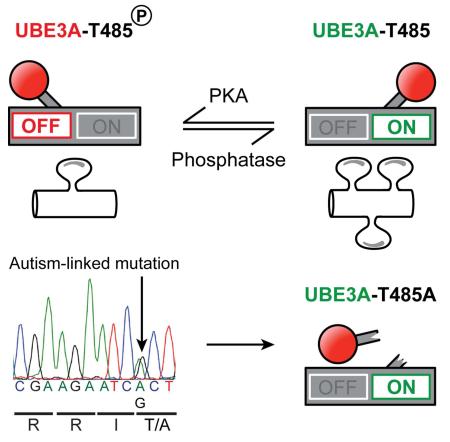
Summary
Deletion of UBE3A causes the neurodevelopmental disorder Angelman syndrome (AS) while duplication or triplication of UBE3A is linked to autism. These genetic findings suggest that the ubiquitin ligase activity of UBE3A must be tightly maintained to promote normal brain development. Here, we found that protein kinase A (PKA) phosphorylates UBE3A in a region outside the catalytic domain, at residue T485, and inhibits UBE3A activity towards itself and other substrates. A de novo autism-linked missense mutation disrupts this phosphorylation site, causing enhanced UBE3A activity in vitro, enhanced substrate turnover in patient-derived cells, and excessive dendritic spine development in the brain. Our study identifies PKA as an upstream regulator of UBE3A activity, and shows that an autism-linked mutation disrupts this phosphorylation control. Moreover, our findings implicate excessive UBE3A activity and the resulting synaptic dysfunction to autism pathogenesis.
Introduction
Autism is a genetically heterogeneous disorder associated with synaptic deficits, social impairment, and stereotyped behaviors. Recent studies indicate that copy number variations and de novo mutations in hundreds of genes can increase the risk for autism (Chen et al., 2015; De Rubeis et al., 2014; Iossifov et al., 2014). While some of these recently identified de novo mutations introduce stop codons and hence likely disrupt gene function, most (79%; 1,500 / 1,891) introduce missense mutations of unknown significance (Iossifov et al., 2014). How these missense mutations—representing the bulk of what has been discovered in exome sequencing studies—affect protein function or contribute to disease is currently unknown.
Intriguingly, we noticed that one of these missense mutations was in UBE3A. UBE3A is associated with cervical cancer and two neurodevelopmental disorders—Angelman syndrome (AS) and autism. Duplication or triplication of maternally inherited 15q11-13, the chromosomal location where UBE3A resides, is one of the most common cytogenetic events associated with autism (Glessner et al., 2009; Hogart et al., 2010). Individuals with one extra maternal copy of 15q11-13 display partial autism penetrance, whereas individuals with two extra copies display almost complete penetrance (Hogart et al., 2010; Urraca et al., 2013). UBE3A is the only gene in this region that is consistently expressed from the maternal, but not paternal, allele in mature neurons (Albrecht et al., 1997; Rougeulle et al., 1997; Vu and Hoffman, 1997), suggesting that abnormally elevated levels of UBE3A contribute to autism in 15q11-13 duplication syndrome. However, UBE3A is not the only gene duplicated in this syndrome, and pathogenicity in individuals with paternal 15q11-13 duplication has been reported, raising the possibility that additional genes in the region might increase autism risk (Germain et al., 2014; Urraca et al., 2013).
There is no doubt that deletion or null mutation of the maternal UBE3A allele causes AS, a disorder characterized by a happy demeanor with frequent smiling, speech impairment, severe intellectual disability, motor dysfunction, and seizures (Jiang et al., 1998; Kishino et al., 1997; Mabb et al., 2011). In mice, deletion of Ube3a impairs synapse development and plasticity, and recapitulates several neurobehavioral symptoms of AS (Greer et al., 2010; Jiang et al., 1998; Margolis et al., 2010; Sato and Stryker, 2010; Wallace et al., 2012; Yashiro et al., 2009).
UBE3A encodes a HECT domain E3 ubiquitin ligase that targets substrate proteins, including itself, for degradation (de Bie and Ciechanover, 2011). Given that loss of UBE3A causes AS, while increases in UBE3A are associated with autism, UBE3A levels and activity are likely to be under tight control during normal brain development. Autoregulation of UBE3A via self-targeted degradation is cited as a mechanism for maintaining UBE3A levels (de Bie and Ciechanover, 2011; Mabb et al., 2011; Nuber et al., 1998). However, such a mechanism is likely to be overly simplistic, as unchecked self-degradation could lead to self-elimination. We thus speculated that additional mechanisms might exist to control UBE3A activity. Here, we systematically examined how a large number of disease-linked UBE3A missense mutations affect protein levels and activity. These analyses revealed that UBE3A is inhibited by PKA phosphorylation at T485, a site that was recently found to be mutated in an autism proband (Iossifov et al., 2014). Mutation of this phosphorylation site abnormally elevates UBE3A activity and increases synapse number in vivo. Our study describes an upstream regulatory mechanism for UBE3A and provides a comprehensive understanding of how missense mutations linked to AS and autism affect UBE3A protein function.
Results
AS-linked missense mutations inactivate UBE3A via distinct mechanisms
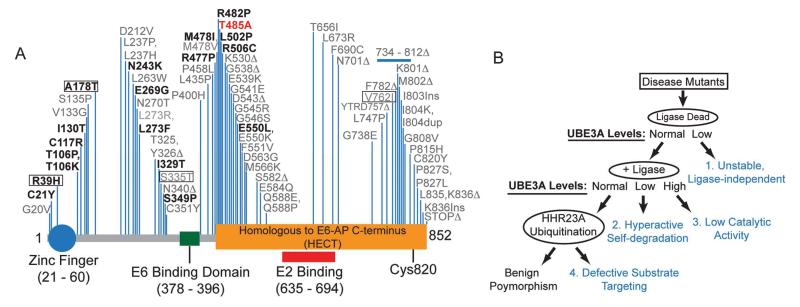
Most cases of AS arise due to a deletion of the maternal copy of UBE3A, but some AS patients (~10%) harbor missense mutations in the coding region of UBE3A (Sadikovic et al., 2014). Some of these AS-linked mutations cluster near the catalytic cysteine (C820) and disrupt the ubiquitin ligase activity of UBE3A (Sadikovic et al., 2014). However, the majority of these mutations are located far from the catalytic site. Precisely how most of these mutations, each of which changes a single amino acid, disrupt UBE3A function has not been resolved. After mapping all reported AS-linked missense mutations relative to the known domains in UBE3A, we noticed that these missense mutations were not randomly distributed, but clustered within distinct regions (Figure 1A). Based on this observation, we hypothesized there might be additional domains within UBE3A that control enzyme activity or stability.
Figure 1. Characterization of UBE3A missense mutations.
(A) Schematic showing human UBE3A with known functional domains, N-terminal zinc finger domain (Lemak et al., 2011), the location of non-truncating AS-linked mutations (black), and the de novo missense mutation identified in an autism proband (red). Mutations tested in our study are shown in bold and include benign variants (boxed).
(B) Approach used to determine how missense mutations disrupt UBE3A function. Mutations were introduced into a ligase-dead (LD, C820A) version of UBE3A. These mutants were expressed in HEK293T cells to identify mutations that were 1) unstable independent of UBE3A activity (all relative to UBE3A-LD). For the remaining stable mutants, ligase activity was restored (+Ligase) and the resulting constructs were transfected into HEK293T cells. This allowed us to identify mutations that 2) resulted in hyperactivity (these mutants would be detected at lower levels relative to WT UBE3A) or 3) reduced catalytic activity (these mutants would be detected at higher levels relative to WT UBE3A). Lastly, any UBE3A mutation (in +Ligase background) that was detected at equal levels relative to WT UBE3A was tested for its ability to target HHR23A for ubiquitination. This allowed us to identify mutations that 4) affect substrate targeting without affecting self-targeted degradation, or are benign polymorphisms.
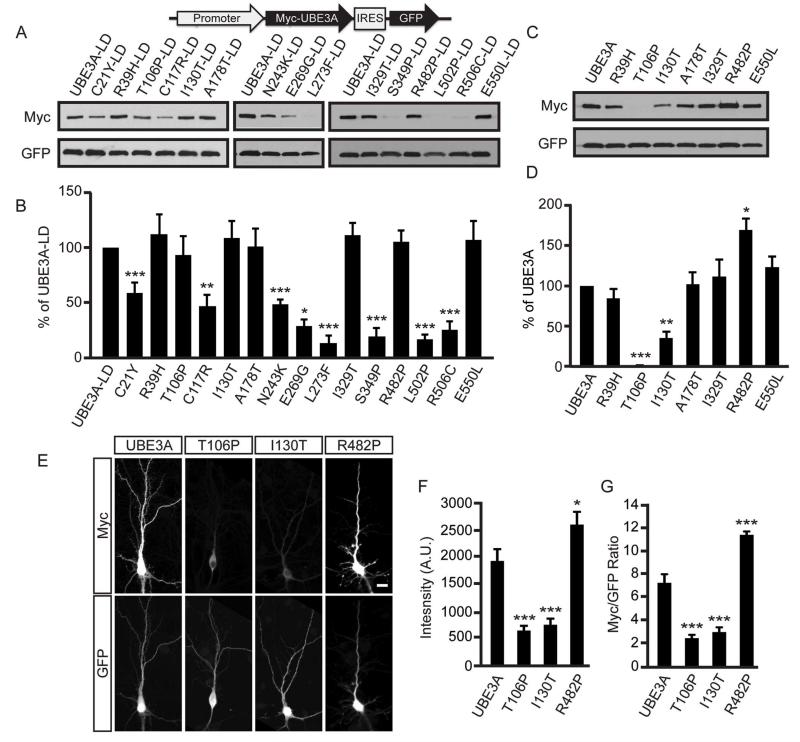
Like most E3 ubiquitin ligases, UBE3A mediates the ubiquitination of target proteins and itself (de Bie and Ciechanover, 2011; Kumar et al., 1999). These missense mutations could thus disrupt UBE3A in four different ways, each of which can be distinguished experimentally (Figure 1B, Table S1): 1) by affecting protein stability independent of ligase activity, 2) by promoting self-targeted degradation, 3) by disrupting the catalytic domain, or 4) by preventing UBE3A from targeting substrates for degradation. To determine if AS-linked missense mutations affect UBE3A stability independent of ligase activity, we introduced missense mutations (Figure 1A; highlighted in bold) into a ligase-dead (LD; C820A) version of human UBE3A, transfected expression constructs into HEK293T cells, and then monitored protein levels by western blotting. Of the 18 mutations tested, the protein levels of 8 were significantly lower than UBE3A-LD control (Figure 2A and 2B; Table S1). These data indicate that many AS-linked missense mutations destabilize UBE3A independent of its ligase activity.
Figure 2. AS-linked missense mutations in UBE3A cause loss-of-function via distinct mechanisms.
(A and B) Representative western blot of protein levels for AS-linked mutations introduced into ligase-dead (LD) UBE3A and quantification (B). All UBE3A constructs were Myc-tagged, contained an IRES-GFP to normalize for expression and transfection efficiency and were transfected into HEK293T cells. Values are shown as the percent ± standard error of UBE3A-LD levels. n=3-6/condition; **p<0.005, ***p<0.0005. See Supplemental Information for detailed statistical methods.
(C and D) Representative western blot of UBE3A mutants that possess ubiquitin ligase activity (+Ligase) and quantification (C). Values are shown as the mean percent ± standard error of WT UBE3A levels, n=4, *p<0.005, ***p<0.0005.
(E - G) Immunofluorescence staining of Myc-tagged UBE3A and mutants in DIV 10 mouse cortical neurons, scale bar, 15 μm. Raw intensity values for Myc immunofluorescence (F) or Myc immunofluorescence normalized to GFP (G) are shown as the mean ± standard error. n=16-19 neurons/condition; *p<0.05, ***p<0.0005.
We then introduced the remaining seven mutations into catalytically active wild-type (WT) UBE3A and found that protein levels of two mutants (T106P and I130T) were significantly lower than WT UBE3A, whereas the R482P mutant was detected at significantly higher levels (Figure 2C and 2D; Table S1). These findings suggested that the T106P and I130T mutations promoted hyperactive self-degradation of UBE3A, a novel gain-of-function mechanism that reduces UBE3A levels, while the R482P mutation impaired UBE3A activity. Endogenous UBE3A activity did not contribute to the effects of these mutants, as endogenous enzyme levels were very low in HEK293T cells (Figure S1A). We tested additional AS-linked missense mutations at T106 or near R482. T106K promoted self-degradation like T106P, while R477P and M478I impaired activity like R482P (Figure S1B-E, Table S1).
In further support that the T106P and I130T mutations promote self-degradation, protein levels of these mutants were rescued (elevated) following proteasomal inhibition with MG-132 (Figure S1F and S1G), and high molecular weight polyubiquitinated conjugates were detected on these mutants only when the ligase domain was intact (Figure S1H). Conversely, polyubiquitinated UBE3A conjugates were not detected on the R482P mutant (Figure S1H) and absent or reduced for the R477P and M478I mutants (Figure S1I), suggesting that these mutants had little or no ubiquitin ligase activity. Likewise, when transfected into primary mouse cortical neuron cultures, the protein levels of the T106P and I130P mutants were lower relative to WT UBE3A, while the R482P mutant was detected at higher levels (Figure 2E – 2G).
We next evaluated the extent to which the T106P, I130T, and R482P mutants targeted the UBE3A substrate HHR23A for degradation in HEK293T cells (Kumar et al., 1999). We found that protein levels of the UBE3A T106P and I130T mutants were low relative to WT UBE3A, and HHR23A levels were unaffected (Figure S1J and S1K). Moreover, the T106P and I130T mutants did not effectively polyubiquitinate HHR23A in cells that were treated with the proteasome inhibitor MG-132 (Figure S1L). These data collectively suggest that the T106P and I130T mutations endowed a gain-of-function that was specific for UBE3A self-degradation, which paradoxically drives UBE3A loss-of-function. In contrast, the UBE3A R482P mutation elevated protein levels of UBE3A and HHR23A (Figure S1J and S1K), further indicating loss-of-function.
The remaining four mutations (R39H, A178T, I329T, E550L) did not significantly alter UBE3A protein levels (Figure 2C and 2D; Table S1). Two of these mutations (R39H, A178T) are now recognized as benign variants and are not associated with AS (Malzac et al., 1998; Sadikovic et al., 2014). The E550L mutant does not effectively target the UBE3A substrate HHR23A for ubiquitination (Cooper et al., 2004), but how the I329T mutation affects UBE3A function is unknown (Camprubi et al., 2009). To determine if the I329T mutant is also defective in targeting substrates, we examined polyubiquitination of HHR23A relative to three controls (R39H, A178T and E550L). As expected, we found that the benign R39H and A178T variants polyubiquitinated HHR23A at levels similar to WT UBE3A (Figure S1M). In contrast, the I329T mutant behaved like the E550L mutant, and did not polyubiquitinate HHR23A to the same extent as WT UBE3A (Figure S1M). This reduction in substrate ubiquitination was reflected in steady-state HHR23A levels, which were elevated when co-expressed with the I329T and E550L mutants (Figure S1N and S1O). Collectively, our data indicate that AS-linked missense mutations inactivate UBE3A via distinct mechanisms: 1) through ligase-independent protein destabilization, 2) through enhanced ligase-dependent degradation of itself only, or 3) through failure to target substrates for ubiquitination. These experiments reveal how numerous AS-linked mutations disrupt UBE3A function and hence contribute to AS.
Phosphorylation inhibits UBE3A
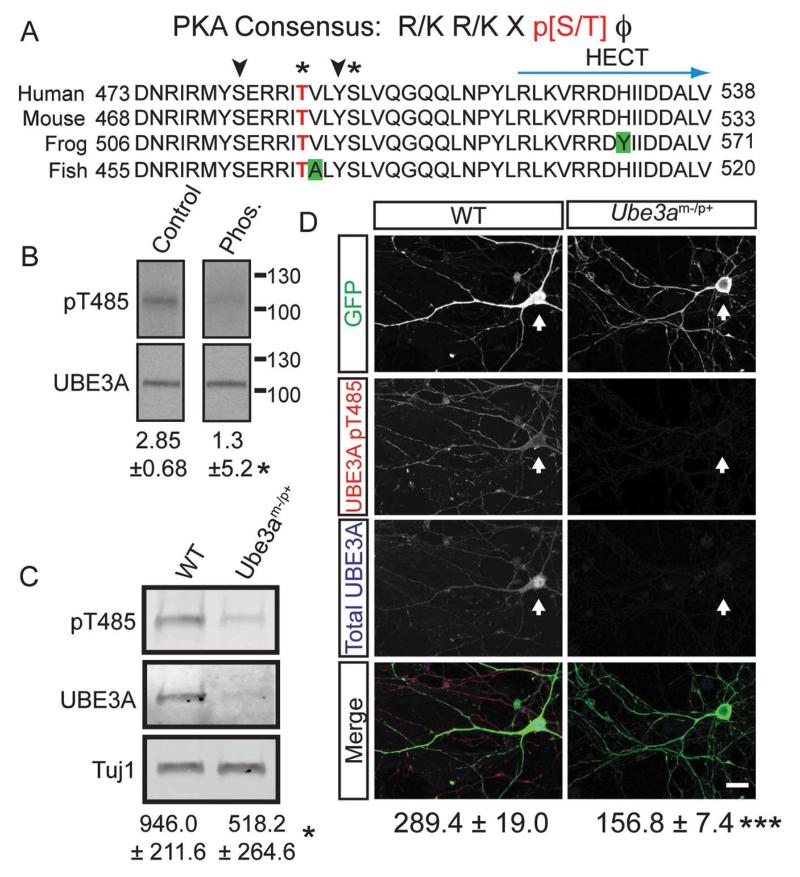
The R477P, M478I, and R482P mutations are tightly clustered with other missense mutations (Figure 1A) and are located within the α1 helix of UBE3A, a highly conserved structural element that controls enzyme activity in other HECT domain ubiquitin ligases (Pandya et al., 2010; Ronchi et al., 2014). In addition, arginine 482 forms a canonical PKA consensus motif (Songyang et al., 1994), with threonine 485 (T485) as the putative phospho-acceptor residue (Figure 3A). Given that other disease-relevant ubiquitin ligases are regulated by phosphorylation (Cohen, 2014; Ko et al., 2010), these observations suggested that phosphorylation at this site might control UBE3A activity. In support of this possibility, we found that T485 was phosphorylated using mass spectrometry (Figure S2A). We then generated a phospho-T485 (pT485) antibody and confirmed its specificity experimentally: 1) this antibody did not recognize UBE3A when T485 was mutated to alanine (T485A), a residue that cannot be phosphorylated (Figure S2B and S2C). 2) The antibody recognized a single band at ~110 kD in whole brain lysates that was sensitive to phosphatase treatment (Figure 3B). 3) This antibody was specific for UBE3A, as staining was detected in cultured cortical neurons from WT mice by western blot (Figure 3C) and by immunocytochemistry (Figure 3D), but was not detected in cultures from UBE3A-deficient (Ube3am−/p+) mice, which model AS. In immature neurons cultured for 10 days in vitro (DIV 10), phospho-UBE3A staining was cytoplasmic, and present in dendritic and axonal projections, but was largely excluded from the nucleus (Figure 3D, Figure S2E and S2F). This contrasted with total UBE3A, which was also in the nucleus (Figure 3D, Figure S2E and S2F). In mature neurons (DIV 18), UBE3A staining was predominantly nuclear, as expected (Sato and Stryker, 2010; Yashiro et al., 2009), while weak phospho-UBE3A immunoreactivity was detected in the cytoplasm and in dendritic spines (Figure S2G and S2H). These data suggest that endogenous UBE3A is phosphorylated at T485 in neurons and is excluded from the nucleus relative to total UBE3A.
Figure 3. UBE3A is phosphorylated at T485 in vivo.
(A) Sequence alignment showing conservation of human UBE3A residues 473 – 538. Green boxes indicate non-conserved residues, asterisks indicate phosphorylation identified by mass spectrometry, T485 highlighted in red, and arrowheads indicate potential phosphorylation sites.
(B) A phosphatase-sensitive UBE3A band is recognized by the UBE3A (pT485) antibody in brain lysates. Numbers indicate the mean ratio of UBE3A pT485 intensity to total UBE3A intensity ± standard error, n=3, *p<0.05.
(C) Protein lysates from DIV 10 WT and Ube3a-deficient (Ube3am−/p+) cortical neuron cultures were probed with the UBE3A pT485 antibody. The small amount of UBE3A remaining originates from non-neuronal cells that biallelically express UBE3A. Phospho-UBE3A levels were normalized to the neuron-specific marker Tuj1 and shown as the mean intensity ± standard error, n=3, *p<0.05.
(D) Confocal projections of UBE3A pT485 and total UBE3A in dissociated DIV10 cortical neurons from WT and Ube3am−/p+ embryos. Cell were transfected with GFP and labeled by immunofluorescence for UBE3A pT485 and total UBE3A. Arrows mark cells transfected with GFP, scale bar, 15 μm. The mean intensity ± standard error for pT485 immunofluorescence is indicated. WT, n = 20; Ube3am−/p+ n = 20, ***p<0.005.
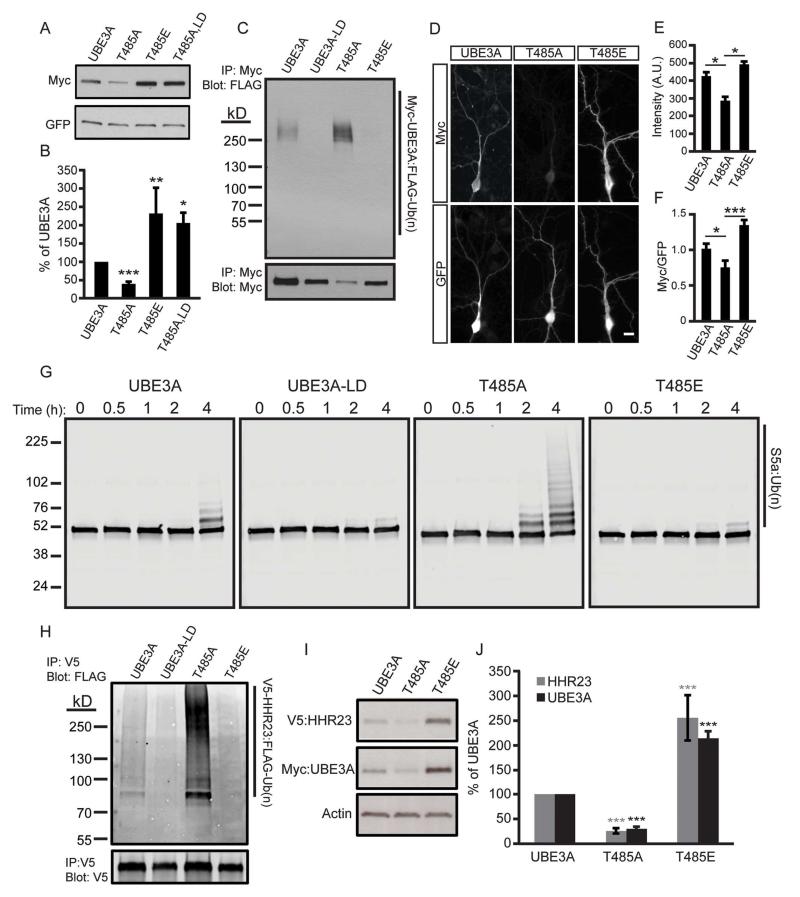
To study the functional significance of T485 phosphorylation, we transfected HEK293T cells with UBE3A constructs harboring the T485A mutation (which cannot be phosphorylated; Figure S2B and S2C), or a phospho-mimetic (T485E) mutation. We found that the T485A mutant was detected at low levels (40.0% ± 4.9), whereas a ligase-dead version of T485A (T485A, LD) was detected at higher levels (204.7% ± 26.5) relative to WT UBE3A (Figure 4A and 4B). The phospho-mimetic mutant (T485E) was detected at higher levels (232.1% ± 20.5) relative to WT UBE3A (Figure 4A and 4B). Furthermore, inhibiting protein synthesis with cycloheximide showed that the phospho-mimetic T485E mutant had a longer protein half-life when compared to the T485A mutant (Figure S3A and S3B). Collectively, these data suggest that phosphorylation at T485 inhibits self-ubiquitination of UBE3A, thereby increasing UBE3A levels, while dephosphorylation of T485 increases UBE3A activity and self-degradation. Indeed, ubiquitin-conjugated forms of UBE3A were observed with the T485A mutant but were not observed with the T485E mutant (Figure 4C). And, levels of T485A were elevated when the proteasome was inhibited with MG-132 (Figure S3C and S3D). Similar results were obtained in cortical neurons—the T485A mutation reduced UBE3A levels in neurons as compared to WT UBE3A, whereas the T485E mutation stabilized UBE3A levels (Figure 4D – 4F).
Figure 4. Phosphorylation at T485 inhibits UBE3A ubiquitin ligase activity.
(A and B) Representative western blot and quantification of HEK293T cells transfected with the indicated constructs. Values are shown as the percent of WT UBE3A levels ± standard error, n=4, *p<0.05, **p<0.005, ***p<0.0005.
(C) HEK293T cells transfected with the indicated Myc-UBE3A and FLAG-ubiquitin constructs were treated with the proteasome inhibitor MG-132 (30 μM, 4 h). UBE3A was immunoprecipitated using an anti-Myc antibody, and western blot probed with an anti-FLAG antibody to detect ubiquitinated UBE3A.
(D - F) Immunofluorescence staining and quantification of WT UBE3A and T485 mutants in DIV 10 mouse cortical neurons, scale bar 15 μm. Raw intensity values for Myc immunofluorescence (E) or Myc immunofluorescence normalized to GFP (F) are shown as the mean intensity ± standard error. T485A, n=14; T485E, n=16, *p<0.05, ***p<0.0005.
(G) In vitro ubiquitination assay was performed using S5a as the substrate and UBE3A mutants expressed and purified from HEK293T cells. Reactions were stopped after 4 h and the formation of high molecular weight polyubiquitinated S5a was monitored by western blot using an S5a antibody.
(H) HEK293T cells were transfected with the indicated Myc-UBE3A, FLAG-ubiquitin, and V5-HHR23A constructs and treated with MG-132. HHR23A was immunoprecipitated and western blot probed with an anti-FLAG antibody.
(I and J) Western blot and quantification of protein lysates from HEK293T cells transfected with the indicated constructs. Values are expressed as the mean percent ± standard error of protein levels in WT UBE3A expressing cells, n=4, ***p<0.0005.
Our mass spectrometry analyses indicated that serine 489 (S489) was also phosphorylated (Figure S2D). And, the neighboring serine 480 (S480) residue is located within a conserved consensus site for Akt phosphorylation (Figure S3E). We mutated each potential phosphorylation site in this region (S489, S480, and Y488) to residues that cannot be phosphorylated (Ala or Phe) then measured UBE3A levels by western blotting. Unlike the T485A mutation, none of these additional phosphorylation site mutations altered UBE3A levels (Figure S3F and S3G), suggesting that T485 is the principal site for posttranslational regulation in this region.
Next, we examined the extent to which the T485A and T485E mutants targeted substrates for ubiquitination. In vitro ubiquitination reactions were performed with S5a, a non-specific model substrate that has been used to assess the activities of various ubiquitin ligases (Jacobson et al., 2014). We performed our reactions with WT or UBE3A mutants expressed and purified from HEK293T cells. We found that the T485A mutant accelerated the formation of S5a ubiquitin conjugates as compared to WT UBE3A, whereas the UBE3A T485E mutant behaved like the ligase-dead UBE3A mutant (Figure 4G). We next monitored ubiquitination in cells using HHR23A. Consistent with our in vitro assays, the T485A mutant was hyperactive, generating more HHR23A-ubiquitin conjugates relative to WT UBE3A, whereas UBE3A-T485E did not ubiquitinate HHR23A (Figure 4H). Moreover, HHR23A levels were directly correlated with UBE3A levels in cells (Figure 4I and 4J). Taken together, these data suggest that phosphorylation at T485 inhibits the ubiquitin ligase activity of UBE3A towards itself and its substrates.
De novo UBE3A T485A mutation in an autism proband
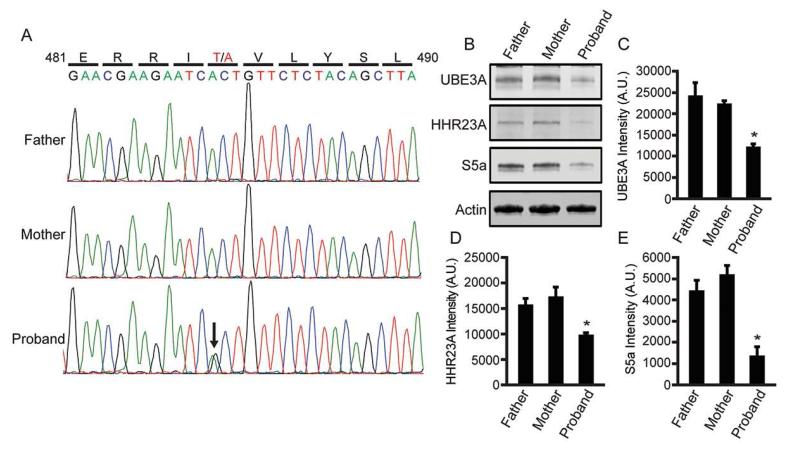
As part of a recent whole exome sequencing study, Iossifov and colleagues identified one autism proband, out of thousands of probands sequenced, with a de novo missense mutation in UBE3A (Iossifov et al., 2014). This one proband was characterized with autistic features, near normal IQ, and possessed an A:G substitution at chromosome 15:25,615,808; generating the same T485A missense mutation that, as we found above, disrupts the PKA phosphorylation site. We obtained immortalized lymphocyte cell lines from the affected child and the parents (Family ID: 13873) and sequenced PCR amplified fragments of genomic DNA encompassing T485. This analysis confirmed that the T485A mutation was present only in the child with autism (Figure 5A).
Figure 5. A de novo A>G (T485A) missense mutation in an autism proband enhances UBE3A substrate turnover.
(A) A genomic region of UBE3A from the father, mother, and the autism proband was amplified and sequenced from immortalized lymphocyte cell lines (the Simon’s Simplex Collection; Family ID: 13873).
(B - E) Endogenous protein levels of (C) UBE3A, (D) HHR23A, and (E) S5a in lymphocyte cell lines were quantified by western blot analysis. Protein levels were normalized to actin and shown as mean intensity ± standard error, n = 3, *p<0.05, A.U., arbitrary units.
Next, we assessed the relative abundance of endogenous substrates (UBE3A, S5a, and HHR23A) in lymphocyte-derived cell lines from the affected child and unaffected parents. Remarkably, all three substrates were significantly reduced in cells from the proband relative to the parents (Figure 5B – E). Collectively, our experiments indicate that T485A disables phosphorylation control over UBE3A ubiquitin ligase activity, leading to hyperactivation of UBE3A and enhanced targeting of itself and other substrates for degradation.
PKA is an upstream regulator of UBE3A
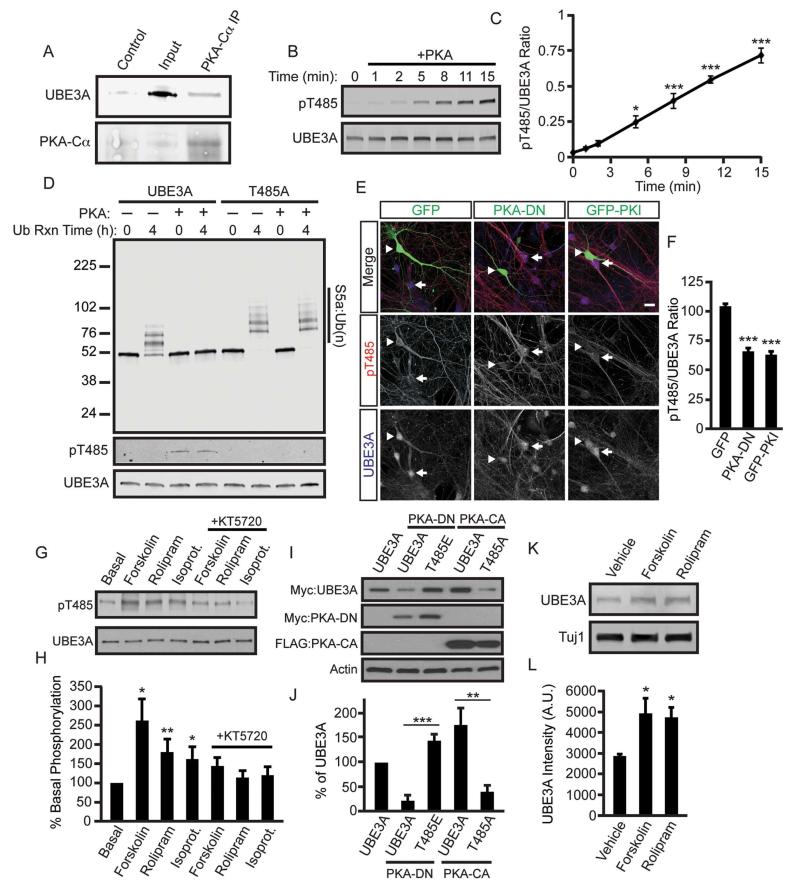
Since T485 is located within a canonical PKA consensus motif, we next evaluated the extent to which alterations in PKA activity influence UBE3A protein levels. We first performed co-immunoprecipitation experiments from cortical neuron lysates derived from postnatal day 1 (P1), a time when UBE3A phosphorylation peaks in the cortex (Figure S4A and S4B). We found that UBE3A can exist in a complex with the catalytic subunit of PKA (Figure 6A). Next, we tested whether UBE3A is a substrate of PKA by performing an in vitro kinase assay with recombinant proteins and monitoring UBE3A phosphorylation using our phospho-specific antibody. We found that PKA phosphorylates UBE3A at T485 in a time-dependent manner, demonstrating that UBE3A is a direct substrate of PKA (Figure 6B and 6C). We next assessed the functional significance of PKA phosphorylation on UBE3A ubiquitin ligase activity. We subjected phosphorylated UBE3A from our in vitro PKA reaction to an in vitro ubiquitination reaction and found that phosphorylation profoundly inhibited UBE3A ubiquitin ligase activity (Figure 6D). This effect was specific to phosphorylation at T485, as PKA was incapable of inhibiting the ubiquitin ligase activity of the T485A mutant (Figure 6D). Likewise, co-expression of WT UBE3A with increasing amounts of constitutively active PKA (PKA-CA) led to increased levels of HHR23A, while HHR23A levels remained low in cells expressing UBE3A-T485A and PKA-CA (Figure S4C, S4D). These data suggest that PKA phosphorylation at T485 inhibits UBE3A ubiquitin ligase activity in cells.
Figure 6. PKA regulates UBE3A activity.
(A) Co-immunoprecipitation from P1 mouse cortical lysate. Lysate was incubated with an anti-PKA-Cα antibody and the immunoprecipitates were analyzed by western blot with an anti-UBE3A antibody or no primary antibody as control.
(B and C) In vitro kinase assay showing phosphorylation of UBE3A by PKA. The reaction was performed with recombinant human PKA (catalytic subunit) and recombinant human UBE3A. Phosphorylation was monitored with the pT485 UBE3A antibody and quantified in (C). Values are shown as the ratio of UBE3A pT485 intensity to total UBE3A intensity ± standard error, n=3, *p<0.05, ***p<0.0005.
(D) An in vitro kinase reaction was performed with or without PKA using WT UBE3A and UBE3A T485A expressed and purified from HEK293T cells, followed by an in vitro ubiquitin ligase reaction with the substrate S5a. Ubiquitination was monitored at time 0 and 4 h by western blot with an anti-S5a antibody.
(E and F) PKA inhibition reduces basal levels of phosphorylated UBE3A in neurons. DIV11 mouse cortical neurons were transfected with GFP, PKA-DN, or GFP fused to the peptide inhibitor PKI (GFP-PKI). After 48 h, cells were fixed and immunostained with antibodies against GFP, UBE3A pT485, and total UBE3A. Arrowheads indicate the transfected GFP-positive neuron and arrows indicate nearby untransfected neurons, scale bar, 25 μm. UBE3A pT485A immunoreactivity is shown as the mean intensity ± standard error in (F). GFP: n=12, PKA-DN: n=16, GFP-PKI: n=16, ***p<0.0005.
(G and H) Phosphorylation of UBE3A T485 in dissociated DIV11 mouse cortical neurons following stimulation with forskolin (15 μM, 1 h), rolipram (75 μM, 1 h), isoproterenol (10 μM, 1 h), and inhibition of PKA with KT5720 (5 μM). (H) Quantification. Values were normalized to total UBE3A levels and are the mean percentage ± standard error of basal UBE3A phosphorylation, n=5, *p<0.05, **p<0.005. Statistical comparisons were made between appropriate agonist and agonist + inhibitor conditions.
(I and J) UBE3A T485 mutants are resistant to changes in PKA activity. Western blot and quantification of the indicated constructs co-transfected into HEK293T cells with DN or CA-PKA. Values are shown as the mean percent ± standard error of WT UBE3A levels, n=6, **p<0.005, ***p<0.0005.
(K and L) Chronic PKA activation increases the UBE3A levels in neurons. DIV 11 neurons were cultured for 48 h in the presence of 1 μM forskolin or 5 μM rolipram, UBE3A levels were analyzed by western blot and (L) quantified. UBE3A values were normalized to Tuj1 and are shown as the mean intensity ± standard error, n=3, *p<0.05.
In neurons, overexpression of a dominant-negative PKA mutant (PKA-DN) or expression of GFP fused to the PKA-specific inhibitory peptide PKI, resulted in markedly reduced basal phospho-UBE3A levels (Figure 6E and 6F). UBE3A phosphorylation at T485 was also reduced by KCl-induced depolarization (Figure S4E and S4F), suggesting strong activation of an upstream UBE3A phosphatase by neuronal activity. In contrast, phosphorylation was induced acutely with pharmacological activators of PKA, including agents that stimulate cyclic-AMP (cAMP) production, prevent cAMP breakdown, and activate Gαs signaling (Figure 6G and 6H). The PKA inhibitor KT5720 (5 μM) blocked all of these effects (Figure 6G and 6H). Moreover, we found that WT UBE3A levels increased when co-transfected with PKA-CA and decreased when co-transfected with PKA-DN, while the protein levels of the T485A mutant remained lower and the T485E mutant remained higher than WT UBE3A (Figure 6I and 6J). Self-degradation of the hyperactive AS disease-causing mutants (T106P and I130T) could likewise be rescued by co-expressing PKA-CA or by introducing the phospho-mimetic T485E mutation (Figure S4G and S4H).
Our findings also suggested that it should be possible to inhibit self-targeted UBE3A degradation by chronically increasing PKA activity. Indeed, long-term treatment (48 h) of neuronal cultures with forskolin or a phosphodieterase-4 inhibitor (rolipram) significantly increased UBE3A protein levels (Figure 6K and 6L). Altogether, these genetic and pharmacological data provide direct evidence that PKA is the predominant upstream protein kinase that phosphorylates UBE3A at T485.
Our findings raise the question of precisely how phosphorylation controls UBE3A ubiquitin ligase activity. Previous mapping experiments identified a roughly ~500 amino acid domain (residues ~280 – 768) in UBE3A that is required for intermolecular self-ubiquitination and interaction with E6-dependent substrates (Huibregtse et al., 1993; Nuber et al., 1998). T485 is located centrally within this domain, thus we tested whether modifications at T485 affect substrate binding. Indeed, the phospho-mutant (T485A) showed strong interaction with itself and with HHR23A in pulldown experiments, while WT UBE3A and the phospho-mimetic mutant (T485E) showed little to no interaction with substrates (Figure S4I and S4J). These data suggest that phosphorylation prevents UBE3A from binding to its substrate, hence preventing UBE3A from marking substrates for degradation.
Phosphorylation does not appear to affect other aspects of UBE3A catalysis. For example, the T485E phospho-mimetic mutant retained its ability to form a thioester bond with ubiquitin (Figure S4K). This suggested that phosphorylation does not inhibit the transfer of ubiquitin from the E2 enzyme, and does not inhibit the conjugation of ubiquitin to the catalytic cysteine of UBE3A. Moreover, the active form of UBE3A has been proposed to exist as a homo-trimeric complex (Ronchi et al., 2014). However, we did not detect homomeric interaction with WT or T485E UBE3A (Figure S4I), and the T485A mutant retained activity (as evidence by lower protein levels relative to WT) when harboring a mutation (F727D) that reportedly destabilizes trimeric UBE3A (Figure S4L and S4M).
Disruption of UBE3A phosphorylation impairs dendritic spine development in vivo
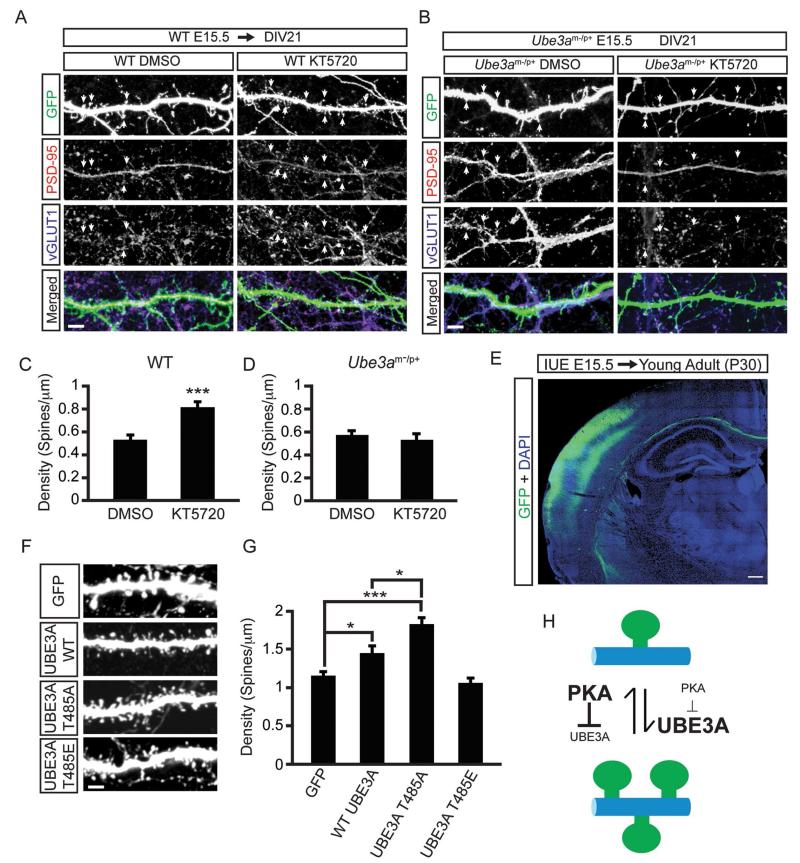
A previous study found that chronic pharmacological inhibition of PKA led to increased dendritic spine densities along with supernumerary synapses (Lu et al., 2011). To determine if this effect was mediated by UBE3A, we chronically treated E15.5 neurons from WT and Ube3a m−/p+ mice with the small molecule inhibitor KT5720 (on DIV 19), then processed cells for the excitatory postsynaptic marker PSD-95 and the excitatory presynaptic marker vGLUT1 (Figure 7A and 7B). Consistent with Lu and colleagues, we also found that WT neurons treated with KT5720 exhibited a significant increase in the densities of spines positive for PSD-95 and vGLUT1 when compared to DMSO (Figure 7A and 7C; WT DMSO, 0.54 ± 0.05; WT KT5720, 0.81 ± 0.05, spines/μm of dendrite). Notably however, this effect was absent in Ube3a m−/p+ neurons (Figure 7B and 7D; Ube3a m−/p+ DMSO 0.56 ± 0.04; Ube3a m−/p+ KT5720, 0.53 ± 0.05, spines/μm of dendrite). For WT neurons, these changes were accompanied by an increase in the number of paired or unpaired vGLUT1 and PSD-95 puncta (Figure S5A and S5B), but they were not accompanied by obvious morphological changes in spines (Fig. S5B). Moreover, chronic KT5720 treatment also reduced levels of phosphorylated UBE3A in spines and dendrites (Figure S5C – S5E). Altogether, these experiments suggest that UBE3A is a principal effector of PKA-dependent dendritic spine development.
Figure 7. UBE3A T485A phospho-mutant abnormally increases dendritic spine formation in vivo.
(A) Confocal projections showing synapse formation in WT neurons treated with vehicle (DMSO; left panels) or 1 μM KT5720 (right panels) for 48 h (DIV 19-21), scale bar, 4 μm.
(B) Confocal projections showing synapse formation in Ube3am−/p+ treated with vehicle (DMSO; left panels) or 1 μM KT5720 (right panels) for 48 h (DIV 19-21), scale bar, 4 μm. GFP is shown in green, PSD-95 immunofluorescence in red, and vGLUT1 immunofluorescence in blue.
(C and D) Quantification showing increased spine densities with KT5720 treatment in WT neurons (C) but not in Ube3am−/p+ neurons (D). Values are shown as the mean spine densities ± standard error. n=25-30 neurons/condition; ***p<0.0005.
(E) Low magnification confocal image showing transfected neurons (green) in the cortex of young adult (P30) animals after in utero electroporation (IUE) at E15.5. Nuclear stain DAPI (blue). Scale bar, 200 μm.
(F and G) Representative images of dendrites showing spine densities in neurons expressing the indicated constructs, and quantification (G); scale bar, 2 μm. Values are shown as mean spine densities ± standard error. n=20-25 neurons/condition, *p<0.05, ***p<0.0005.
(H) Model of PKA and UBE3A signaling in spine growth. Text size is proportional to enzyme activity level.
Lastly, we investigated whether phosphorylation at T485 affected spine formation in vivo by introducing GFP, WT UBE3A, the T485A, or the T485E mutants through in utero electroporation into the cerebral cortex of E15.5 animals. We then analyzed spines on primary basal dendrites of layer 2/3 pyramidal neurons in the somatosensory cortex of young adult (postnatal day 30; P30) mice (Figure 7E). Animals overexpressing WT UBE3A showed a modest increase in dendritic spine densities relative to the GFP control while spine density increased by ~58% in mice expressing the hyperactive T485A mutant (Figure 7F and 7G; GFP, 1.16 ± 0.05; UBE3A WT, 1.45 ± 0.08; UBE3A-T485A, 1.83 ± 0.08 spines/μm dendrite). In addition, dendritic head widths for both WT UBE3A and the T485A mutant were significantly decreased (Figure S5F and S5G), indicating subtle alterations to spine morphologies in addition to changes in density. In contrast, expression of the inactive phospho-mimetic T485E mutant had no effect on spine density (1.06 ± 0.06 spines/μm dendrite) or morphology relative to GFP only controls (Figure 7F and 7G, Figure S5F and S5G). Thus, expression of a constitutively active, phosphorylation defective version of UBE3A (T485A) promoted excessive spine formation that persisted into adulthood, while expression of the inactive phospho-mimetic mutant did not alter spine number relative to the control. These data collectively suggest that PKA dependent phosphorylation of UBE3A controls proper spine formation during brain development (Figure 7H).
Discussion
Phosphorylation at T485 inhibits UBE3A ubiquitin ligase activity
We noticed that numerous disease-linked missense mutations in UBE3A were distant from the active site and clustered together. At the epicenter of one of these mutational hotspots was a PKA phosphorylation site (T485). We found that this site can be phosphorylated in vitro, in cells, in neurons, and in the brain, and that phosphorylation of this site inhibits UBE3A ubiquitin ligase activity towards itself and its substrates. Our in vitro and in vivo experiments, including experiments with lymphocytes from an autism proband harboring a phospho-mutant T485A missense mutation, shows that T485 phosphorylation serves as a master switch—disengaging UBE3A from substrates and hence blocking UBE3A enzymatic activity. Our study shows that UBE3A is regulated by phosphorylation, and that an autism-linked mutation disables this phosphorylation control and impairs synapse formation in vivo.
Two additional de novo missense variants were identified in the autism proband we studied (Family ID: 13873). These variants include a V49I substitution in histone H2B type 1-J (HIST1H2BJ) and an E90K substitution in laminin α4 (LAMA4) (Iossifov et al., 2014). Although both variants await experimental characterization, existing data do not support a role for these variants in autism pathology. V49I is a conservative substitution, and neither HIST1H2BJ nor LAMA4 are expressed in the human brain (Su et al., 2004).
Collectively, our findings suggest that abnormal elevation of UBE3A activity, caused by the T485A missense mutation, contributes to autism pathology in this individual. Identifying additional autism-linked mutations that disrupt UBE3A T485 phosphorylation will strengthen this assertion. Moreover, our work provides further evidence that excess UBE3A activity in the brain increases risk for autism. Future studies will be needed to assess phenotypic similarities between this genetically precise T485A point mutation that hyperactivates UBE3A, 15q11-13 duplication that elevates UBE3A and other genes, and a recently identified family with a 15q11.2 duplication that encompasses only UBE3A and that segregates with autistic features, developmental delay, depression and schizophrenia (Noor et al., 2015).
AS-linked mutations destabilize UBE3A via distinct loss- and gain-of-function mechanisms
We found that most AS-linked missense mutations destabilize UBE3A independent of ubiquitin ligase activity, making them classic loss-of-function mutations. Our work also identified a cluster of mutations (R477P, M478I, R482P) that were distant from the active site that blocked UBE3A substrate targeting. The R477P, M478I, and R482P mutations all reside near the T485 phosphorylation site, and these mutations inhibited UBE3A in a manner analogous to phosphorylation at T485. Given the importance of this region for substrate and homomeric interactions (Figure S4I and S4J), these mutations likely act independent of UBE3A phosphorylation and disrupt the docking of substrates to UBE3A, thereby causing a loss-of-function. In support of this mechanism, we found that the R482P mutation dominantly inhibited the hyperactive T485A mutation (Figure S5H and S5I).
Our study also identified mutations that differentially affect UBE3A targeting of itself and other substrates. One cluster of mutations (T106P, T106K, I130T) enhanced self-targeting without affecting the targeting of other substrates. The converse was found with mutations that flank the α1 helical region (I329T, E550L). These mutations blocked substrate targeting without affecting self-targeting. Collectively, our study highlights how a rigorous characterization of disease-linked missense mutations can provide fundamental new insights into enzyme catalysis and enzyme regulation under normal and pathological conditions.
Hyperactive UBE3A drives abnormal spine formation in the brain
We found that phosphorylation of UBE3A peaks in the first week of postnatal life in the mouse cerebral cortex, which corresponds to a developmental window when synaptic plasticity is driven primarily by PKA signaling (Lu et al., 2007; Yasuda et al., 2003). PKA might thus regulate UBE3A activity within the first ~7 days of life to allow for the proper progression of synapse development. In support of this idea, we found that chronic PKA inhibition failed to increase dendritic spine density in Ube3a-deficient neurons, and overexpression of UBE3A-T485A, a mutant that cannot be phosphorylated, profoundly increased dendritic spine density in vivo. This increase in spine density persisted into young adulthood (P30), suggesting that the hyperactive UBE3A T485A mutant promotes long-lasting structural changes in the brain. The T485A mutation was identified in a child with autism, a disorder associated with increased spine number (Hutsler and Zhang, 2010; Piochon et al., 2014; Tang et al., 2014), further suggesting the hyperactive UBE3A mutant contributes to autism pathology. In light of our findings, a reduction in PKA activity, which reduces phosphorylation at T485 and elevates UBE3A activity (Figure 7H), might contribute to autism in a larger number of patients than presently recognized.
Lastly, our findings have therapeutic implications. We found that chronic treatment of neurons with pharmacological agents that stimulate PKA can turn down UBE3A activity (Figure 6K and 6L). This suggests it may be possible to reduce UBE3A activity in individuals with 15q11-13 duplication/triplication forms of autism using such agents. Excessive UBE3A activity is also associated with the vast majority of all cervical cancers (Pisani et al., 1999; Walboomers et al., 1999). Our findings thus raise the possibility that targeting upstream regulators of UBE3A could provide a general strategy for treating neurological disorders as well as cancers linked to excessive UBE3A activity.
Experimental Procedures
Animals
C57BL/6 WT and Ube3am−/p+ mouse lines and associated genotyping procedures have been previously described (Huang et al., 2012; Yashiro et al., 2009). All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill, and in accordance with NIH guidelines. Embryonic day 0.5 (E0.5) was defined as noon following initiation of timed mating.
Molecular biology
The nomenclature used in this study is based on the amino acid sequence of human UBE3A isoform I (accession: NP_570853.1). Detailed methods can be found in the Supplemental Information.
Primary Neuron Cultures
Neuronal cultures were prepared from E13.5 to E15.5 C57BL/6 WT or Ube3am−/p+ mice as previously described (Huang et al., 2012). Mouse cortices were dissected and trypsinized at 37°C for 10 min, and dissociated with a fire-polished Pasteur pipette in plating medium (Neurobasal medium with 5% fetal bovine serum, Glutamax, B27 (Life Technologies)), and Antibiotic-Antimycotic. Dissociated neurons were seeded onto 12 or 24-well plates with or without coverslips coated with poly-D-lysine (0.1 mg/ml) at a density of about 526 cells/mm2. Cultured neurons were maintained at 37°C with 5% CO2 and supplemented with Neurobasal medium containing 4.84 mg/ml Uridine 5′-triphosphate (Sigma), 2.46 mg/ml 5-fluoro 2-deoxyuridine (Sigma), Glutamax, B27, and 1× Antibiotic-Antimycotic at days in vitro (DIV) 3 and 9.
In utero electroporation
Embryos (E15.5) from timed-pregnant CF-1 females were isolated and their lateral ventricles injected with 1-2 μg of plasmid DNA. Five electrical pulses were delivered at 30V (50 ms duration) with a 950 ms interval using 5 mm paddle electrodes. The embryos were then placed back into the female. After birth, the neonates were transferred on postnatal day 1 (P1) to foster mothers. At P30, mice were sacrificed and their brains processed for analysis.
Statistical Analysis
Statistical analyses were performed using the GraphPad Prism software. Statistical treatments for each experiment can be found in the Supplemental Information.
Supplementary Material
Acknowledgments
We thank Gabriela Salazar and Jayalakshmi Miriyala for technical assistance, Bonnie Taylor-Blake for help with confocal imaging, Yaohong Wu for help with in utero electroporations, David Smalley and Lee Graves for performing mass spectrometry analysis, Lei Xing for cortical lysates, and Rong Mao and Tatiana Tvrdik from The University of Utah and ARUP Laboratories for disclosing the UBE3A c.349T>C; p.C117R mutation. Immortalized lymphocytes from the Simons Simplex Collection were acquired from the NIMH Center for Genetic Studies via the Rutgers University Cell and DNA Repository. This work was supported by grants from the Angelman Syndrome Foundation (M.J.Z., B.D.P.), a Grant-In-Aid from the Foundation for Angelman Syndrome Therapeutics (M.J.Z.), The Simons Foundation (SFARI Award 274426, B.D.P.), the NIMH (R01MH093372; M.J.Z., B.D.P.), NINDS (R01NS085093, B.D.P.), a NIH Pioneer Award from The National Institutes of Health (DP1ES024088; M.J.Z.), the NIGMS (P01-GM103723; K.M.H.), and a Basic and Clinical Grant (#7760) from Autism Speaks (K.M.H.). J.J.Y. is a recipient of the Christina Castellana postdoctoral fellowship of the Foundation for Angelman Syndrome Therapeutics and was supported by NICHD (T32HD040127). The confocal imaging core was funded by grants from NINDS (P30NS045892) and NICHD (U54HD079124).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information includes Extended Experimental Procedures, one supplemental Table and five supplemental figures.
References
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- Camprubi C, Guitart M, Gabau E, Coll MD, Villatoro S, Oltra S, Rosello M, Ferrer I, Monfort S, Orellana C, et al. Novel UBE3A mutations causing Angelman syndrome: different parental origin for single nucleotide changes and multiple nucleotide deletions or insertions. Am J Med Genet A. 2009;149A:343–348. doi: 10.1002/ajmg.a.32659. [DOI] [PubMed] [Google Scholar]
- Chen JA, Penagarikano O, Belgard TG, Swarup V, Geschwind DH. The emerging picture of autism spectrum disorder: genetics and pathology. Annu Rev Pathol. 2015;10:111–144. doi: 10.1146/annurev-pathol-012414-040405. [DOI] [PubMed] [Google Scholar]
- Cohen P. Immune diseases caused by mutations in kinases and components of the ubiquitin system. Nat Immunol. 2014;15:521–529. doi: 10.1038/ni.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper EM, Hudson AW, Amos J, Wagstaff J, Howley PM. Biochemical analysis of Angelman syndrome-associated mutations in the E3 ubiquitin ligase E6-associated protein. J Biol Chem. 2004;279:41208–41217. doi: 10.1074/jbc.M401302200. [DOI] [PubMed] [Google Scholar]
- de Bie P, Ciechanover A. Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 2011;18:1393–1402. doi: 10.1038/cdd.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain ND, Chen PF, Plocik AM, Glatt-Deeley H, Brown J, Fink JJ, Bolduc KA, Robinson TM, Levine ES, Reiter LT, et al. Gene expression analysis of human induced pluripotent stem cell-derived neurons carrying copy number variants of chromosome 15q11-q13.1. Mol Autism. 2014;5:44. doi: 10.1186/2040-2392-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, Griffith EC, Waldon Z, Maehr R, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogart A, Wu D, LaSalle JM, Schanen NC. The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol Dis. 2010;38:181–191. doi: 10.1016/j.nbd.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, Sciaky N, Dutton JW, Jr., Lee HM, Chen X, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481:185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010;1309:83–94. doi: 10.1016/j.brainres.2009.09.120. [DOI] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson AD, MacFadden A, Wu Z, Peng J, Liu CW. Autoregulation of the 26S proteasome by in situ ubiquitination. Mol Biol Cell. 2014;25:1824–1835. doi: 10.1091/mbc.E13-10-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Ko HS, Lee Y, Shin JH, Karuppagounder SS, Gadad BS, Koleske AJ, Pletnikova O, Troncoso JC, Dawson VL, Dawson TM. Phosphorylation by the c-Abl protein tyrosine kinase inhibits parkin’s ubiquitination and protective function. Proc Natl Acad Sci U S A. 2010;107:16691–16696. doi: 10.1073/pnas.1006083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Talis AL, Howley PM. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem. 1999;274:18785–18792. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- Lemak A, Yee A, Bezsonova I, Dhe-Paganon S, Arrowsmith CH. Zn-binding AZUL domain of human ubiquitin protein ligase Ube3A. J Biomol NMR. 2011;51:185–190. doi: 10.1007/s10858-011-9552-y. [DOI] [PubMed] [Google Scholar]
- Lu Y, Allen M, Halt AR, Weisenhaus M, Dallapiazza RF, Hall DD, Usachev YM, McKnight GS, Hell JW. Age-dependent requirement of AKAP150-anchored PKA and GluR2-lacking AMPA receptors in LTP. EMBO J. 2007;26:4879–4890. doi: 10.1038/sj.emboj.7601884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Zha XM, Kim EY, Schachtele S, Dailey ME, Hall DD, Strack S, Green SH, Hoffman DA, Hell JW. A kinase anchor protein 150 (AKAP150)-associated protein kinase A limits dendritic spine density. J Biol Chem. 2011;286:26496–26506. doi: 10.1074/jbc.M111.254912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabb AM, Judson MC, Zylka MJ, Philpot BD. Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes. Trends Neurosci. 2011;34:293–303. doi: 10.1016/j.tins.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzac P, Webber H, Moncla A, Graham JM, Kukolich M, Williams C, Pagon RA, Ramsdell LA, Kishino T, Wagstaff J. Mutation analysis of UBE3A in Angelman syndrome patients. Am J Hum Genet. 1998;62:1353–1360. doi: 10.1086/301877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis SS, Salogiannis J, Lipton DM, Mandel-Brehm C, Wills ZP, Mardinly AR, Hu L, Greer PL, Bikoff JB, Ho HY, et al. EphB-mediated degradation of the RhoA GEF Ephexin5 relieves a developmental brake on excitatory synapse formation. Cell. 2010;143:442–455. doi: 10.1016/j.cell.2010.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A, Dupuis L, Mittal K, Lionel AC, Marshall CR, Scherer SW, Stockley T, Vincent JB, Mendoza-Londono R, Stavropoulos DJ. 15q11.2 Duplication Encompassing only the UBE3A Gene is Associated with Developmental Delay and Neuropsychiatric Phenotypes. Hum Mutat. 2015 doi: 10.1002/humu.22800. doi: 10.1002/humu.22800. [DOI] [PubMed] [Google Scholar]
- Nuber U, Schwarz SE, Scheffner M. The ubiquitin-protein ligase E6-associated protein (E6-AP) serves as its own substrate. Eur J Biochem. 1998;254:643–649. doi: 10.1046/j.1432-1327.1998.2540643.x. [DOI] [PubMed] [Google Scholar]
- Pandya RK, Partridge JR, Love KR, Schwartz TU, Ploegh HL. A structural element within the HUWE1 HECT domain modulates self-ubiquitination and substrate ubiquitination activities. J Biol Chem. 2010;285:5664–5673. doi: 10.1074/jbc.M109.051805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piochon C, Kloth AD, Grasselli G, Titley HK, Nakayama H, Hashimoto K, Wan V, Simmons DH, Eissa T, Nakatani J, et al. Cerebellar plasticity and motor learning deficits in a copy-number variation mouse model of autism. Nat Commun. 2014;5:5586. doi: 10.1038/ncomms6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisani P, Parkin DM, Bray F, Ferlay J. Estimates of the worldwide mortality from 25 cancers in 1990. Int J Cancer. 1999;83:18–29. doi: 10.1002/(sici)1097-0215(19990924)83:1<18::aid-ijc5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Ronchi VP, Klein JM, Edwards DJ, Haas AL. The active form of E6-associated protein (E6AP)/UBE3A ubiquitin ligase is an oligomer. J Biol Chem. 2014;289:1033–1048. doi: 10.1074/jbc.M113.517805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- Sadikovic B, Fernandes P, Zhang VW, Ward PA, Miloslavskaya I, Rhead W, Rosenbaum R, Gin R, Roa B, Fang P. Mutation Update for UBE3A Variants in Angelman Syndrome. Hum Mutat. 2014;35:1407–1417. doi: 10.1002/humu.22687. [DOI] [PubMed] [Google Scholar]
- Sato M, Stryker MP. Genomic imprinting of experience-dependent cortical plasticity by the ubiquitin ligase gene Ube3a. Proc Natl Acad Sci U S A. 2010;107:5611–5616. doi: 10.1073/pnas.1001281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83:1131–1143. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urraca N, Cleary J, Brewer V, Pivnick EK, McVicar K, Thibert RL, Schanen NC, Esmer C, Lamport D, Reiter LT. The interstitial duplication 15q11.2-q13 syndrome includes autism, mild facial anomalies and a characteristic EEG signature. Autism Res. 2013;6:268–279. doi: 10.1002/aur.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu TH, Hoffman AR. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat Genet. 1997;17:12–13. doi: 10.1038/ng0997-12. [DOI] [PubMed] [Google Scholar]
- Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Wallace ML, Burette AC, Weinberg RJ, Philpot BD. Maternal loss of Ube3a produces an excitatory/inhibitory imbalance through neuron type-specific synaptic defects. Neuron. 2012;74:793–800. doi: 10.1016/j.neuron.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro K, Riday TT, Condon KH, Roberts AC, Bernardo DR, Prakash R, Weinberg RJ, Ehlers MD, Philpot BD. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009;12:777–783. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda H, Barth AL, Stellwagen D, Malenka RC. A developmental switch in the signaling cascades for LTP induction. Nat Neurosci. 2003;6:15–16. doi: 10.1038/nn985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.