Abstract
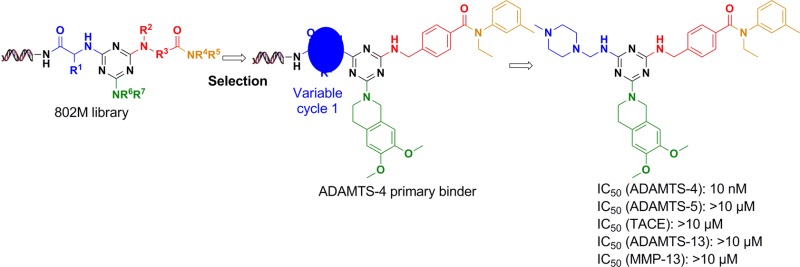
The aggrecan degrading metalloprotease ADAMTS-4 has been identified as a novel therapeutic target for osteoarthritis. Here, we use DNA-encoded Library Technology (ELT) to identify novel ADAMTS-4 inhibitors from a DNA-encoded triazine library by affinity selection. Structure–activity relationship studies based on the selection information led to the identification of potent and highly selective inhibitors. For example, 4-(((4-(6,7-dimethoxy-3,4-dihydroisoquinolin-2(1H)-yl)-6-(((4-methylpiperazin-1-yl)methyl)amino)-1,3,5-triazin-2-yl)amino)methyl)-N-ethyl-N-(m-tolyl)benzamide has IC50 of 10 nM against ADAMTS-4, with >1000-fold selectivity over ADAMT-5, MMP-13, TACE, and ADAMTS-13. These inhibitors have no obvious zinc ligand functionality.
Keywords: DNA-encoded library, ADAMTS-4, inhibitors
Type II collagen and aggrecan are two major components of cartilage matrix.1 Disregulation of proteolytic enzymes can result in cartilage destruction, leading to joint injury and diseases such as rheumatoid and osteoarthritis.2 Currently NSAIDS (nonsteroidal anti-inflammatories) are used to alleviate the symptoms of osteoarthritis. However, this treatment offers little, if any, effect on the progress of the disease. One expectation is that inhibiting aggrecanase should directly slow cartilage breakdown as compared to current treatment for joint disease.3,4 One choice for such therapeutic intervention is the aggrecanase family of proteins (ADAMTS). This family of zinc metalloproteases is characterized by degradation of aggrecan.5 Two closely related members, ADAMTS-4 (Aggrecanase-1) and ADAMTS-5 (Aggrecanase-2), cleave aggrecan at the same site (Glu373-Ala374), which makes them attractive targets for the treatment of osteoarthritis.6
Most metalloprotease (MMP) inhibitors were reported to contain moieties that bind to the zinc, such as carboxylate, hydroxamate, hydantoin, and sulfhydryl groups.7 Even though such MMP inhibitors like Marimastat, Batimastat, CGS-27023A, and Prinomastat demonstrated significant preclinical efficacy in vivo, they exhibited musculoskeletal side-effects in humans.8 The identification of selective and potent inhibitors, preferably without obvious metal chelating groups, was our goal. Unfortunately there are limited examples of ADAMTS-4 inhibitors with potent activity and high selectivity reported in the literature.9,10
DNA-encoded library technology has been developed over a decade both in academia and industry. It has been successfully applied in the discovery of small molecule hits for various targets.11−16 This makes it possible to create and screen a library containing large numbers of compounds through affinity binding and provides an alternative solution in drug discovery along with current techniques such as high throughput screening. Many potent inhibitors for various targets have been successfully discovered through this technology, including inhibitors for ADAMTS-5.17−21 This Letter will discuss the discovery of low nanomolar, highly selective inhibitors of ADAMTS-4 using ELT.
As we reported earlier, a DNA-encoded library based on a triazine scaffold and containing 800 million compounds has been synthesized and has already delivered potent inhibitors for both Aurora A kinase and p38 MAP kinase.12 In this library, 192 Fmoc amino acids were employed in chemistry cycle 1, followed by installation of triazine with cyanuric chloride. Then 32 amino acids were incorporated in cycle 2, followed by substitution of chlorine with 340 amines in cycle 3. In cycle 4, 384 amines were acylated with cycle 2 acid, yielding a 4-cycle library size of 802,160,640 (Figure 1a). Short double-stranded DNA tags encoding each building block were ligated to the opposite side of the headpiece at each stage of the small molecule chemistry.
Figure 1.
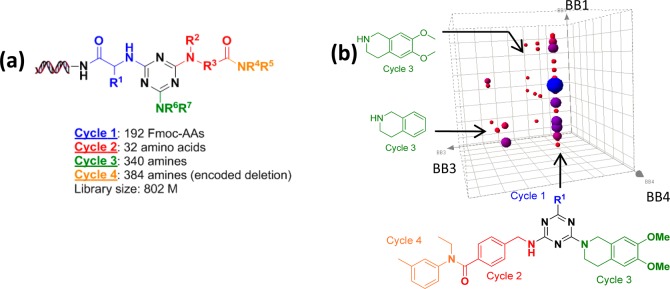
Selection of library against ADAMTS-4. (a) The triazine library. (b) The cube corresponding to 4-(aminomethyl)benzoic acid as cycle 2 is shown in detail. The copy number of the selected species is indicated by the continuous color and size of the points (darker and bigger points indicate higher copy number). The structure shown at the bottom represents the family of compounds defined by a line along the BB1 axis (arrow).
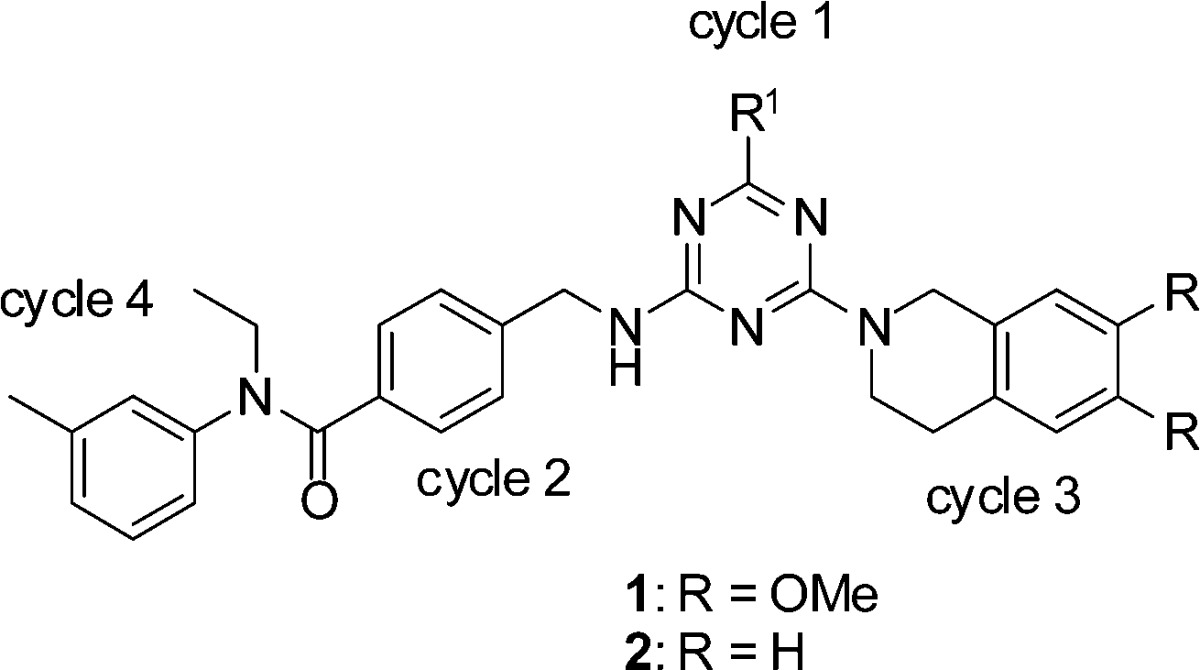
The affinity selection was performed on His-6 labeled ADAMTS-4 protein immobilized on nickel resin. The library was passed over the bound protein for 1 h and the nonbinders were washed away. The protein was denatured, and the binding population was recovered. Two additional rounds of selection were then performed, using fresh protein at each round. After final PCR amplification and DNA sequencing, we obtained 88 242 sequences, which encompassed 36 061 unique structures. After removing all sequences that occurred only once, the remaining families of chemotypes all contained 4-(aminomethyl)benzoic acid as selected synthon at cycle 2. Preferences of the other chemistry cycles can be visualized in a 3D scatter plot (Figure 1b). There was a highly populated plane, which corresponds to 6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline along the cycle 3 axis. Within this plane, there was a line corresponding to a family defined by the presence of N-ethyl-3-methylaniline as the cycle 4 synthon, with no observable preference for cycle 1. Notably, there was another less populated plane, which corresponds to 1,2,3,4-tetrahydroisoquinoline along the cycle 3 axis. The similarity between these two selected cycle 3 synthons provided additional support to the authenticity of the chemotype.
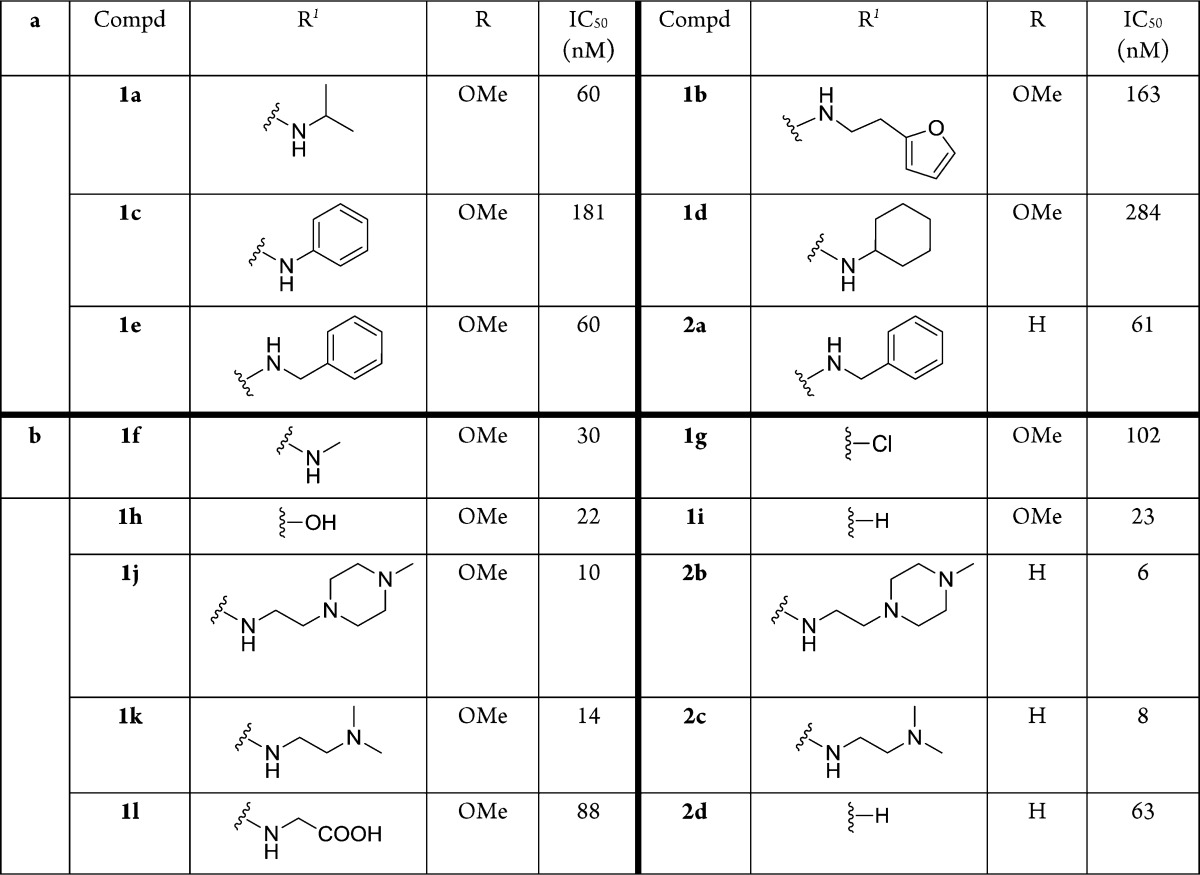
To confirm the selected features, a representative set of the fully elaborated library warhead molecules were synthesized (Table 1a) using the protocol in Scheme 1. The building blocks at cycle 1 were the decarboxy analogues of the cycle 1 amino acids. The biochemical assay showed these compounds (1a–1e, 2a) had activity ranging from 60 to 284 nM.
Table 1. (a) Full-Length Off-DNA Molecules and Their Activities; (b) SAR at Cycle 1 Position.
Scheme 1. Synthesis of ADAMTS-4 Off-DNA Hits.
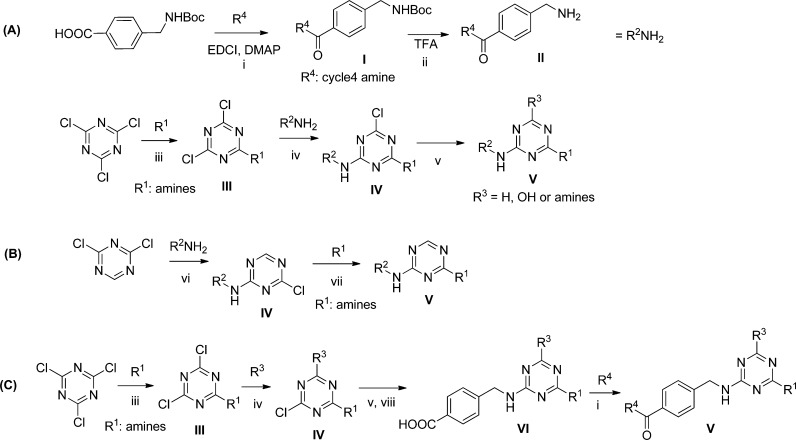
Reagents and conditions: (i) amine (1 equiv), EDCI (1.25 equiv), DMAP (0.2 equiv), CH2Cl2, 0 °C – rt; (ii) TFA (50% in CH2Cl2), room temperature, 20 min; (iii) amine (1 equiv), CH3CN/H2O (1/1), pH 9–10, 0 °C; (iv) amine (R2NH2 or R3) (1 equiv), CH3CN/H2O (1/1), pH 9–10, room temperature overnight; (v) R3 = H: Pd/C, H2, room temperature 1 h; R3 = OH/HCl (6 N, 10% v/v), CH3CN/H2O (1/1), 80 °C overnight; R3 = amines/amine (5–10 equiv), CH3CN/H2O (1/1), 80 °C; (vi) amine (1 equiv), DIEA (2.5 equiv), NMP, 0 °C; (vii) amine (5 equiv), NMP, 80 °C for 1 h; (viii) 1 N NaOH, room temperature overnight.
Additional truncations and modifications were performed at the cycle 1 position on triazine (Table 1b). In the case of 6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline derivative, truncation of the R1 to methyl amine improved activity (1f). The chlorine intermediate (1g) also showed good activity. Both the hydrolyzed byproduct (1h) and the complete deletion of cycle 1 (1i) yielded better activity than the amine derivatives (1a–1e). To increase the solubility of the compounds, we installed diamines and glycine at the cycle 1 position, and they all showed potent activity (1j, 1k, 1l). In the case of 1,2,3,4-tetrahydroisoquinoline as cycle 3 synthon, both the diamine derivatives (2b, 2c) and the complete deletion of cycle 1 (2d) yielded similar or better activity compared to the 6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline derivatives. All of the compounds confirmed the selection result that the cycle 1 was not necessary for the activity. Since cycle 1 was also the position linked to the DNA, we proposed that it might be the position exposed to the solvent and, as such, provide a handle that could be used to improve the physicochemical properties of the inhibitors.
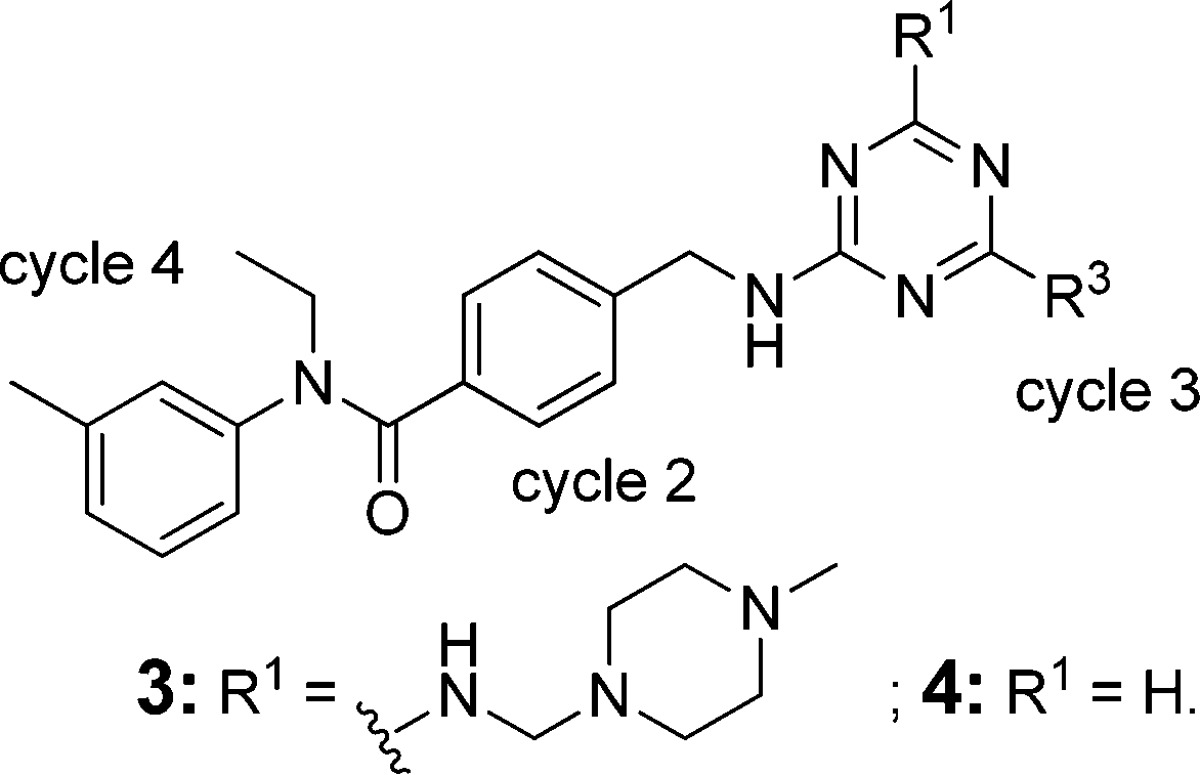
We next turned our attention to the cycle 3 position on triazine (Table 2). Changing tetrahydroisoquinoline to tetrahydroquinoline (3) and isoindoline (4c), the activity dramatically dropped from 10 nM to 5–6 μM. Alternative substituents, such as F, at 6-position (4a) and 7-position (4b) lost about 30-fold activity. A simple piperidine derivative (4e), which was included in the library, but was not selected, resulted in a complete loss of activity. However, with (3,4-dimethoxyphenyl)methanamine at R3 position, the compound still maintained excellent activity (4d).
Table 2. SAR at Cycle 3 Position.
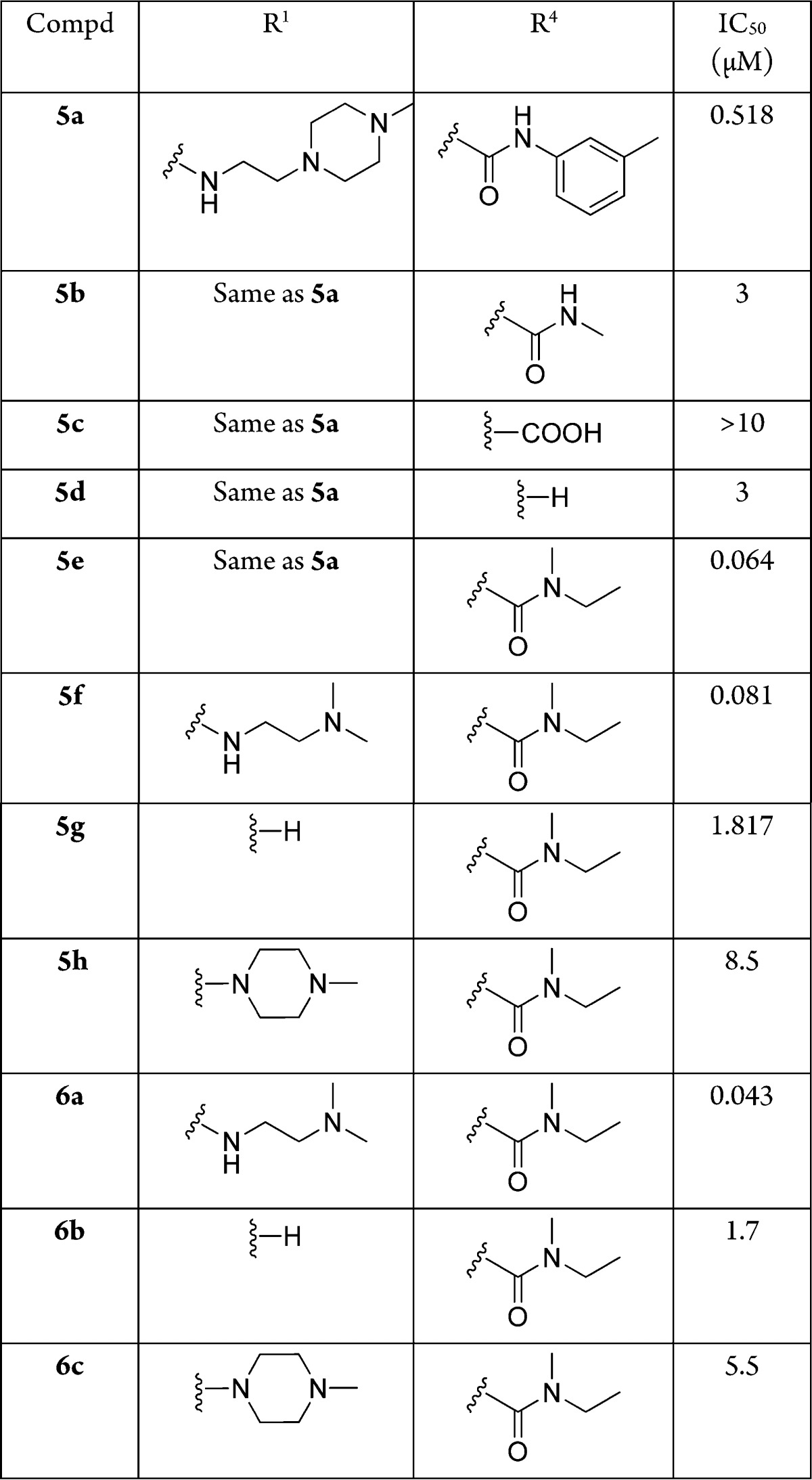
After exploring cycle 1 and cycle 3, we next turned our attention to cycle 4 (Table 3). From the selection data, besides N-ethyl-3-methylaniline, which was strongly selected for that cycle, we also observed other secondary amines, including both aniline derivatives and aliphatic secondary amines. To understand why primary amines were not selected at this position, we synthesized the compound with m-toluidine as cycle 4 synthon (5a). The activity dropped about 50-fold. Further truncation to methylamide (5b), free carboxylic acid (5c), and hydrogen (5d) led to even more loss of the activity. When we truncated the cycle 4 position to methylethylamide (5e), the activity was improved about 45-fold compared with methylamide (5b). Though its activity was about 6-fold less compared with N-ethyl N-(m-tolyl) amide (1j), N-ethyl N-methyl amide (5e) had better molecular properties with respect to both molecular weight and clogP. Since cycle 1 was the most tolerant position, next, we explored varying cycle 1 while fixing cycle 4 as N-ethyl N-methyl amide. With N1,N1-dimethylethane-1,2-diamine at cycle 1, the activity of the molecule (5f) was similar to that of compound 5e. However, without an R1 substituent, the activity dropped to 1.8 μM (5g). Substituting R1 with a tertiary amine, the activity dropped dramatically (5h). Replacing 6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline with 1,2,3,4-tetrahydroisoquinoline at cycle 3, all the N-ethyl N-methyl amide derivatives with diversity at the cycle 1 position yielded similar activity (6a–6c). The above data showed that, with a fine-tuning at the R1 position, it is possible to truncate the m-tolyl group while retaining potency. It should be noted that the optimized compounds with N-ethyl N-methyl amide at cycle 4 did not originate from the library itself, but were identified under the guidance of the selection structure–activity relationship (SAR).
Table 3. SAR at Cycle 4 Position.
The illustrated off-DNA compounds were synthesized via route A, B, or C in Scheme 1. In route A, the cycle 2,4 disynthon was synthesized through the acylation between Boc-protected cycle 2 amino acids and cycle 4 amine, followed by deprotection of the Boc group. Starting from cyanuric chloride, two chlorines were substituted in turn with amine and cycle 2,4 disynthon under increasing temperature yielding the monochloro intermediate. Final displacement of the last chlorine was conducted under various conditions to yield the desired substituted compounds or dechloro compound. Similar to route A, route B started with 2,4-dichloro-1,3,5-triazine, followed by subsequent substitution of two chlorines with amines. In route C, cyanuric chloride was subsequently substituted with two amines and an aminoester. After hydrolysis of the ester, the corresponding acid was acyclated with amine. Displacement of the last chlorine yielded the desired final compounds.
The selection data suggested that the cycle 1 synthon was not selected and the cycle 2, 3, and 4 synthons were highly selected against ADAMTS-4. The synthesis of off-DNA small molecules confirmed the selection data. Furthermore, under the guidance of the selection data, we found that the cycle 4 can tolerate some level of truncation, as long as it is a tertiary amide. Simple methylethyl amide is enough for the potent activity. We propose that the tertiary amide may have participated in the interaction with the target. Though the R1 position was the most tolerant position, while truncating R4 to smaller amides, there is a preference for some substituents, such as secondary amine instead of tertiary amine.
Finally, we assessed the selectivity of these triazine hits against other related zinc metalloproteinases. The compound 1j was measured against ADAMTS-5, MMP13, ADAMTS-13, and TACE (Table 4). For ADAMTS-5, we only observed about 30% inhibition at a concentration of 10 μM. For all the other targets, the compound was inactive at 10 μM. Overall, the compound had >1000-fold selectivity for inhibition of ADAMTS-4 over ADAMTS-5, MMP-13, ADAMTS-13, and TACE. A few other compounds 1b, 1d, 1e, and 1f also had excellent selectivity for inhibition of ADAMTS-4 over TACE (see Supporting Information). Compound 1j does not contain any obvious zinc ligand functionality, like hydroxamate, which may explain its specific activity toward ADAMTS-4.
Table 4. Selectivity Data for Compound 1j.
| IC50 (μM) | fold of selectivity | |
|---|---|---|
| ADAMTS-4 | 0.01 | |
| ADAMTS-5 | >10 | >1000 |
| ADAMTS-13 | >10 | >1000 |
| MMP-13 | >10 | >1000 |
| TACE | >10 | >1000 |
In summary, we have described the discovery of a novel and potent class of inhibitors for ADAMTS-4 using DNA-encoded library technology. The selection data identified numerous obligatory structural components such as the N-alkyl amide and the tetrahydroisoquinoline, which were confirmed by off-DNA synthesis. In general, we did not observe a clear correlation between the number of copies of individual warheads sequenced and their corresponding off-DNA potency. This could result from a number of factors, including differing potency of “on” and “off” DNA, building block-dependent variation in the chemistry during library synthesis, and low (∼1%) sequencing coverage of the library selection output. Nonetheless, taken in aggregate, the selection-based enrichment profile is predictive of off-DNA SAR, enabling additional chemistry and optimization. The ELT selection output generated ADAMTS-4 inhibitors that were highly selective with respect to ADAMTS-5 and other MMPs and without any obvious zinc ligand functionality. Further optimization of these hits will be published elsewhere in the future.
Glossary
ABBREVIATIONS
- ELT
encoded library technology
- DEL
DNA-encoded library
- TACE
tumor necrosis factor α-converting enzyme
- MMP
matrix metalloproteinase
- SAR
structure–activity relationship
Supporting Information Available
Synthesis and characterization data for the new compounds. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.5b00138.
The authors declare no competing financial interest.
Supplementary Material
References
- Tortorella M. D.; Malfait A. M. Will the Real Aggrecanase(s) Step Up: Evaluating the Criteria That Define Aggrecanase Activity in Osteoarthritis. Curr. Pharm. Biotechnol. 2008, 9, 16–23. 10.2174/138920108783497622#sthash.Wj45IQRp.dpuf. [DOI] [PubMed] [Google Scholar]
- Rienzo F. D.; Saxena P.; Filomia F.; Caselli C.; Colace F.; Stasi L.; Giordani A.; Menziani M. C. Progress Towards the Identification of New Aggrecanase Inhibitors. Curr. Med. Chem. 2009, 16, 2395–2415. 10.2174/092986709788682092. [DOI] [PubMed] [Google Scholar]
- Malfait A. M.; Liu R. Q.; Ijiri K.; Setsuro K.; Tortorella M. Inhibition of ADAM-TS4 and ADAM-TS5 Prevents Aggrecan Degradation in Osteoarthritic Cartilage. J. Biol. Chem. 2002, 277, 22201–22208. 10.1074/jbc.M200431200. [DOI] [PubMed] [Google Scholar]
- Fajardo M.; Di Cesare P. E. Disease-Modifying Therapies for Osteoarthritis: Current Status. Drugs Aging 2005, 22, 141–164. 10.2165/00002512-200522020-00005. [DOI] [PubMed] [Google Scholar]
- Porter S.; Clark I. M.; Kevorkian L.; Edwards D. R. The ADAMTS Metalloproteinases. Biochem. J. 2005, 386, 15–27. 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R.-H.; Tortorella M. D.; Malfait A. M.; Alston J. T.; Yang Z.; Arner E. C.; Griggs D. W. Aggrecan Degradation in Human Articular Cartilage Explants Is Medicated by Both ADAMTS-4 and ADAMTS-5. Arthritis Rheum. 2007, 56, 575–585. 10.1002/art.22334. [DOI] [PubMed] [Google Scholar]
- Gilbert A. M.; Bikker J. A.; O’Neil S. V. Advances in the Development of Novel Aggrecanase Inhibitors. Expert Opin. Ther. Pat. 2011, 21 (1), 1–12. 10.1517/13543776.2011.539204. [DOI] [PubMed] [Google Scholar]
- Renkiewicz R.; Qiu L.; Lesch C.; Sun X.; Devalaraja R.; Cody T.; Kaldjian E.; Welgus H.; Baragi V. Broad-Spectrum Matrix Metalloproteinase Inhibitor Marimastat-Induced Musculoskeletal Side Effects in Rats. Arthritis Rheum. 2003, 48, 1742–49. 10.1002/art.11030. [DOI] [PubMed] [Google Scholar]
- Hopper D. W.; Vera M. D.; How D.; Sabatini J.; Xiang J. S.; Ipek M.; Thomason J.; Hu Y.; Feyfant E.; Wang Q.; Georgiadis K. E.; Reifenberg E.; Sheldon R. T.; Keohan C. C.; Majumdar M. K.; Morris E. A.; Skotnicki J.; Sum P. Synthesis and Biological Evaluation of ((4-Keto)-phenoxy)methylbiophenyl-4-sulfonamides: A Class of Potent Aggrecanase-1 Inhibitors. Bioorg. Med. Chem. Lett. 2009, 19, 2487–2491. 10.1016/j.bmcl.2009.03.056. [DOI] [PubMed] [Google Scholar]
- Durham T. B.; Klimkowski V. J.; Rito C. J.; Marimuthu J.; Toth J. L.; Liu C.; Durbin J. D.; Stout S. L.; Swearingen C.; Lin C.; Chambers M. G.; Thirunavukkarasu K.; Wiley M. R. Identification of Potent and Selective Hydantoin Inhibitors of Aggrecanase-1 and Aggrecanase-2 That Are Efficacious in Both Chemical and Surgical Models of Osteoarthritis. J. Med. Chem. 2014, 57 (24), 10476–10485. 10.1021/jm501522n. [DOI] [PubMed] [Google Scholar]
- Clark M. A.; Acharya R. A.; Arico-Muendel C. C.; Belyanskaya S. L.; Benjamin D. R.; Carlson N. R.; Centrella P. A.; Chiu C. H.; Creaser S. P.; Cuozzo J. W.; Davie C. P.; Ding Y.; Franklin G. J.; Franzen K. D.; Gefter M. L.; Hale S. P.; Hansen N. J. V.; Israel D. I.; Jiang J.; Kavarana M. J.; Kelley M. S.; Kollmann C. S.; Li F.; Lind K.; Mataruse S.; Medeiros P. F.; Messer J. A.; Myers P.; O’Keefe H.; Oliff M. C.; Rise C. E.; Satz A. L.; Skinner S. R.; Svendsen J. L.; Tang L.; Vloten K. V.; Wagner R. W.; Yao G.; Zhao B.; Morgan B. A. Design, Synthesis and Selection of DNA-Encoded Small-Molecule Libraries. Nat. Chem. Biol. 2009, 5, 647–654. 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- Brenner S.; Lerner R. A. Encoded Combinatorial Chemistry. Proc. Natl. Acad. Sci. U. S. A. 1992, 89, 5381–5383. 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner R. E.; Dumelin C. E.; Liu D. R. Small-Molecule Discovery from DNA-Encoded Chemical Libraries. Chem. Soc. Rev. 2011, 40, 5707–5717. 10.1039/c1cs15076f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannocci L.; Leimbacher M.; Wichert M.; Scheuermann J.; Neri D. 20 Years of DNA-Encoded Chemical Libraries. Chem. Commun. 2011, 47, 12747–12753. 10.1039/c1cc15634a. [DOI] [PubMed] [Google Scholar]
- Halpin D. R.; Harbury P. B. DNA Display II. Genetic Manipulation of Combinatorial Chemistry Libraries for Small-molecule Evolution. PLoS Biol. 2004, 2, e174. 10.1371/journal.pbio.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connors W. H.; Hale S. P.; Terrett N. K. DNA-Encoded Chemical Libraries of Macrocycles. Curr. Opin. Chem. Biol. 2015, 26, 42–47. 10.1016/j.cbpa.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Deng H.; O’Keefe H.; Davie C. P.; Lind K. E.; Acharya R. A.; Franklin G. J.; Larkin J.; Matico R.; Neeb M.; Thompson M. M.; Lohr T.; Gross J. W.; Centrella P. A.; O’Donovan G. K.; Bedard K. L.; Vloten K.-v.; Mataruse S.; Skinner S. R.; Belyanskaya S. L.; Carpenter T. Y.; Shearer T. W.; Clark M. A.; Cuozzo J. W.; Arico-Muendel C. C.; Morgan B. A. Discovery of Highly Potent and Selective Small Molecule ADAMTS-5 Inhibitors That Inhibit Human Cartilage Degradation via Encoded Library Technology (ELT). J. Med. Chem. 2012, 55, 7061–7079. 10.1021/jm300449x. [DOI] [PubMed] [Google Scholar]
- Disch J. S.; Evindar G.; Chiu C. H.; Blum C. A.; Dai H.; Jin L.; Schuman E.; Lind K. E.; Belyanskaya S. L.; Deng J.; Coppo F.; Aquilani L.; Graybill T. L.; Cuozzo J. W.; Lavu S.; Mao C.; Vlasuk G. P.; Perni R. B. Discovery of Thieno[3,2-d]pyrimidine-6-carboxamides as Potent Inhibitors of SIRT1, SIRT2, and SIRT3. J. Med. Chem. 2013, 56, 3666–3679. 10.1021/jm400204k. [DOI] [PubMed] [Google Scholar]
- Thalji R. K.; McAtee J. J.; Belyanskaya S.; Brandt M.; Brown G. D.; Costell M. H.; Ding Y.; Dodson J. W.; Eisennagel S. H.; Fries R. E.; Gross J. W.; Harpel M. R.; Holt D. A.; Israel D. I.; Jolivette L. J.; Krosky D.; Li H.; Lu Q.; Mandichak T.; Roethke T.; Schnackenberg C. G.; Schwartz B.; Shewchuk L. M.; Xie W.; Behm D. J.; Douglas S. A.; Shaw A. L.; Marino J. P. Jr. Discovery of 1-(1,3,5-Triazin-2-yl)piperidine-4-carboxamides as Inhibitors of Soluble Epoxide Hydrolase. Bioorg. Med. Chem. Lett. 2013, 23, 3584–3588. 10.1016/j.bmcl.2013.04.019. [DOI] [PubMed] [Google Scholar]
- Encinas L.; O’Keefe H.; Neu M.; Remuiñán M. J.; Patel A. M.; Guardia A.; Davie C. P.; Pérez-Macías N.; Yang H.; Convery M. A.; Messer J. A.; Pérez-Herrán E.; Centrella P. A.; Álvarez-Gómez D.; Clark M. A.; Huss S.; O’Donovan G. K.; Ortega-Muro F.; McDowell W.; Castañeda P.; Arico-Muendel C. C.; Pajk S.; Rullás J.; Angulo-Barturen I.; Álvarez-Ruíz E.; Mendoza-Losana A.; Pages L. B.; Castro-Pichel J.; Evindar G. Encoded Library Technology as a Source of Hits for the Discovery and Lead Optimization of a Potent and Selective Class of Bactericidal Direct Inhibitors of Mycobacterium tuberculosis InhA. J. Med. Chem. 2014, 57, 1276–1288. 10.1021/jm401326j. [DOI] [PubMed] [Google Scholar]
- Kollmann C. S.; Bai X.; Tsai C.-H.; Yang H.; Lind K. E.; Skinner S. R.; Zhu Z.; Israel D. I.; Cuozzo J. W.; Morgan B. A.; Yuki K.; Xie C.; Springer T. A.; Shimaoka M.; Evindar G. Application of Encoded Library Technology (ELT) to a Protein–protein Interaction Target: Discovery of a Potent Class of Integrin Lymphocyte Function-associated Antigen 1 (LFA-1) Antagonists. Bioorg. Med. Chem. 2014, 22, 2353–2365. 10.1016/j.bmc.2014.01.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.