neurons (Cao and Nitabach, 2008; Colwell, 2011; Kuhlman and McMahon, 2006; Park and Griffith, 2006; Schaap et al., 2003; Sheeba et al., 2008), indicating that DN1p analysis will be useful to define the mechanisms for clock control of membrane excitability. Given the role of the DN1p in morning and evening behaviors (Zhang et al., 2010a; Zhang et al., 2010b), these activity measurements suggest that DN1p activity in the morning can drive locomotor activity while the relative silence of the DN1p in the evening may have a permissive role on other cells controlling evening behavior.
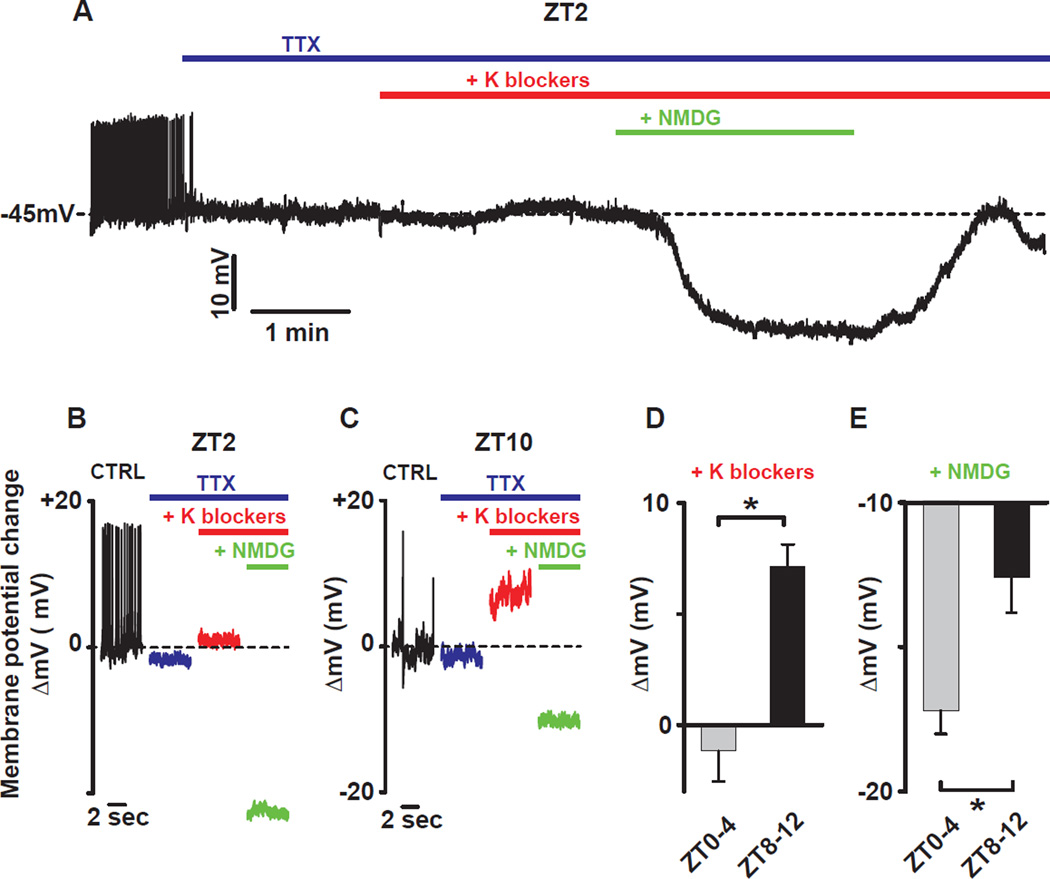
To identify ionic conductances responsible for the resting membrane potential (RMP) rhythm, we blocked action potential firing using the voltage-dependent sodium channel blocker, tetrodotoxin (TTX, 10µM), and then applied a cocktail of potassium (K) channel inhibitors (10mM TEA, 5mM 4AP, and 2mM CsCl) to block both voltage-dependent and voltage-independent (leak) K conductances (Fogle et al., 2011). We subsequently used N-Methyl-D-glucamine (NMDG) substitution of extracellular sodium to block sodium leak currents (Jackson et al., 2004; Lu et al., 2007; Raman et al., 2000) at different times of day. As in mammals (Kuhlman and McMahon, 2004) and mollusks (Michel et al., 1993), the effect of blocking K leak conductances in Drosophila was dependent on time-of-day, producing little change in the morning (Fig. 2A–B-D) but a sizable depolarization in the evening (Fig. 2C–D), indicating that rhythmic resting K conductance is conserved between flies and mammals (Kuhlman and McMahon, 2006). In contrast to K blockade, we discovered that blockade of resting sodium leak produced a larger hyperpolarization in the morning (Fig. 2A–B-E) than in the evening (Fig. 2C–E). Such time-of-day dependent effects of sodium channel blockade have not been previously reported. Notably, this time-of-day dependent effect on membrane potential of sodium blockade (Δ∼7 mV morning vs evening) is roughly equal to that of potassium blockade, suggesting that each makes a comparable contribution to daily excitability rhythms. As these rhythms are observed during network silencing from TTX, this suggests that changes in RMP are not driven by synaptic inputs but are intrinsic to the cells. Taken together, our results demonstrate that time-of-day-dependent sodium and K conductances, in the morning and evening, respectively, may underlie RMP rhythms.
Figure 2. Time of day dependent effects of resting K and sodium leak conductance blockade on membrane potential in DN1p neurons.
(A) Representative current clamp recording at ZT2 showing the effect of K and sodium conductance blockers on membrane potential. Bars indicate when drugs were applied (blue: TTX 10µM, red: TEA 10mM, 4-AP 5mM, CsCl 2mM and green: NMDG to replace the sodium from the extracellular solution). The effect of K blockers and sodium replacement on the membrane potential at different times of day are shown in (B) for ZT2 and (C) for ZT10. (D) Averaged changes of the membrane potential by K blockers (10mM TEA, 5mM 4-AP and 2mM CsCl): −1.2±1.4mV, n=5 between ZT0-4 and 7.1±1mV, n=5 between ZT8–12 and (E) sodium replacement with NMDG: −17.2±0.8mV, n=5 between ZT0-4 and − 12.6±1.2mV, n=5 between ZT8–12. Results are expressed as mean±SEM. Asterisks indicate statistical significance (p<0.05) from t-test.
The ion channel NARROW ABDOMEN controls Drosophila circadian pacemaker rhythms
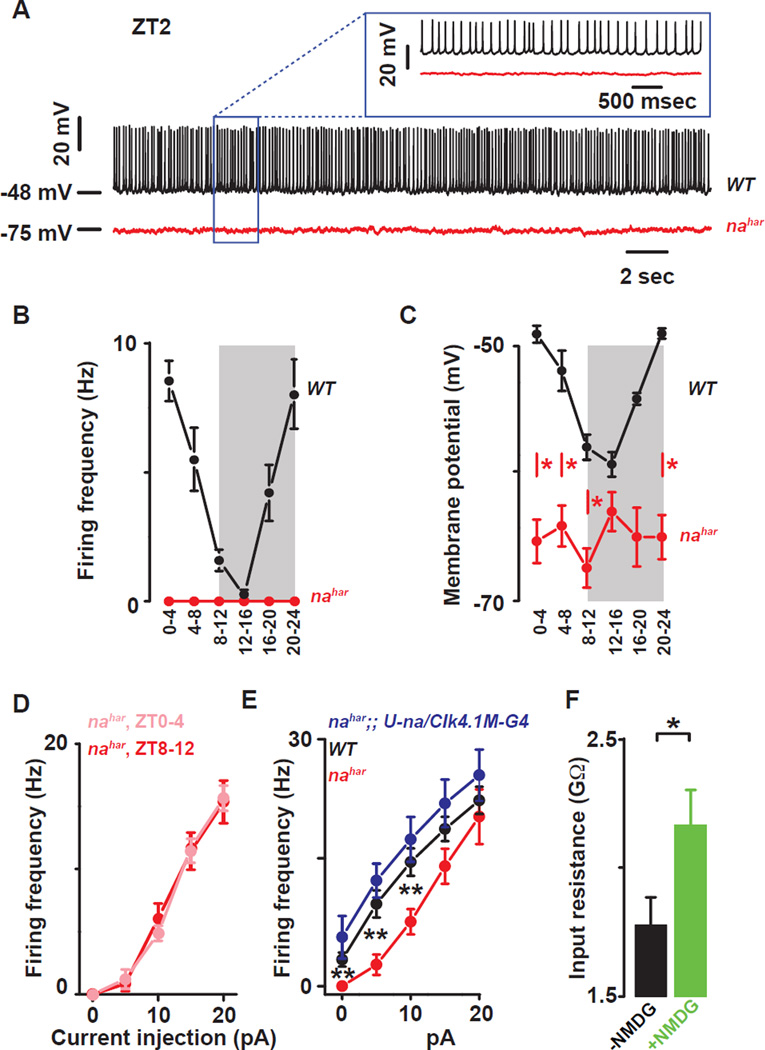
A candidate mediator of resting sodium conductances in clock neurons and circadian behavior is the NARROW ABDOMEN (NA) ion channel (Lear et al., 2005; Nash et al., 2002). NALCN, the closely conserved mammalian homolog of NA, has been characterized as a voltage-independent mixed cation channel important for setting RMP and mediating resting leak sodium current (Lu et al., 2007; Swayne et al., 2009). This current is not blocked by TTX but can be reduced by either Gd3+ or replacement of extracellular sodium with NMDG (Lu et al., 2007). In a 12 hours light: 12 hours dark (LD) cycle, increases in locomotor activity in advance of lights-on (i.e., morning anticipation) and lights-off (i.e., evening anticipation) are suppressed in nahar mutants (Lear et al., 2005; Nash et al., 2002). Although NA expression in the DN1p can rescue morning and, to a lesser extent, evening phenotypes (Zhang et al., 2010a), it remains unclear whether NA is a rhythmic mediator of resting membrane potential of circadian clock neurons. We therefore examined clock neuron excitability in na mutant DN1p neurons. Strikingly, nahar mutant DN1p neurons were completely silent (Fig. 3A–B) and remained hyperpolarized throughout the whole day (Fig. 3C and table S.1B). No daily rhythm in cellular excitability was detected in nahar (Fig. 3D and Table S.2C; p>0.35). Positive current injections show that nahar mutant neurons fire fewer action potentials compared to controls, indicating that these neurons are healthy and can still generate action potentials but require more depolarizing current to fire at the same rate as WT neurons (Fig. 3D,E). Wild type membrane excitability can be restored by inducing NA expression only in the DN1p in the mutant, confirming that these effects are due to na and likely cell autonomous (Fig. 3E and Table S.2D). NMDG substitution induces an increase in the input resistance indicating that NA is open at rest (Fig. 3F).
Figure 3. The ion channel NARROW ABDOMEN controls Drosophila circadian pacemaker neuronal rhythms.
(A) Representative current clamp recordings at ZT2 showing that the nahar DN1ps neurons (red) are hyperpolarized and silent compared to WT DN1p neurons (black). Statistical analysis comparing the firing frequency (B) and membrane potential (C) of the WT (black) and nahar (red) DN1p neurons. Red asterisks indicate statistical significance between WT and nahar neurons (p<0.05, from a one-way ANOVA, Tukey’s post-hoc test). (Data for WT neurons are also depicted in Fig 1B and 1C). (D) Depolarizing current injections confirm the lack of detectable rhythms in cellular excitability in the nahar neurons (light red: ZT0-4, dark red ZT8–12, p>0.35). (E) The decrease in cellular excitability can be restored by rescuing the expression of NA only in the DN1p in the mutant: WT (black), nahar (red) and nahar;; U-na/Clk4.1M–G4 (blue) DN1ps neurons. (F) Histograms showing that sodium substitution with NMDG induces an increase in the input resistance indicating that NA is open at rest (black and green columns are before and after NMDG substitution, respectively). Results are expressed as mean±SEM. Asterisks indicate statistical significance (t-test, p<0.05). See also Tables S.1–2.
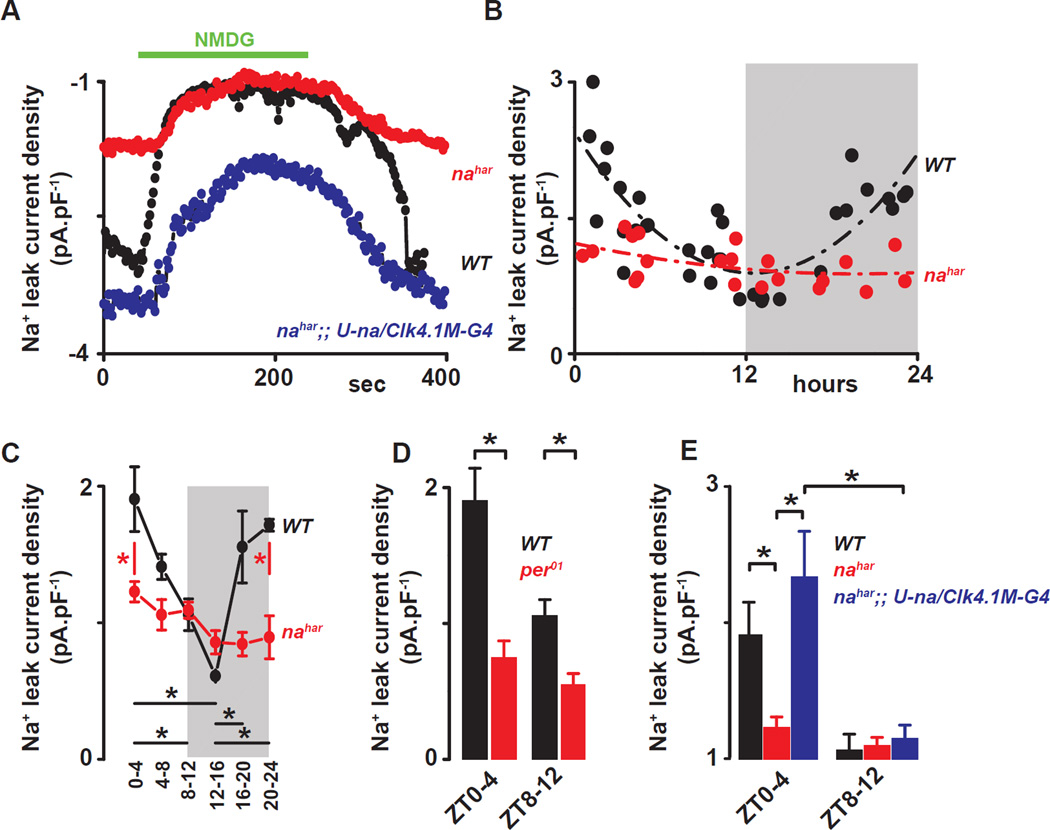
We next directly measured voltage-clamped NA dependent current (INA) at different times of day. A voltage ramp protocol (from –113 mV to +87 mV) was used to measure the inward current at −113mV, in the presence of TTX. Replacing the sodium from the extracellular solution with NMDG reveals the sodium leak current (Fig. 4A). Consistent with the sodium leak current being driven specifically by NA, the observed current is reduced in the nahar mutant neurons and can be restored by rescuing the expression of NA in the mutant (Fig. 4A). Measuring INA at different times of day reveals a diurnal modulation of current density: it is higher in the morning and lower in the evening (Fig. 4B–C and table S.1D). No rhythm is detected in the nahar (Fig. 4B–C and table S.1D) or in per01 mutants (Fig. 4D, p=0.21), the latter indicating core clock control. Further, the rhythm in NA conductance was evident even after Clk4.1M-GAL4 driven rescue (Fig. 4E). Given GAL4 stability, any promoter driven transcriptional rhythms may not be evident as GAL4 protein rhythms and thus GAL4 induced transcription of na may not be rhythmic (Kaneko et al., 2000), suggesting that NA current rhythms do not require na transcript rhythms. Taken together these results indicate that the clock control of sodium leak current through NA mediates rhythms of resting membrane potential.
Figure 4. The sodium leak current is under clock control in Drosophila circadian pacemaker neurons.
(A) Representative time courses showing the sodium leak current (INA) recorded at −113mV from a ramp protocol in WT (black), nahar (red) and nahar;; U-na/Clk4.1M–G4 (blue) DN1p neurons. (B) All recorded WT neurons (black dots) and nahar neurons (red dots) are plotted against time of day for sodium leak current (INA). (C) Quantification and statistical analysis are shown. Grey areas represent the dark phase of the LD cycle. Red asterisks indicate statistical significance between WT and nahar neurons, and black asterisks indicate statistical significance between different time points in WT neurons (p<0.05) from a one-way ANOVA, Tukey’s post-hoc test. (D) Histograms showing the NA current in WT (black) and per01 (red) DN1ps recorded at different times of day ZT0-4 vs ZT8–12 (for per01, INA=0.7±0.2pA.pF−1, n=8 at ZT0-4 and 0.5±0.1pA.pF−1, n=7 at ZT8–12. Asterisks indicate statistical difference between WT and per01, p<0.05 from t-test). (E) Histograms showing the sodium leak current in WT (black), nahar (red) and nahar;; U-na/Clk4.1M–G4 (blue) DN1p neurons at different times of day (ZT0-4 vs ZT8–12) (for nahar;; U-na/Clk4.1M–G4, INA=2.3±0.3pA.pF−1, n=4 at ZT0-4 and 1.1±0.1pA.pF−1, n=4 at ZT8–12). Results are expressed as mean±SEM. Asterisks indicate statistical significance (p<0.05) from a t-test.
Nlf-1 expression is time-dependent and is required for locomotor activity rhythms and NA leak current
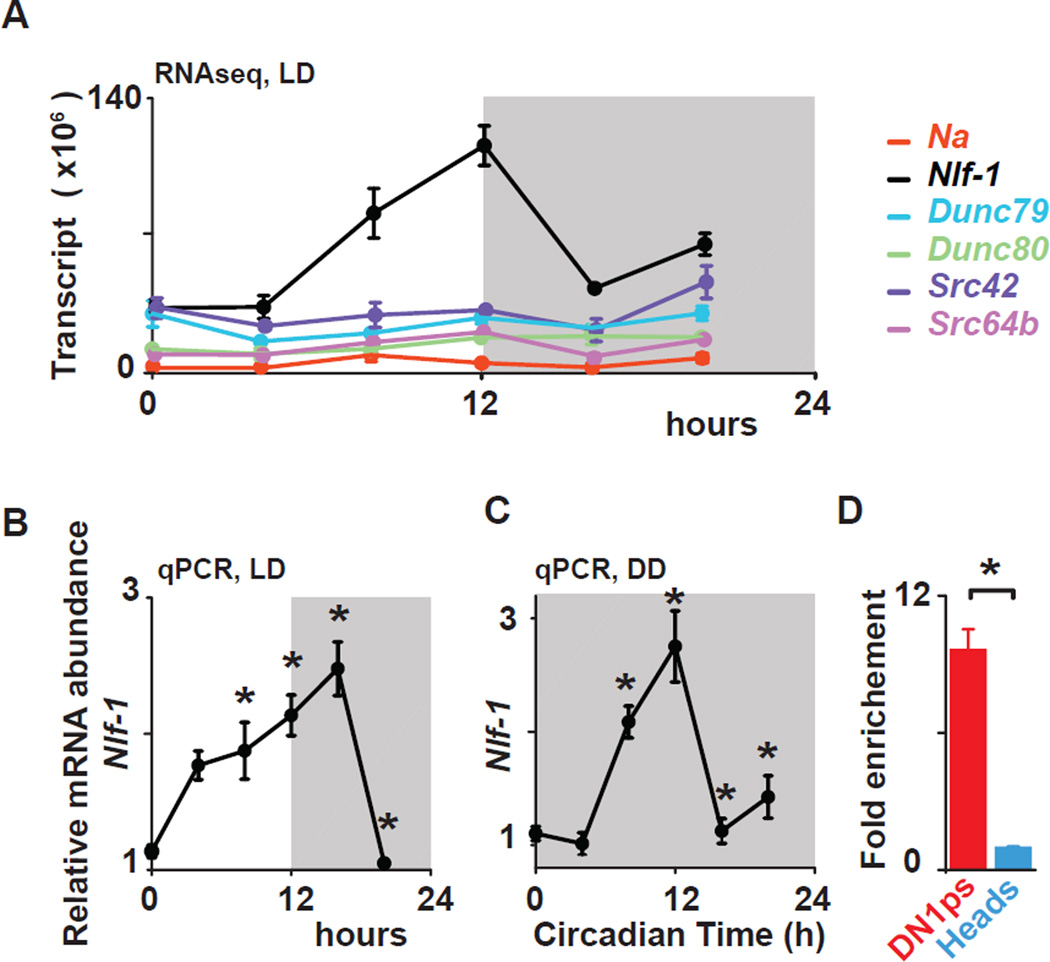
To identify molecular links between core clocks and membrane excitability, we employed fluorescence-activated cell sorting of GFP-labeled DN1p and performed RNA-Seq at distinct times during the LD cycle. Using empirical JTK_CYCLE (Hutchison et al., 2015), an updated version of JTK_CYCLE (Hughes et al., 2010), to detect rhythmic transcripts at a false discovery rate of 5% (Benjamini-Hochberg corrected p<0.05), we observed robust 24 h rhythms in CG33988, the fly ortholog of the NCA Localization Factor-1 (NLF-1) but not in na itself nor its regulatory subunits unc79, unc80 (Lear et al., 2013) and NALCN-activators, Src family kinases (Lu et al., 2009), Src42 and Src64b in flies (Fig. 5A). NLF-1 has been previously shown to interact with NA orthologs in worms (NCA-1 and −2) and mammals (Xie et al., 2013). NLF-1 protein is expressed in the endoplasmic reticulum and is required for the proper axonal localization of NCA-1 and −2 (Xie et al., 2013). Rhythmic expression of CG33988/Nlf-1 transcript was further confirmed with quantitative PCR (Fig. 5B) and consistent with clock control, CG33988/Nlf-1 transcript is rhythmic in the DN1p in constant darkness (DD) (Fig 5C). Nlf-1 transcript is also highly enriched in the DN1p clock neurons in comparison to whole heads (Fig. 5D). Chromatin immunoprecipitation experiments indicate that the core clock transcription factor CLOCK rhythmically binds the Nlf-1 genomic locus suggesting a direct biochemical link to the core clock (Abruzzi et al., 2011). Taken together, this suggests that Nlf-1 is a key mediator of NA rhythms that couples the transcriptional oscillator to membrane potential rhythms.
Figure 5. Nlf-1 is rhythmically expressed in DN1p neurons.
(A) Nlf-1 mRNA shows rhythmic expression using RNA-seq data from FACS sorted DN1p neurons in LD (for isoform RB- shown in graph, BH corrected p=0.005). na, Unc79, Unc80, Src64B, and Src42a are not robustly cycling (graph shows isoforms with highest expression : BH= 0.28 for na-RF, 0.2 for Dunc79-RE, 0.85 for Dunc80-RE, 0.71 for Src42a–RA and 0.07 for Src64B–RJ). Nlf-1 cycles under LD (B) and during the first day of constant darkness (DD1) conditions (C) in DN1ps using qPCR. Based on two independent experiments, an asterisk indicates differences statistically significant one-way ANOVA, Tukey’s post-hoc test, LD ZT0 vs ZT12 p= 0.0011, ZT0 vs ZT16, p= 0.000142, ZT4 vs ZT16 p=0.029, ZT0 vs ZT8 p=0.022, ZT12 vs ZT20, p= 0.000441, ZT16 vs ZT20 p= 0.000136. DD1 CT0 vs CT8 p= 0.01081, CT0 vs CT12 p= 0.000142, CT12 vs CT16 p= 0.000145 and CT12 vs CT20 p= 0.000459. (D) Nlf-1 expression is enriched in the DN1ps vs whole head (t-test, p<0.02). Results are expressed as mean±SEM.
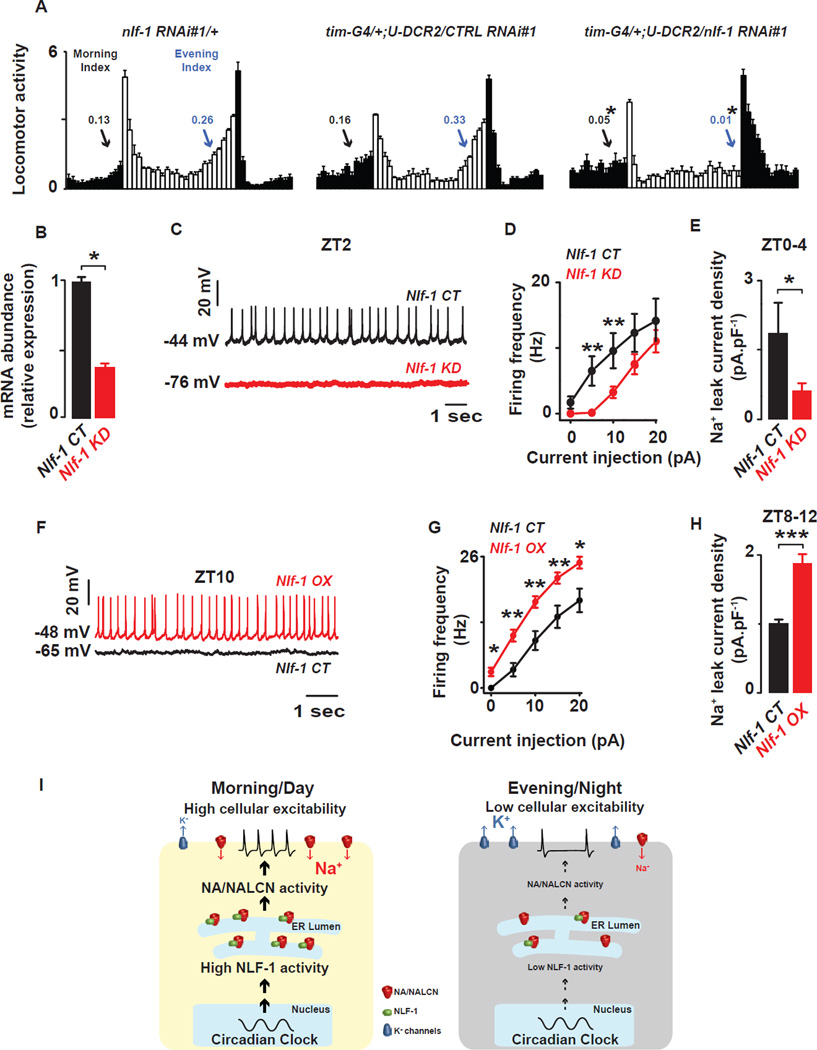
To assess the function of NLF-1 in circadian behavior, we knocked down its transcript levels using three independent transgenic dsRNA and shRNA lines in combination with the broad circadian driver tim-GAL4. In contrast to the previously reported weak effect of CG33988 RNAi knockdown on evening behavior (Ghezzi et al., 2014), we found dramatic reductions in rhythmic strength in DD (3/3 lines) and reduced anticipation of lights-on (2/3 lines) and lights-off transitions (3/3 lines) under LD conditions (Fig. 6A, Fig. S.3). These effects are comparable to those observed in loss-of-function na alleles (Lear et al., 2005) and knockdown of na using RNAi (Fig. S.3 and Tables S.3–4). Restricting Nlf-1 knockdown to non-PDF clock neurons (tim-GAL4, pdf-GAL80) also caused reduced morning and evening anticipation as well as reduced rhythmicity (Tables S.3–4), consistent with prior na rescue studies (Lear et al., 2005). Further restricting Nlf-1 knockdown to the DN1p using Clk4.1M–GAL4 resulted in reduced DD rhythmicity (Table S.3).
Figure 6. Nlf-1 is required for anticipatory behavior and NA current.
(A) Nlf-1 RNAi expressing flies (tim-G4/+; U-Dcr2/Nlf-1 RNAi#1) show reduced morning anticipation (Morning Index) and evening anticipation (Evening Index) under LD conditions when compared to genetic controls (Nlf-1 RNAi#1/+ and tim-G4/+; U-Dcr2/CTRL RNAi#1), (t-test, p<0.05). (B) Nlf-1 expression is reduced in the DN1ps of Nlf-1 RNAi expressing flies (t-test, p<0.05) (C) Representative current clamp recordings at ZT2 showing that the Nlf-1 knockdown DN1p neurons (red) are hyperpolarized and silent compared to control DN1p neurons (black). (D) Depolarizing current injections confirm the decrease in cellular excitability in Nlf-1 knockdown neurons (red) vs control (black) (p<0.05). (E) Sodium leak current density is dramatically reduced in the Nlf-1 knockdown neurons (red) vs control neurons (black) (1.9±0.7pA.pF−1, n=4 in Nlf-1 CT and 0.6±0.2pA.pF−1, n=5 in Nlf-1 KD, measured at ZT0-4, p<0.05). (F) Representative current clamp recordings at ZT10 showing that the Nlf-1 overexpressing DN1p neurons (red) are depolarized and more active compared to control DN1p neurons (black). (G) Depolarizing current injections confirm the increase in cellular excitability in Nlf-1V5 overexpressing neurons (red) vs control (black) (p<0.05). (H) Sodium leak current density is also increased in the Nlf-1V5 overexpressing neurons (red) vs control neurons (black) (1±0.05pA.pF−1, n=4 in Nlf-1 CT and 1.9±0.1pA.pF−1, n=5 in Nlf-1 OX, measured at ZT8–12, p<0.05). Results are expressed as mean±SEM. Asterisks indicate statistical significance (p<0.05 from a t-test). A summary cartoon depicting the conserved bicycle model for controlling membrane excitability of circadian pacemaker neurons is shown in (I). In the morning/day, the molecular clock drives high NLF-1 activity increasing the sodium leak activity and K conductances are reduced thus increasing cellular excitability. In the evening/night, the sodium leak is decreased and, in parallel, K conductances are high, thus silencing the neurons. This dual regulation of the conductances responsible for the membrane properties is critical for driving high amplitude rhythmic oscillations of cellular excitability. See also Figures. S.3–5 and Tables S.2–4.
The role of Nlf-1 extends to PDF neurons. Restricting Nlf-1 knockdown to PDF neurons, using two different pdf-GAL4 drivers (pdf-GAL4 and pdf0.5-GAL4 (Park et al., 2000), dramatically reduces free running rhythms (Table S.3), consistent with highly enriched Nlf-1 transcript observed in larval PDF+ sLNv neurons (Nagoshi et al., 2010) and with the described role of NA in PDF neurons (Lear et al., 2005). In addition, we extended our patch clamp analysis to the large LNv neurons (Fig. S.4A). Here we observed clock-dependent rhythms in membrane properties as previously observed (Fig. S.4B–C) (Cao and Nitabach, 2008; Sheeba et al., 2008). In addition, we found clock-dependent NA current rhythms similar to those we observed for the DN1p with peak levels in the morning (Fig. S.4D). Thus our findings in DN1p extend to other circadian neurons.
We then tested whether Nlf-1 is important for NA current levels, which may reflect the proper channel localization to the cell membrane. Knockdown of Nlf-1 expression was confirmed in the DN1p with quantitative PCR (Fig. 6B). We find that knockdown in the DN1p results in a similar phenotype to that observed for na mutants with cells becoming hyperpolarized and silent (Fig. 6C). Cellular excitability is also decreased in the Nlf-1 knockdown, as the neurons are less responsive to depolarizing currents (Fig. 6D and Table S.2E). NA-dependent current was also strongly suppressed after Nlf-1 knockdown (Fig. 6E).
To further examine the mechanism by which NLF-1 might regulate NA, we assayed NA protein expression after Nlf-1 knockdown. Nlf-1 knockdown with a broad neuronal driver (elav-G4) also results in strong reductions in rhythmic strength in DD and reduced morning and evening anticipation (Fig. S.5A, Tables S.3–4). Surprisingly, NA protein levels were dramatically reduced in these flies (Fig. S.5B). We also observed lower NA expression (∼50% reduction) when na was driven transgenically in the DN1p of Nlf-1 knockdown flies (Fig. S.5C–D). In part due to the small soma and limited expression in projections, we could not reliably assess cell membrane or axonal localization. Yet Nlf-1 knockdown does not reduce DN1p na transcript levels (Fig. S.5E). Nlf-1 knockdown in the DN1p phenocopies a na mutant suggesting NA current is nearly abolished (Fig. 6E) yet transgenic NA is reduced by just ∼50%. Thus, we favor the view that strong effects of Nlf-1 knockdown on NA current are only in part due to changes in NA levels.
If the oscillation of Nlf-1 transcript is critical to setting NA levels and DN1p membrane excitability, we would predict that Nlf-1 overexpression would increase NA current at evening time points when NA current is typically at trough levels. We observed that in the evening (ZT8–12) NLF-1 overexpression depolarizes membrane potential, elevates firing rates (Fig. 6F) and cellular excitability (Fig. 6G and Table S.2.F), and, most importantly, increases NA current (Fig. 6H) at a time when each of those parameters is near their daily trough in wild-type flies. Indeed, sodium leak current density in the evening in Nlf-1 overexpression flies (∼2pA.pF−1) is comparable to that seen at peak levels in wild-type flies in the morning. Taken together, these results indicate Nlf-1 expression is rhythmic and mediates NA activity rhythms. This demonstrates a molecular mechanism linking the core clock to membrane excitability via the rhythmic transcription of a factor important for ion channel function in Drosophila circadian neurons (Fig. 6I).
NALCN current is under clock control in mammalian SCN pacemaker neurons
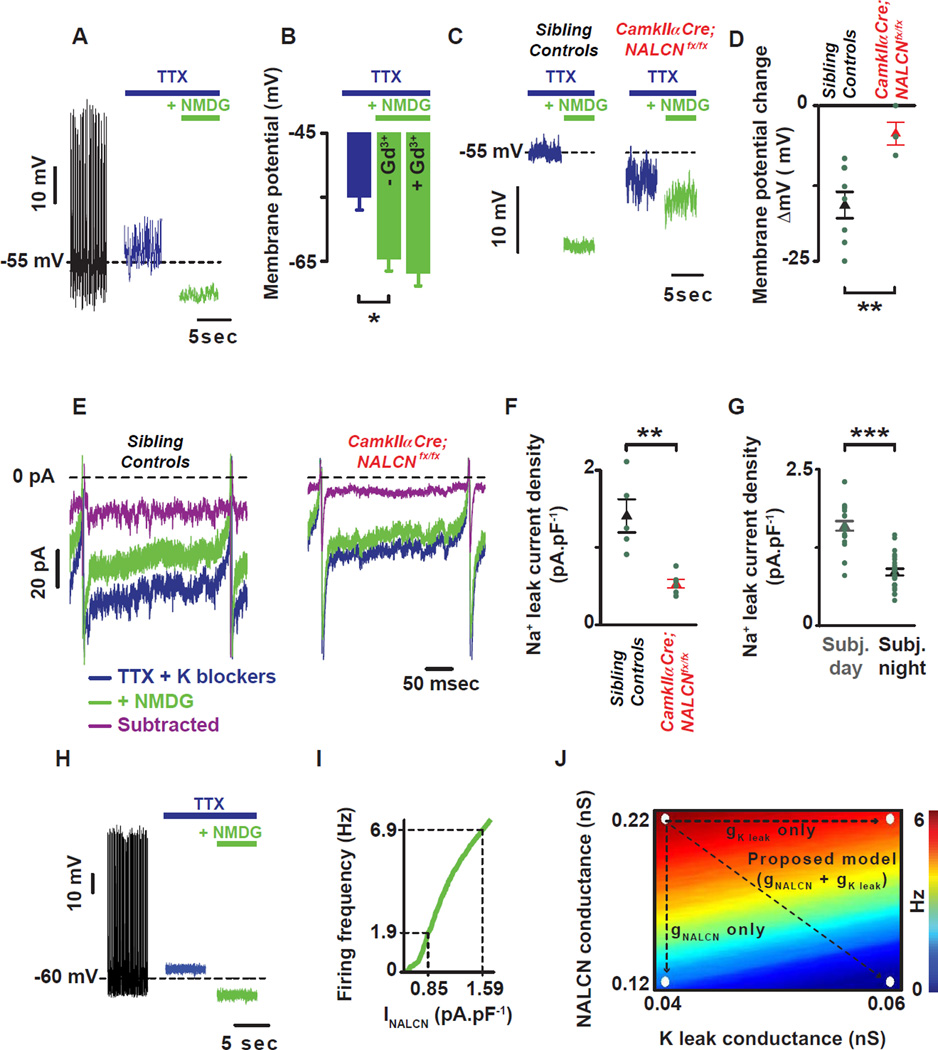
Although we demonstrated a rhythmic function for resting sodium leak in Drosophila clock neurons, rhythmic resting sodium conductances have yet to be described in mammalian clock neurons. Previous patch clamp analyses of dissociated SCN neurons demonstrated the presence of a NALCN-like current (TTX-resistant, NMDG-sensitive, voltage-independent sodium conductance termed Ibackground) that is largely responsible for the initial phase of the depolarizing drive during the interspike interval (Jackson et al., 2004). To determine whether this activity is rhythmic in mammalian circadian pacemaker neurons, we performed voltage clamp analysis during subjective day and night from organotypic slices containing the SCN from mice entrained for two weeks in LD and then maintained under constant darkness conditions for at least three weeks. Rhythms in firing frequency, membrane potential and input resistance were observed, thus validating the preparation (Fig. S.6A–C). In the presence of TTX to block action potentials, the NALCN blockers, NMDG (Fig. 7A) or Gd3+ (Fig. S.6D), induce a hyperpolarization, while no additional effect of applying Gd3+ after sodium replacement with NMDG was observed (Fig. 7B). Importantly, in hippocampal neurons the vast majority of current with this pharmacological profile is mediated by NALCN (Lu et al., 2007).
Figure 7. NALCN current is under clock control in mammalian SCN pacemaker neurons.
(A) Representative current clamp recording showing the role of the TTX-resistant sodium leak (difference between green and blue) in setting the membrane potential of mammalian SCN neurons. (B) NMDG hyperpolarizes the cell with no additional effect in the presence of Gd3+. (C) NMDG-evoked hyperpolarization was reduced in a brain specific knockout of NALCN. (D) Quantification and statistical analysis of the NMDG-evoked hyperpolarization are shown: −15.9±2.0mV, n=9 in controls (black triangle) and −4.5±1.7mV, n=4 (red triangle). Asterisks indicate statistical significance (t-test, p=0.005). (E) Action potential clamp recordings showing the sodium leak flowing during the interspike interval in SCN neurons from sibling control (left) and CamkIIa-Cre;NALCNfx/fx animals (right). In the presence of TTX and K blockers (blue trace), the sodium leak current flowing during the interspike interval (INALCN) was reduced after sodium substitution with NMDG (green trace). The sodium leak current (INALCN = subtracted = purple trace) was revealed by subtracting the inward current in the presence of NMDG from the inward current present with TTX and K blockers. (F) INALCN was reduced in CamkIIa-Cre;NALCNfx/fx compared to sibling controls animals (0.5±0.1pA.pF−1, n=6 in CamkIIa-Cre;NALCNfx/fx (red triangle) and 1.4±0.2pA.pF−1, n=5 in sibling controls (black triangle)). Asterisks indicate statistical significance (t-test, p=0.002). (G) Circadian variation of INALCN: 1.6±0.1pA.pF−1, n=25 during the subjective day (grey columns) and 0.8±0.1pA.pF−1, n=23 during the subjective night (black columns). Asterisks indicate statistical significance (t-test, p<0.001). Green dots represent individual cells. (H) Simulations showing the role of TTX resistant sodium leak in setting the membrane potential using a mathematical model of SCN membrane excitability. Voltage traces from control simulation (gNa = 229 nS, gNALCN = 0.22 nS) and simulated application of TTX (gNa = 0 nS) and NMDG (gNALCN = 0 nS). (I) The model predicts the magnitude of change in firing rate as a function of magnitude of change in NALCN current density (gNALCN = 0.12 to 0.22 nS). A decrease of 0.74 pA.pF−1 in INALCN (observed between the subjective day and night (G)) leads to a 5 Hz decrease in firing rate. (J) Firing rate as a function of gNALCN and gKleak in a model SCN neuron. Arrows: decreasing gNALCN alone reduces firing rate from 7 Hz to 2 Hz, whereas increasing gKleak reduces firing rate from 7 Hz to 6 Hz. Concurrently decreasing gNALCN and increasing gKleak reduces firing rate from 7 Hz to 0.5 Hz. Results are expressed as mean±SEM. See also Figures. S.6–7.
To confirm the molecular identity of the sodium leak in the SCN, we generated a forebrain specific knockout of NALCN with a CamkIIα-cre driver. With this driver, CRE expression mimics the endogenous expression of CamkIIα (enriched in the forebrain, neuron specific (Casanova et al., 2001)). CamkIIα expression is also highly enriched in the SCN and loss of circadian rhythms in mice with a CamkIIα specific knockout of a core clock gene (Bmal1) is observed (Izumo et al., 2014). We first confirmed that the NMDG evoked hyperpolarization (Fig 7A) is greatly reduced in the CamkIIα–Cre;NALCNfx/fx animals compared to age-matched sibling controls (Fig. 7C–D, p=0.005). Consistent with a role of NALCN in controlling the membrane potential and firing frequency of neurons, CamkIIα–Cre;NALCNfx/fx SCN neurons were hyperpolarized and silent (Fig. S.6E–F, p<0.007). Importantly, a small depolarization current injected into the CamkIIα–Cre;NALCNfx/fx SCN neurons was able to evoke strong firing (Fig. S.7G) indicating that cells were healthy but required a greater depolarizing input to evoke action potentials. We further confirmed the presence of NALCN current during the interspike interval in organotypic slices containing the SCN with pharmacology: NMDG and Gd3+ sensitive. No additional block by Gd3+ was observed after NMDG application (Fig. S.6H–I). This NMDG sensitive inward current was greatly reduced in the CamkIIα-Cre;NALCNfx/fx SCN neurons (Fig. 7E–F, p=0.002). Taken together, these data indicate that the vast majority of the sodium leak flowing during the interspike interval in SCN neurons is carried by NALCN (INALCN) consistent with other mammalian neurons (Lu et al., 2007). We then assessed INALCN at different times of day and found that it was significantly larger during the subjective day than subjective night consistent with a control by the circadian clock (Fig. 7G, p<0.001).
To determine the impact of a day-night change in INALCN (∼0.7 pA.pF−1) on firing frequency and membrane potential, we simulated sodium leak current modulation using an updated version of a mathematical model of SCN membrane excitability (Diekman et al., 2013) (see Experimental Procedures). This model accurately captures the effect of NALCN blockers on the membrane potential of SCN neurons (Fig. 7H). According to this model, modest daily changes in sodium leak conductance comparable to those observed experimentally can have sizable effects on neuronal firing rates (Fig. 7I). To explore the contributions of both sodium and potassium leak currents to the daily variation of firing rate in SCN neurons, we simulated concurrent modulation of these two conductances. Beginning from a subjective day firing rate of 7 Hz, reducing sodium leak conductance by the amount suggested by our experimental measurements (∼0.7 pA.pF−1, Fig. 7G) decreases firing rate to 2 Hz. Experimentally, we observed that during the subjective day, INALCN is, in fact, positively correlated with firing frequency (Fig. S.6J), suggesting that INALCN significantly impacts neuronal physiology. Even lower firing rates that are characteristic of subjective night (0.5 Hz) can be achieved by increasing potassium leak conductance in conjunction with this reduction in sodium leak (Fig. 7J, Fig. S.7). Thus, elevated sodium leak during the day and elevated potassium leak at night can recapitulate the experimentally observed daily variations in SCN firing rate through relatively modest changes in these leak currents.
Discussion
Taken together, our work defines a conserved mechanism for the maintenance of circadian oscillations necessary for robust daily behaviors (Fig. 6I) that we term the “bicycle” model. Membrane oscillations are driven by two cycles with opposite temporal phases analogous to cycling bicycle pedals. During the morning/day, sodium leak mediated by NA/NALCN is elevated while resting K currents are reduced depolarizing the neuron to promote elevated firing rates. During the evening/night, sodium leak is low and resting K currents are elevated, hyperpolarizing the cell to suppress firing rates. The clock-controlled transcript Nlf-1 drives the rhythm of NA/NALCN current, linking the core clock to ion channel activity.
While Drosophila has been a well-established model for defining molecular genetic mechanisms, relatively little is known about the specific ionic currents that underlie fly pacemaker neuron excitability rhythms due to the small size of Drosophila soma. Most cellular electrophysiological analyses have focused on the largest cells, the large ventral lateral neurons (Cao and Nitabach, 2008; Fogle et al., 2015; Fogle et al., 2011; Sheeba et al., 2008). Yet, even in these neurons, the specific ionic currents under clock control have yet to be defined. Using whole cell patch clamp electrophysiology of DN1p pacemaker neurons, we found high amplitude oscillations of spontaneous firing rates and basal membrane potential that are comparable to those observed in mammalian SCN clock neurons. Moreover, we demonstrate clock control of both resting sodium leak conductance as well as resting potassium conductance. Our data suggest that the patch-clamp analysis of the DN1p will be valuable in defining the ionic currents that mediate clock control of neuronal excitability.
Our data indicate that the daily changes in membrane excitability that we observe are cell autonomous. The observed cycles of resting currents are evident even in the presence of TTX. Bath application of TTX silences neurons and thus would block firing-dependent neurotransmitter release. Moreover, cycling sodium leak currents are driven by the transcriptional oscillation of Nlf-1, providing a cell autonomous mechanism for clock control. We propose that cell autonomous clock regulation collaborates with rhythmic network inputs, such as PDF, which likely act in the morning to excite DN1p neurons (Kunst et al., 2014; Seluzicki et al., 2014). In turn DN1p excitation drives waking behavior in the morning (Kunst et al., 2014; Zhang et al., 2010a; Zhang et al., 2010b) and free running rhythmicity, perhaps via the DH44 neurons in the pars intercerebralis (Cavanaugh et al., 2014). As the DN1p are also important for evening behavior (Zhang et al., 2010a), evening silencing may permit other neurons (e.g. the LNd) to drive evening behavior.
The clock control of membrane potential has largely focused on modulation of resting potassium conductance in the SCN (Kuhlman and McMahon, 2004) as well as in Bulla photoreceptors (Michel et al., 1993; Michel et al., 1999). Surprisingly, we observed rhythms of sodium leak conductance in the fly DN1ps and l-LNvs, as well as in mammalian SCN, that are mediated by the NA/NALCN channel. This sodium leak exhibits the pharmacological sensitivity previously defined for the NALCN current (Lu et al., 2007), most notably NMDG+ and Gd3+ block. In addition, the current is reduced in na mutant fliesand in mice with a brain specific knockout of NALCN. Clock modulation of this sodium current also likely impacts neurophysiology. Loss of function na mutants and NALCN knockout result in silent and hyperpolarized neurons. Computational modeling of SCN neurons demonstrates that the modest daily rhythm of sodium leak can significantly impact overall firing rates. Thus our work defines a molecular mechanism for clock control of membrane excitability.
Using a combination of genomics, electrophysiology and behavior, our work reveals a molecular pathway that links the transcriptional clock to these sodium current rhythms. The mechanisms of clock control of membrane excitability have largely focused on the direct clock control of ion channel transcripts (Itri et al., 2005; Kudo et al., 2011; Meredith et al., 2006; Nahm et al., 2005; Pennartz et al., 2002; Pitts et al., 2006). Using RNA-seq and qPCR validation on FACS sorted DN1p neurons, we identify robust rhythms of Nlf-1, an ER protein that is important for the localization of NALCN and its orthologs (Xie et al., 2013). Moreover, RNAi knockdown of Nlf-1 results in suppression of behavioral rhythms, NA expression and related current. Conversely, NLF-1 overexpression increases NA current, firing frequency and membrane potential in the evening when these parameters are typically at their troughs in wild-type flies, suggesting that Nlf-1 controls activity and/or localization of NA. Chromatin immunoprecipitation has demonstrated rhythmic CLK binding at the Nlf-1 locus (Abruzzi et al., 2011). Our cell-specific knockdown experiments indicate that Nlf-1 functions broadly within the clock network to control morning and evening anticipation as well as DD rhythms, suggesting this mechanism is widely applied. Future work will be required to determine if Nlf-1/NA rhythms in morning and evening cells have distinct phases. Nonetheless, we have defined a molecular pathway that directly links CLK-driven transcriptional oscillations to NA current and behavioral rhythms.
This mechanism may not only be operating in clock neurons but may be broadly involved in rhythmic changes in brain states. For instance, NALCN is critical to the maintenance of respiratory rhythms (Lu et al., 2007). Both fly and worm na/nca loss-of-function results in disrupted locomotion as well as altered sensitivity to general anesthetics (Humphrey et al., 2007). na mutant flies also show altered behavioral state transitions related to sleep and anesthesia (Joiner et al., 2013). More generally, the NA current here shows the identical electrophysiological profile to the tonic cation current required for regular firing in neurons of the mouse cerebellar nuclei (Raman et al., 2000).
Our work also demonstrates that, like the core molecular clock, clock control of membrane potential is also widely conserved in neurons important for sleep and wake. We hypothesize that the common ancestor of the mouse and the fly had master circadian pacemaker neurons that drove its daily behavior. Moreover, these clock neurons employed daily anti-phase sodium and potassium conductances to drive their rhythmic activity. Thus, our finding suggests an ancient strategy governing neuronal activity important for driving daily cycles of sleep and wake.
Experimental Procedures
Please see Supplemental Experimental procedures for detailed protocols.
Electrophysiological recordings from Drosophila neurons
Whole brain electrophysiology experiments were performed with pipettes (10–14 MΩ) filled with internal solution. The sodium leak current (INa) was examined in the presence of TTX (10µM), TEA (10mM), 4-AP (5mM) and CsCl (2mM), and was revealed by replacing the extracellular sodium with NMDG (Lu et al., 2007). All recordings were corrected for liquid junction potential (13mV). For analysis, cells with high series resistance or with low membrane resistance (<1GΩ) were discarded.
RNA isolation, amplification, and sequencing
RNA was isolated and amplified as previously described (Kula-Eversole et al., 2010). The quality and quantity of dsDNA was assessed on Bioanalyzer (Agilent). After quality control, libraries were generated using TruSeq Sample Preparation Guide (following manufacturer’s protocol (Illumina)). The RNA-Seq was performed on HiSeq2000 (Illumina). Bowtie (Langmead et al., 2009) was used to align short-read aligner to references (obtained from flybase.org). Quantification was performed using eXpress (Roberts et al., 2011). Rhythmic transcripts were detected using empirical JTK_CYCLE (Hutchison et al., 2015), transcripts were considered robustly rhythmic when the Benjamini-Hochberg corrected p value or false discovery rate<0.05. Empirical JTK_CYCLE derives p values empirically considering asymmetric waveforms.
Mathematical modeling
Simulations of a Hodgkin-Huxley type model of SCN membrane excitability were performed using MATLAB R2012b (Mathworks, Natick, MA, USA). The model was fit to experimental data from SCN neurons and consists of a system of ordinary differential equations for membrane potential (V) and six ionic gating variables (m, h, n, r, f, and b):
with parameter values C=5.7 pF, Iapp=0 pA, gNa=229 nS, gK=14 nS, gCa=65 nS, gBK=10 nS, gCl=0.3 nS, gK-leak=0.04 nS, ENa=45 mV, EK=−97 mV, ECa=64 mV, ECl=−60 mV, and ENALCN=−20 mV.
Our SCN model extends previously published versions (Diekman and Forger, 2009; Sim and Forger, 2007) by separating the leak current into sodium, potassium, and chloride components (with parameter values chosen based on our measurements of sodium leak current density and RMP), and incorporating a large-conductance calcium-activated potassium (BK) current. BK channels are voltage and calcium-gated. However, since the nanodomain calcium concentration sensed by the channel reaches equilibrium very quickly, we follow (Tomaiuolo et al., 2012) and model the gating as a purely voltage-dependent process. The differential equations were solved using the Euler-Maruyama method, with a time step of 0.01ms and standard Gaussian distributed voltage noise (µ=0, σ=0.5).
Supplementary Material
Acknowledgments
We thank B. White and H-S. Li for communicating results prior to publication, M. Vitaterna, K. Shimomura, A. Para, E. Zaharieva, D. Wokosin, C. Olker, Jordan I. Robson for technical assistance, J. Gu, R. Scott and B. White for the cloning and generation of UAS-NAHA flies, Dr. Jean Richa (Transgenic and Chimera Mouse Facility at the University of Pennsylvania) for ES cell injection and M. Gallio for reagents and comments on a draft of the manuscript. We also thank Bloomington Drosophila stock center, the National Institute of Genetics (NIG), Vienna Drosophila RNAi center (VDRC) for reagents and the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks This work was supported by National Institutes of Health (NIH) grants R01NS052903 (to R.A), NS055293 and NS074257 (to D.R.), R00GM080107 (to B.C.L.), National Science Foundation (NSF) Division of Mathematical Science grant (DMS1412877 to C.O.D). This effort was in part sponsored by the Defense Advanced Research Projects Agency (DARPA) (D12AP00023, to R.A.); the content of the information does not necessarily reflect the position or the policy of the government, and no official endorsement should be inferred. This work was also supported by the Northwestern University Flow Cytometry Facility, by a Cancer Center Support Grant (NCI CA060553).The two photon microscope was supported by NINDS (NS054850). A.L.H. is a trainee of the NIH Medical Scientist Training program at the University of Chicago (NIGMS T32GM07281). This work made use of the Open Science Data Cloud (OSDC), which is an Open Cloud Consortium (OCC)-sponsored project. This work was supported in part by grants from Gordon and Betty Moore Foundation, the NSF and major contributions from OCC members like the University of Chicago. Our work is in memory of Howard Nash who re-discovered narrow abdomen and inspired our pursuit of studies of this channel.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
R.A., I.M.R. and M.F. designed the experiments; M.F. performed experiments and analyses related to Fig. 1, 2, 3, 4, 6 B-H, 7 A-G, S.1, S.2, S.5C–D, S.6 and Tables S.1–2; E.K.E. designed, performed and analyzed experiments related to Fig 5, 6A–B, S.3, S.5A, S.5E and Tables S.3–4; A.L.H., K.P.W. and A.R.D. performed RNAseq analyses related to Fig 5A; C.O.D. developed the mathematical model related to Fig 7H–J and S.7; T.H.H performed electrophysiological recordings and analyses on lLNvs neurons related to Fig. S.3, D.L.M. and B.C.L. performed experiments and analyses related Fig. S.5B; K.A. and D.R. generated the NALCNfx/fx mice; M.F. and R.A. wrote the manuscript; COD wrote sections related to the mathematical model. I.M.R. edited the manuscript.
References
- Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes & development. 2011;25:2374–2386. doi: 10.1101/gad.178079.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annual review of physiology. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Nitabach MN. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci. 2008;28:6493–6501. doi: 10.1523/JNEUROSCI.1503-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova E, Fehsenfeld S, Mantamadiotis T, Lemberger T, Greiner E, Stewart AF, Schutz G. A CamKIIalpha iCre BAC allows brain-specific gene inactivation. Genesis. 2001;31:37–42. doi: 10.1002/gene.1078. [DOI] [PubMed] [Google Scholar]
- Cavanaugh DJ, Geratowski JD, Wooltorton JR, Spaethling JM, Hector CE, Zheng X, Johnson EC, Eberwine JH, Sehgal A. Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell. 2014;157:689–701. doi: 10.1016/j.cell.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS. Linking neural activity and molecular oscillations in the SCN. Nature reviews Neuroscience. 2011;12:553–569. doi: 10.1038/nrn3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jeu M, Hermes M, Pennartz C. Circadian modulation of membrane properties in slices of rat suprachiasmatic nucleus. Neuroreport. 1998;9:3725–3729. doi: 10.1097/00001756-199811160-00028. [DOI] [PubMed] [Google Scholar]
- Diekman CO, Belle MD, Irwin RP, Allen CN, Piggins HD, Forger DB. Causes and consequences of hyperexcitation in central clock neurons. PLoS computational biology. 2013;9:e1003196. doi: 10.1371/journal.pcbi.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekman CO, Forger DB. Clustering predicted by an electrophysiological model of the suprachiasmatic nucleus. J Biol Rhythms. 2009;24:322–333. doi: 10.1177/0748730409337601. [DOI] [PubMed] [Google Scholar]
- Flourakis M, Allada R. Patch-clamp electrophysiology in Drosophila circadian pacemaker neurons. Methods in enzymology. 2015;552:23–44. doi: 10.1016/bs.mie.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Fogle KJ, Baik LS, Houl JH, Tran TT, Roberts L, Dahm NA, Cao Y, Zhou M, Holmes TC. CRYPTOCHROME-mediated phototransduction by modulation of the potassium ion channel beta-subunit redox sensor. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2245–2250. doi: 10.1073/pnas.1416586112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogle KJ, Parson KG, Dahm NA, Holmes TC. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science. 2011;331:1409–1413. doi: 10.1126/science.1199702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi A, Liebeskind BJ, Thompson A, Atkinson NS, Zakon HH. Ancient association between cation leak channels and Mid1 proteins is conserved in fungi and animals. Frontiers in molecular neuroscience. 2014;7:15. doi: 10.3389/fnmol.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Cerullo I, Chen X, Rosbash M. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. eLife. 2014:3. doi: 10.7554/eLife.02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE. Molecular genetic analysis of circadian timekeeping in Drosophila. Advances in genetics. 2011;74:141–173. doi: 10.1016/B978-0-12-387690-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C. Neurobiology of the fruit fly’s circadian clock. Genes, brain, and behavior. 2005;4:65–76. doi: 10.1111/j.1601-183X.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey JA, Hamming KS, Thacker CM, Scott RL, Sedensky MM, Snutch TP, Morgan PG, Nash HA. A putative cation channel and its novel regulator: cross-species conservation of effects on general anesthesia. Curr Biol. 2007;17:624–629. doi: 10.1016/j.cub.2007.02.037. [DOI] [PubMed] [Google Scholar]
- Hutchison AL, Maienschein-Cline M, Chiang AH, Tabei SM, Gudjonson H, Bahroos N, Allada R, Dinner AR. Improved statistical methods enable greater sensitivity in rhythm detection for genome-wide data. PLoS computational biology. 2015;11:e1004094. doi: 10.1371/journal.pcbi.1004094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itri JN, Michel S, Vansteensel MJ, Meijer JH, Colwell CS. Fast delayed rectifier potassium current is required for circadian neural activity. Nature neuroscience. 2005;8:650–656. doi: 10.1038/nn1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumo M, Pejchal M, Schook AC, Lange RP, Walisser JA, Sato TR, Wang X, Bradfield CA, Takahashi JS. Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. eLife. 2014:3. doi: 10.7554/eLife.04617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AC, Yao GL, Bean BP. Mechanism of spontaneous firing in dorsomedial suprachiasmatic nucleus neurons. J Neurosci. 2004;24:7985–7998. doi: 10.1523/JNEUROSCI.2146-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner WJ, Friedman EB, Hung HT, Koh K, Sowcik M, Sehgal A, Kelz MB. Genetic and anatomical basis of the barrier separating wakefulness and anesthetic-induced unresponsiveness. PLoS genetics. 2013;9:e1003605. doi: 10.1371/journal.pgen.1003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Park JH, Cheng Y, Hardin PE, Hall JC. Disruption of synaptic transmission or clock-gene-product oscillations in circadian pacemaker cells of Drosophila cause abnormal behavioral rhythms. J Neurobiol. 2000;43:207–233. doi: 10.1002/(sici)1097-4695(20000605)43:3<207::aid-neu1>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Kudo T, Loh DH, Kuljis D, Constance C, Colwell CS. Fast delayed rectifier potassium current: critical for input and output of the circadian system. J Neurosci. 2011;31:2746–2755. doi: 10.1523/JNEUROSCI.5792-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman SJ, McMahon DG. Rhythmic regulation of membrane potential and potassium current persists in SCN neurons in the absence of environmental input. The European journal of neuroscience. 2004;20:1113–1117. doi: 10.1111/j.1460-9568.2004.03555.x. [DOI] [PubMed] [Google Scholar]
- Kuhlman SJ, McMahon DG. Encoding the ins and outs of circadian pacemaking. Journal of biological rhythms. 2006;21:470–481. doi: 10.1177/0748730406294316. [DOI] [PubMed] [Google Scholar]
- Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13497–13502. doi: 10.1073/pnas.1002081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst M, Hughes ME, Raccuglia D, Felix M, Li M, Barnett G, Duah J, Nitabach MN. Calcitonin gene-related Peptide neurons mediate sleep-specific circadian output in Drosophila. Curr Biol. 2014;24:2652–2664. doi: 10.1016/j.cub.2014.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear BC, Darrah EJ, Aldrich BT, Gebre S, Scott RL, Nash HA, Allada R. UNC79 and UNC80, putative auxiliary subunits of the NARROW ABDOMEN ion channel, are indispensable for robust circadian locomotor rhythms in Drosophila. PLoS One. 2013;8:e78147. doi: 10.1371/journal.pone.0078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear BC, Lin JM, Keath JR, McGill JJ, Raman IM, Allada R. The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron. 2005;48:965–976. doi: 10.1016/j.neuron.2005.10.030. [DOI] [PubMed] [Google Scholar]
- Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- Lu B, Su Y, Das S, Liu J, Xia J, Ren D. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell. 2007;129:371–383. doi: 10.1016/j.cell.2007.02.041. [DOI] [PubMed] [Google Scholar]
- Lu B, Su Y, Das S, Wang H, Wang Y, Liu J, Ren D. Peptide neurotransmitters activate a cation channel complex of NALCN and UNC-80. Nature. 2009;457:741–744. doi: 10.1038/nature07579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith AL, Wiler SW, Miller BH, Takahashi JS, Fodor AA, Ruby NF, Aldrich RW. BK calcium-activated potassium channels regulate circadian behavioral rhythms and pacemaker output. Nature neuroscience. 2006;9:1041–1049. doi: 10.1038/nn1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel S, Geusz ME, Zaritsky JJ, Block GD. Circadian rhythm in membrane conductance expressed in isolated neurons. Science. 1993;259:239–241. doi: 10.1126/science.8421785. [DOI] [PubMed] [Google Scholar]
- Michel S, Manivannan K, Zaritsky JJ, Block GD. A delayed rectifier current is modulated by the circadian pacemaker in Bulla. J Biol Rhythms. 1999;14:141–150. doi: 10.1177/074873099129000533. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Takahashi JS. Cell autonomy and synchrony of suprachiasmatic nucleus circadian oscillators. Trends in neurosciences. 2011;34:349–358. doi: 10.1016/j.tins.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Sugino K, Kula E, Okazaki E, Tachibana T, Nelson S, Rosbash M. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nature neuroscience. 2010;13:60–68. doi: 10.1038/nn.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahm SS, Farnell YZ, Griffith W, Earnest DJ. Circadian regulation and function of voltage-dependent calcium channels in the suprachiasmatic nucleus. J Neurosci. 2005;25:9304–9308. doi: 10.1523/JNEUROSCI.2733-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash HA, Scott RL, Lear BC, Allada R. An unusual cation channel mediates photic control of locomotion in Drosophila. Curr Biol. 2002;12:2152–2158. doi: 10.1016/s0960-9822(02)01358-1. [DOI] [PubMed] [Google Scholar]
- Park D, Griffith LC. Electrophysiological and anatomical characterization of PDF-positive clock neurons in the intact adult Drosophila brain. J Neurophysiol. 2006;95:3955–3960. doi: 10.1152/jn.00117.2006. [DOI] [PubMed] [Google Scholar]
- Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila free-running rhythms require intercellular communication. PLoS Biol. 2003;1:E13. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennartz CM, de Jeu MT, Bos NP, Schaap J, Geurtsen AM. Diurnal modulation of pacemaker potentials and calcium current in the mammalian circadian clock. Nature. 2002;416:286–290. doi: 10.1038/nature728. [DOI] [PubMed] [Google Scholar]
- Pitts GR, Ohta H, McMahon DG. Daily rhythmicity of large-conductance Ca2+ -activated K+ currents in suprachiasmatic nucleus neurons. Brain research. 2006;1071:54–62. doi: 10.1016/j.brainres.2005.11.078. [DOI] [PubMed] [Google Scholar]
- Raman IM, Gustafson AE, Padgett D. Ionic currents and spontaneous firing in neurons isolated from the cerebellar nuclei. J Neurosci. 2000;20:9004–9016. doi: 10.1523/JNEUROSCI.20-24-09004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome biology. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap J, Pennartz CM, Meijer JH. Electrophysiology of the circadian pacemaker in mammals. Chronobiology international. 2003;20:171–188. doi: 10.1081/cbi-120019311. [DOI] [PubMed] [Google Scholar]
- Seluzicki A, Flourakis M, Kula-Eversole E, Zhang L, Kilman V, Allada R. Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol. 2014;12:e1001810. doi: 10.1371/journal.pbio.1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Rosbash M, Truman JW. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci. 2002;22:5946–5954. doi: 10.1523/JNEUROSCI.22-14-05946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Gu H, Sharma VK, O’Dowd DK, Holmes TC. Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol. 2008;99:976–988. doi: 10.1152/jn.00930.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim CK, Forger DB. Modeling the electrophysiology of suprachiasmatic nucleus neurons. J Biol Rhythms. 2007;22:445–453. doi: 10.1177/0748730407306041. [DOI] [PubMed] [Google Scholar]
- Swayne LA, Mezghrani A, Varrault A, Chemin J, Bertrand G, Dalle S, Bourinet E, Lory P, Miller RJ, Nargeot J, et al. The NALCN ion channel is activated by M3 muscarinic receptors in a pancreatic beta-cell line. EMBO reports. 2009;10:873–880. doi: 10.1038/embor.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaiuolo M, Bertram R, Leng G, Tabak J. Models of electrical activity: calibration and prediction testing on the same cell. Biophysical journal. 2012;103:2021–2032. doi: 10.1016/j.bpj.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Gao S, Alcaire SM, Aoyagi K, Wang Y, Griffin JK, Stagljar I, Nagamatsu S, Zhen M. NLF-1 delivers a sodium leak channel to regulate neuronal excitability and modulate rhythmic locomotion. Neuron. 2013;77:1069–1082. doi: 10.1016/j.neuron.2013.01.018. [DOI] [PubMed] [Google Scholar]
- Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Yao Z, Shafer OT. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science. 2014;343:1516–1520. doi: 10.1126/science.1251285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Chung BY, Lear BC, Kilman VL, Liu Y, Mahesh G, Meissner RA, Hardin PE, Allada R. DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr Biol. 2010a;20:591–599. doi: 10.1016/j.cub.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, Emery P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol. 2010b;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.