Significance
Insulators are genome sequence elements that help to organize eukaryotic genomes into coherent regulatory domains. Insulators can encode both enhancer-blocking activity, which prevents the interaction between enhancers and promoters located in distinct regulatory domains, and/or chromatin barrier activity that helps to delineate active and repressive chromatin domains. The origins and functional characteristics of insulator sequence elements are important, open questions in molecular biology and genomics. This report provides insight into these questions by demonstrating the origins of a number of human insulator sequences from a family of transposable-element–derived repetitive sequence elements: mammalian-wide interspersed repeats (MIRs). Human MIR-derived insulators are characterized by distinct sequence, expression, and chromatin features that provide clues as to their potential mechanisms of action.
Keywords: transposable elements, insulators, chromatin, gene regulation, genomics
Abstract
Insulators are regulatory elements that help to organize eukaryotic chromatin via enhancer-blocking and chromatin barrier activity. Although there are several examples of transposable element (TE)-derived insulators, the contribution of TEs to human insulators has not been systematically explored. Mammalian-wide interspersed repeats (MIRs) are a conserved family of TEs that have substantial regulatory capacity and share sequence characteristics with tRNA-related insulators. We sought to evaluate whether MIRs can serve as insulators in the human genome. We applied a bioinformatic screen using genome sequence and functional genomic data from CD4+ T cells to identify a set of 1,178 predicted MIR insulators genome-wide. These predicted MIR insulators were computationally tested to serve as chromatin barriers and regulators of gene expression in CD4+ T cells. The activity of predicted MIR insulators was experimentally validated using in vitro and in vivo enhancer-blocking assays. MIR insulators are enriched around genes of the T-cell receptor pathway and reside at T-cell–specific boundaries of repressive and active chromatin. A total of 58% of the MIR insulators predicted here show evidence of T-cell–specific chromatin barrier and gene regulatory activity. MIR insulators appear to be CCCTC-binding factor (CTCF) independent and show a distinct local chromatin environment with marked peaks for RNA Pol III and a number of histone modifications, suggesting that MIR insulators recruit transcriptional complexes and chromatin modifying enzymes in situ to help establish chromatin and regulatory domains in the human genome. The provisioning of insulators by MIRs across the human genome suggests a specific mechanism by which TE sequences can be used to modulate gene regulatory networks.
Insulators are regulatory sequence elements that help to organize eukaryotic chromatin into functionally distinct domains (1, 2). Insulators can encode two different functions: enhancer-blocking activity and chromatin barrier activity. Enhancer-blocking insulators prevent the interaction of enhancer and promoter elements located in distinct domains, and chromatin barrier insulators, also known as boundary elements (3, 4), protect active chromatin domains by blocking the spread of repressive chromatin. These two functional roles are not mutually exclusive; compound insulators may encode both enhancer-blocking and chromatin barrier activities (5).
Transposable element sequences are known to provide a variety of regulatory sequences to eukaryotic genomes (6), and there are several examples of transposable element (TE)-derived insulators. The best studied TE insulator comes from the Drosophila gypsy element (7–10). Gypsy is a long terminal repeat retrotransposon that contains an insulator sequence in its 5′ untranslated region. The gypsy insulator interacts with the suppressor of hairy wing [su(Hw)] and modifier of mdg4 [mod(mdg4)] proteins to block regulatory interactions between distal enhancer and proximal promoter sequences. This same insulator can also protect transgenes from position effects, indicating that it encodes chromatin barrier activity as well.
More recently, TE-derived insulator sequences have been discovered in mammalian genomes. The short interspersed nuclear element (SINE) B1 has insulator activity that is mediated by the binding of specific transcription factors along with the insulator associated protein CCCTC-binding factor (CTCF) (11). A genome-wide analysis of CTCF binding sites in the human and mouse genomes discovered that many CTCF binding sites are derived from TE sequences (12), and a survey of six mammalian species revealed that lineage-specific expansions of retrotransposons have contributed numerous CTCF binding sites to their genomes (13). A number of these TE-derived CTCF binding sites in the mouse and rat genomes are capable of segregating domains enriched or depleted for acetylation of histone 2A lysine 5 (H2AK5ac), suggesting that they may encode insulator function. Interestingly, this same analysis did not detect retrotransposon-driven expansion of CTCF binding sites in the human genome (13).
Whereas subsets of CTCF binding sites are known to be associated with insulators, numerous insulators can function in a CTCF-independent manner. An important example comes from a mouse TE, the SINE B2 element, which serves as a developmentally regulated compound insulator, encoding both enhancer-blocking and chromatin barrier activity, at the growth hormone locus (14). B2 is a tRNA-derived SINE that encodes the B-box promoter element, which is bound by RNA polymerase III (RNA Pol III). The connection to tRNAs/Pol III binding is intriguing, given the fact that tRNA gene sequences/Pol III binding have been shown to encode insulators in yeast (15–18), mouse (19), and human (20, 21). The association of insulators to the binding of RNA Pol III, or transcription factor III C (TFIIIC) specifically, to B-box elements is widely observed in multiple species, suggesting that Pol III-related machinery represents another insulator mechanism in addition to CTCF binding. Because the human genome is made up of a substantial fraction of TE sequences, including numerous tRNA-derived SINE retrotransposons (22), it is highly possible that subsets of these tRNA-derived SINE sequences encode insulator functions. The discovery and characterization of such TE-derived insulators will help to augment the currently sparse insulator annotations in the human genome and also provide additional evidence regarding Pol III-related mechanisms of insulator activity.
Mammalian-wide interspersed repeats (MIRs) are an ancient family of TEs (23) that bear several features, suggesting that they may serve as genome regulators in general and insulators in particular. First of all, a number of noncoding MIR sequences were found to be highly conserved, indicative of some functional, presumably regulatory, role (24). Later, it was shown that MIRs are enriched for open chromatin sites (25), encode regulatory RNAs (26), host gene promoters (27) and enhancers (28), and are also associated with tissue-specific expressed genes (29). Finally, and most importantly, MIRs are tRNA-derived SINEs (30) and their sequences include recognizable regulatory motifs, such as the promoter B-box element for Pol III binding, which are thought to be important for insulator activity.
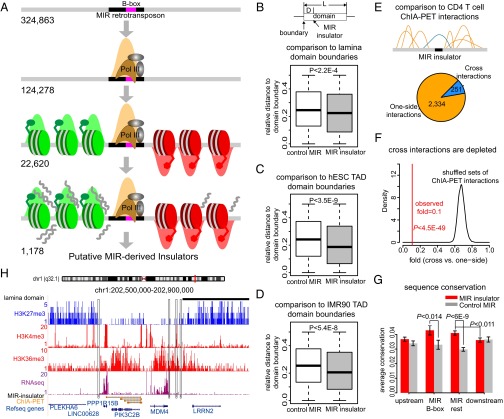
In light of these known MIR regulatory sequence characteristics, particularly the link to Pol III binding, along with their enrichment at chromatin domain boundaries (Results), we sought to evaluate whether MIR elements can encode insulator activity in the human genome. To do this, we used a bioinformatics screen of genome sequence and functional genomic data to identify a subset of MIR sequences that possess insulator-like features. The screen was applied to datasets from human CD4+ T cells and could also be used for other data sources to discover MIR-derived insulators that function in different tissues. The insulator-like features include the presence of intact B-box sequences, occupancy by RNA Pol III, and the partitioning of active and repressive chromatin domains (Fig. 1A). This screen procedure resulted in the identification of 1,178 putative MIR-derived insulator sequences in human CD4+ T cells, which were validated computationally, followed by experimental validation for a subset of the elements, and then evaluated with respect to a number of functional properties (SI Appendix, Fig. S1).
Fig. 1.
Bioinformatic screen and validation of MIR insulators. (A) Scheme of bioinformatic screen used to predict MIR insulators. Predicted MIR insulators contain intact B-box promoter sequences, are bound by RNA Pol III, segregate active (green) versus repressive (red) chromatin, segregate expressed versus silent genomic regions, and are located in intergenic genomic regions and distant (>10 kb) from each other. (B) Relative distances (normalized by domain sizes) between MIR insulators and lamina-associated domain boundaries (gray). Randomly selected B-box–containing MIR sequences are shown as controls (white). (C) Relative distances between MIR insulators and topologically associated domain boundaries in hESC. (D) Relative distances between MIR insulators and topologically associated domain boundaries in IMR90 cells. (E) Local ChIA–PET interactions flanking MIR insulators classified into one-side interactions (orange) and cross-interactions (blue). (F) Depletion of cross-interactions around MIR insulators. The observed fold between cross versus one-side interactions (red line) is compared with the distribution of folds from locally shuffled ChIA–PET interactions. (G) Sequence conservation of MIR insulators (divided into B-box and the remaining parts). Average (±SE) conservation levels of MIR insulators and 100-bp upstream/downstream sequences (red bars) are compared with randomly selected B-box–containing MIR sequences (gray bars). (H) Examples of predicted MIR insulators (black boxes) on a locus of chromosome 1. Repressive histone marks (blue), active histone marks (red), lamina domain (black bar), RNA-seq signals (purple), ChIA–PET interactions (orange) and genes are shown.
Results
Bioinformatic Screen and Validation.
To prioritize a specific B-box–containing TE family for the search of insulators, we first analyzed the enrichment of different TE families in chromatin domain boundaries, which we previously defined as transition regions between repressive and active chromatin domains (21). Whereas several TE families are enriched in domain transition regions (SI Appendix, Fig. S2A), MIRs represent the only B-box–containing TE family that is specifically enriched in domain boundaries compared with flanking regions (SI Appendix, Fig. S2B). This observation led us to focus on MIR sequences to evaluate candidate insulators.
We developed and applied a bioinformatic screen to search for human MIR sequences that may encode insulator activity (Fig. 1A). To do this, we evaluated human genome sequence data along with functional genomic data from CD4+ T cells (Materials and Methods). CD4+ T cells were chosen, owing to their importance as a model system for immunology and for the abundance of available functional genomic data that exist for this cell type. The genome sequence data analyzed consisted of TE and gene annotations, and the functional genomic data included RNA-sequence (RNA-seq) and microarray expression data along with ChIP-seq data for RNA Pol III binding and 39 histone modifications.
First, all MIR sequences in the human genome that contain intact B-boxes and are bound by RNA Pol III in CD4+ T cells were identified. Then, these MIRs were evaluated for their ability to partition active versus repressive chromatin using a previously described approach (21) that segregates histone modifications associated with expressed (active) versus silent (repressive) genomic regions. Broad genomic distributions of 39 histone modifications, with 34 characterized as active and 5 characterized as repressive, were evaluated to detect large contiguous regions (domains) of active and repressive chromatin. The B-box–containing and RNA Pol III-bound MIR elements found to be located between adjacent active versus repressive were then used for further analysis (SI Appendix, SI Methods). RNA-seq was then used to further reduce the list of putative MIR insulators to those that delineate high- versus low-expressed genomic regions. Finally, only MIR insulators that are located in intergenic regions and distant (>10 kb) from each other were selected to reduce the ambiguity. This procedure resulted in the identification of 1,178 putative MIR-derived insulators (0.36% of B-box containing MIR sequences) across the human genome in CD4+ T cells (Fig. 1A and Dataset S1). As a negative control comparison, we also applied the same screen procedure on Alu sequences that contain B-boxes (31, 32), and found a lower fraction (SI Appendix, Fig. S3) of insulator-like Alu sequences (0.15% of B-box–containing Alu sequences). Although the potential insulator function of a subset of Alu sequences is also interesting, this observation indicates that MIR sequences might have higher likelihood of having insulator function. To evaluate the performance of this pipeline, we carried out a series of multidimensional statistical analysis on histone modification ChIP-seq signals around MIR-derived insulators (SI Appendix, Fig. S4 and SI Methods). The negative correlations of individual histone mark signals in upstream versus downstream regions (SI Appendix, Fig. S5A and Table S1), distinct clusters of active and repressive histone mark profiles (SI Appendix, Fig. S5B), and groups of active and repressive histone marks observed in reduced dimensional space of ChIP-seq profiles across MIR-derived insulators (SI Appendix, Fig. S6) suggest that our screen pipeline can efficiently pinpoint MIR sequences that significantly block individual histone modifications and partition active versus repressive histone marks. Significant differences of proximal gene expression on the active chromatin side versus the repressive side of MIR-derived insulators were also confirmed (SI Appendix, Fig. S7).
The predicted MIR-derived insulators were first computationally validated with respect to their domain barrier and enhancer-blocking activities based on independent functional datasets (Materials and Methods). As insulators are expected to partition consecutive chromatin domains, we calculated the relative distances (i.e., normalized by domain sizes) between MIR-derived insulators and lamina-associated domain (LAD) boundaries (33). We found that the predicted MIR insulators are significantly closer to LAD boundaries compared with randomly selected B-box–containing MIR sequences (P < 2.2E-4, Mann–Whitney test) (Fig. 1B). Similarly, we integrated topologically associated domain (TAD) boundaries inferred from Hi-C chromatin contact maps (34) and found that the MIR insulators are also closer to TAD boundaries (P < 5.4E-8, Mann–Whitney test) (Fig. 1 C and D). These findings suggest the predicted MIR insulators have domain barrier function. In addition, we used the ChIA–PET interaction data from human CD4+ T cells (35) to test whether the predicted MIR insulators can potentially block enhancer–promoter interactions. We focused on ChIA–PET interactions between enhancers and promoters that are proximal (<500 kb) to MIR-derived insulators and classified them into one-side interactions, i.e., the interaction’s anchors are restricted to one-side of a MIR insulator, and cross-interactions, i.e., the interaction’s anchors are separated by a MIR insulator (Fig. 1E). Whereas there are 2,334 one-side interactions, only 251 cross-interactions are found (fold = 0.1) (Fig. 1E). The observed depletion of cross-interactions is statistically significant (P < 4.5E-49, Z test) compared with randomly shuffled ChIA–PET interactions (fold ∼0.7) (Fig. 1F and Materials and Methods), supporting the enhancer-blocking activity of the predicted MIR insulators. As a positive control, we carried out the same analyses on a set of CTCF barriers in CD4+ T cells (36), the canonical insulator associated sites, and got similar observations (SI Appendix, Fig. S8). As a negative control, we randomly selected B-box–containing MIR sequences that are located around genomic regions (±500 kb) with ChIA–PET interactions. The cross-interactions are not depleted compared with one-side interactions around control MIRs (fold = 0.53, P = 0.067) and further suggest potential insulator functions of the predicted MIR insulators (SI Appendix, Fig. S8). We also investigated the sequence conservation patterns of MIR insulators and their flanking sequences. We observed significantly higher conservation levels of MIR insulators compared with flanking sequences (P < 0.011, t test) (Fig. 1G). Both the B-box region (P < 0.014, t test) and the remaining part of the MIR insulators (P < 6E-9, t test) show higher conservation levels compared with randomly selected B-box–containing MIR sequences, and the B-box sequences show the highest average conservation level (Fig. 1G).
Examples of MIR insulators are shown along with LAD, histone modifications, RNA-seq, and ChIA–PET interactions (Fig. 1H). In this example, whereas a single MIR insulator on the Left is precisely located at the transition point between repressive and active chromatin domains, a cluster of three MIR insulators on the Right are located at the region where active and repressive histone marks gradually switch. One of the three MIR insulators on the Right is also located at the LAD boundary. High-level RNA-seq signals and multiple ChIA–PET interactions are restricted within the active chromatin region enclosed by the MIR insulators (Fig. 1H).
Experimental Validation.
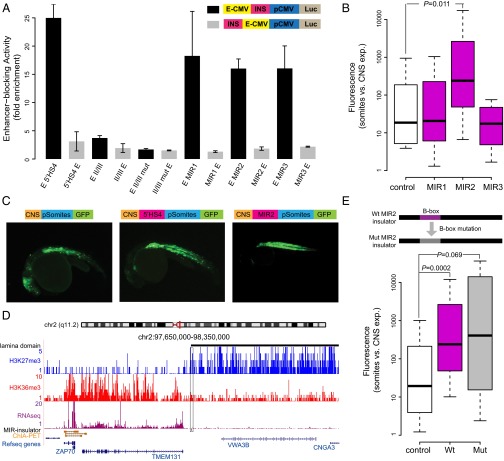
We next sought to experimentally validate the enhancer-blocking activity for a subset of the MIR insulators predicted here using previously described human and zebrafish enhancer-blocking assays (EBAs) (11, 14, 37, 38). For the human EBA, a luciferase reporter construct transfected in human HEK 293 cells (Materials and Methods) was used to evaluate three predicted MIR insulators (SI Appendix, Table S2). All three MIR insulators tested here showed enhancer-blocking activity comparable to the 5′ HS4 positive control (Fig. 2A) and much larger insulator activity than the second positive control, i.e., the minimal insulator sequence motif of 5′ HS4 (II/III). These consistently observed enhancer-blocking activities strongly support the potential insulator function of MIR sequences.
Fig. 2.
Enhancer-blocking assays (EBAs) for predicted MIR insulators. (A) Human EBA. Enhancer-blocking activity levels (fold enrichment) are normalized relative to the empty vector. Average enhancer-blocking activity levels (±SE) for positive (5′ HS4 and II/III) and negative (II/III mutated) controls along with results for three predicted MIR insulators MIR1, MIR2, and MIR3 (located on chromosomes 1, 2, and 11, respectively) are shown. For each sequence analyzed, inserts were cloned upstream of the enhancer (negative control site, gray bars) and between the enhancer and promoter (test site, black bars). (B) Enhancer-blocking activity in zebrafish. Negative (empty vector, white) control sequences along with predicted MIR insulators (purple) were inserted between the CNS enhancer and the somite promoter. The ratio of GFP expression in somites versus CNS indicates relative enhancer-blocking activity. The observed ratio for MIR2 is significantly different from control zebrafish (P = 0.011), whereas control versus MIR1 (P = 1.000) and versus MIR3 (P = 0.934) are not. (C) Zebrafish EBA. Representative pictures of GFP expression in zebrafish somites and CNS generated from negative and positive (5′ HS4) controls and the MIR insulator located on chromosome 2. (D) The MIR insulator active in zebrafish EBA. Histone marks, lamina domains, RNA-seq, and ChIA–PET interactions are shown as in Fig. 1H. (E) B-box mutation of MIR insulator. B-box mutated MIR insulator is tested in zebrafish EBA (gray) and compared with the wild-type MIR insulator (purple) and the negative control (white). The observed ratio for MIR2 is significantly different from control zebrafish (P = 0.0002), whereas the equivalent ratio for the mutated-MIR2 versus control (P = 0.069) is not.
The same MIR-insulator sequences were further tested in a zebrafish EBA using a GFP reporter construct transiently transfected in embryos. This EBA tests the ability of putative insulator sequences to block interaction of a central nervous system (CNS) enhancer with a somite promoter driving the expression of GFP (Materials and Methods). Whereas two MIR sequences (MIR1 and MIR3) do not show enhancer-blocking activity compared with the negative control, one of the MIR insulators (MIR2) is able to efficiently block the CNS enhancer and cause the statistically significant loss of GFP expression (P = 0.011, median test) in zebrafish midbrain (Fig. 2 B and C). Considering the large species differences between human and zebrafish, the significant insulator function seen in zebrafish of this MIR sequence is intriguing. We thus focused on this MIR insulator (MIR2) in more detail. This MIR insulator is located in the transition point between active and repressive chromatin domains as expected (Fig. 2D). In addition, it is located precisely at the boundary of a lamina-associated domain (Fig. 2D), suggesting that it may participate in large-scale genome organization. The blocking of the repressive chromatin domain also appears to be functionally important to CD4+ T cells because one of the proximal genes in the adjacent active chromatin domain, i.e., gene ZAP70, is part of the T-cell receptor pathway and its promoter is involved with multiple active ChIA–PET interactions (Fig. 2D).
To further test whether the B-box is a major factor of the enhancer-blocking activity of this MIR insulator, we specifically mutated two nucleotides of its B-box (Materials and Methods) and repeated the zebrafish EBA (Fig. 2E). The two nucleotides (T45/C) were selected based on previous observations that point mutations at those sites cause loss of barrier function of tRNA genes in yeast (15, 16, 39, 40). Although the median enhancer-blocking activity in zebrafish of the mutated MIR insulator is slightly higher, the activity variance largely increased, and thus the insulator function of the mutated MIR sequence is no longer significantly distinct (P = 0.069, median test) from the negative control (Fig. 2E). The increase in variance may be attributed to the fact that the two mutated B-box sites are only critical for some of the insulator function, and other remaining sites in the B-box may provide additional important sequence context. The variance may also be due to a reduction in the stability of the enhancer blocking activity conferred by the MIR insulators. In any case, this result indicates that the B-box sequence is an important component that is likely to be necessary, but not alone sufficient, for MIR-insulator function.
MIR Insulator Chromatin Features.
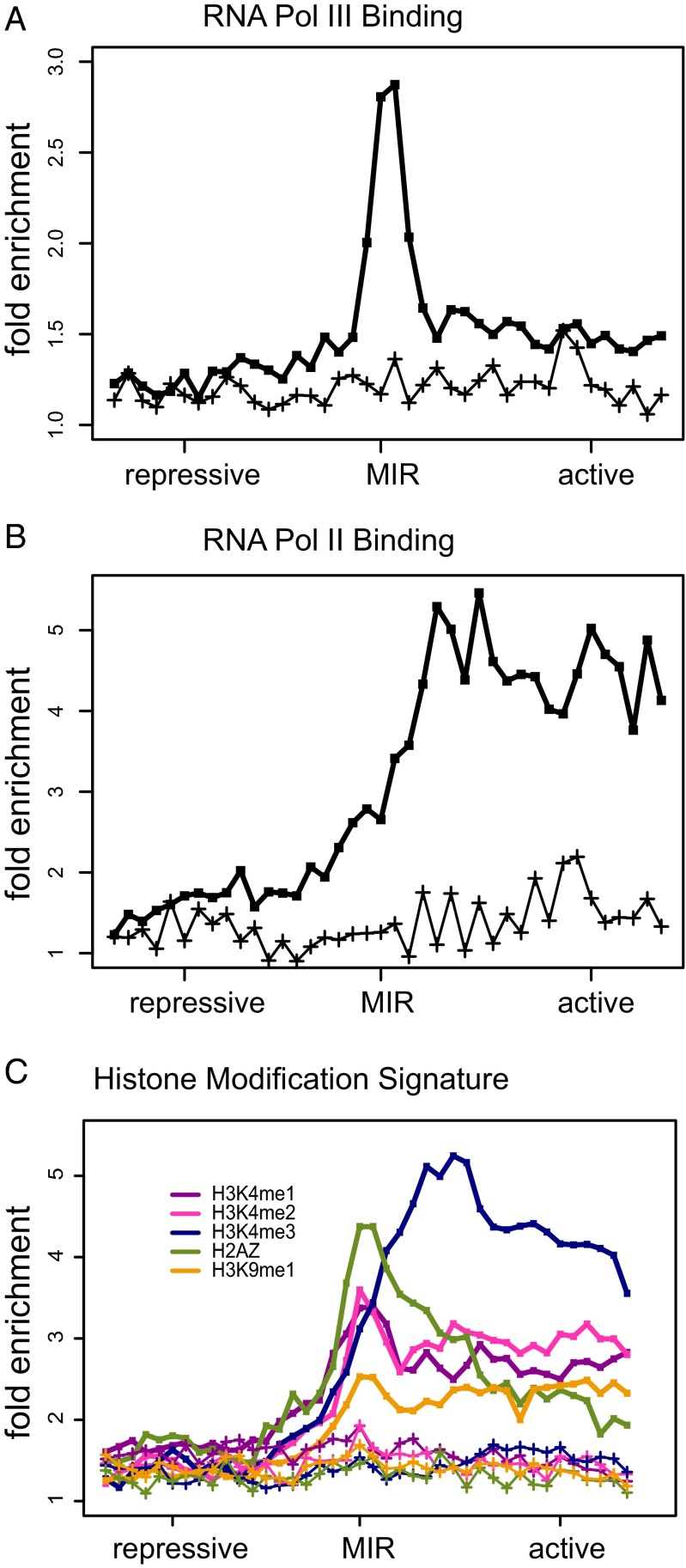
Having established the chromatin barrier and enhancer-blocking activity of predicted MIR insulators, we performed a series of enrichment analyses to characterize the local chromatin environment at and around these insulators. Here we focused on small-scale genomic regions around MIR insulators (∼8 kb) to search for specifically localized enrichment signatures that are not seen at the domain-scale level. The aggregate RNA Pol III occupancy levels peak at MIR-insulator sequences (Fig. 3A), which is consistent with the initial bioinformatic screen used for their identification. Nevertheless, the distinct RNA Pol III peak at MIR insulators differs from the previously observed broad genomic distribution of RNA Pol III binding (41), suggesting the possibility that MIR insulators are activated via specific recruitment of RNA Pol III. In addition, the negative control, performed on a randomly selected set of B-box–containing MIRs, shows that specific RNA Pol III binding is not a generic feature of MIRs across the genome. RNA Pol II levels, on the other hand, increase steadily from the MIR-insulator region into the flanking active chromatin environment (Fig. 3B), consistent with their role as barriers against the spread of repressive chromatin. In contrast to the RNA Pol III enrichment peak, transcription levels do not show enrichment at MIR insulators (SI Appendix, Fig. S9), suggesting transcription may not be involved with MIR-insulator activities. On the other hand, the lack of transcription enrichment is also possibly due to the experimental bias of the RNA-seq library, which is not efficient to capture the short noncoding transcripts.
Fig. 3.
Specific enrichment signature of chromatin features around predicted MIR insulators. The 8-kb windows centered on predicted MIR insulators were evaluated for the fold enrichment (compared with genomic background) of (A) RNA Pol III binding, (B) RNA Pol II binding, and (C) levels of five histone modifications. For each enrichment curve, a corresponding negative control (lower lines marked with crosses) is shown based on a randomly selected set of B-box–containing MIR sequences of the same size.
MIR insulators show a characteristic histone modification signature with distinctive peaks of the H2AZ histone variant, H3K4me1, H3K4me2, and H3K9me1 (Fig. 3C). Such peaked patterns cannot be expected based on the approach used to detect putative MIR insulators because the algorithm evaluates broad distributions of active versus repressive histone modifications over 100-kb windows surrounding the MIRs. H3K4me3 levels peak adjacent to the locations of the MIR insulators on the active chromatin side and remain high across the local active chromatin domain. Most of these marks are associated with active chromatin and transcriptional initiation, suggestive of the recruitment of chromatin-modifying complexes to MIR insulators resulting in the local opening of chromatin and priming for gene expression. Consistent with this possibility, MIR insulators are much closer to the nearest gene transcription start site (TSS) on the active chromatin side than on the repressive side (SI Appendix, Fig. S10). H3K4me1 modifications are often associated with enhancer sequences, raising the possibility of some mechanistic overlap between MIR insulators and enhancers, as has been previously suggested (1).
For the purposes of comparison, the same enrichment analyses were applied on insulator-like Alu sequences generated by our bioinformatics screen pipeline and CTCF barriers produced independently before (36). The insulator-like Alu sequences are less enriched with RNA Pol III binding compared with MIR insulators and do not show distinctive peaks of histone modifications (SI Appendix, Fig. S11). Thus, the unique signatures of MIR insulators described above further suggest them as functional regulatory elements. As the positive control, the canonical insulators, i.e., CTCF barrier sites, show similar enrichment signatures as MIR insulators with even higher peaks (SI Appendix, Fig. S11). The unexpected RNA Pol III enrichment at CTCF barriers also suggest potential interplay between these two DNA binding factors to establish insulator activity (42).
The similarity of chromatin signatures suggests that MIR insulators may overlap with CTCF sites. Although there is mild enrichment of CTCF binding signals around MIR sequences, the peak is not distinctive from flanking regions (SI Appendix, Fig. S12A). Indeed, only 6 of the 1,178 MIR insulators overlap with previously characterized CTCF barrier elements (36). The number of overlaps only increases to 25 when the MIR insulators are extended by 4 kb on each side (SI Appendix, Fig. S12B). The lack of overlap indicates that MIR insulators are likely to be largely CTCF independent.
Tissue-Specific Chromatin Barrier Functions of MIR Insulators.
We also evaluated the role that the putative MIR insulators play in regulating tissue-specific gene expression by measuring the differences in expression levels, across 79 human tissues, for genes that flank the insulators on the active sides versus repressive sides. Genes that flank MIR insulators show greater differences in expression, between the active and repressive sides of the insulators, for CD4+ T cells than seen for the other human tissues (SI Appendix, Fig. S13). This finding suggests a role for the insulators in establishing tissue-specific chromatin domains, consistent with the previously observed tissue specificity of CTCF barriers (36). It also indicates that additional MIR insulators specifically active in other tissues may be identified by applying our screen pipeline on different tissues when more functional genomic datasets become available.
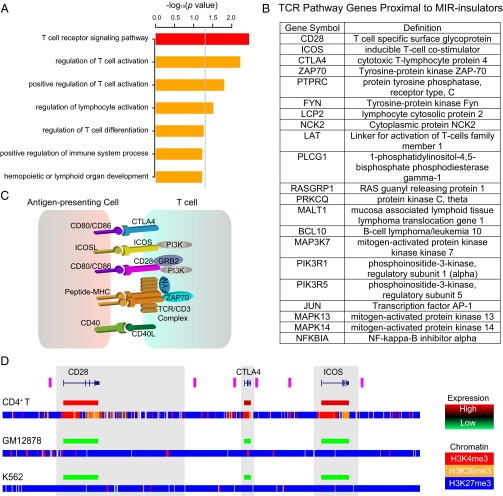
We sought to further evaluate the possible tissue-specific functional roles played by the MIR insulators predicted here (Materials and Methods). To do this evaluation, we performed an analysis of the gene ontology (GO) and pathway (Kyoto Encyclopedia of Genes and Genomes, KEGG) annotations of the proximal genes located on the active chromatin sides of the MIR insulators. These genes are enriched for a number of GO/KEGG functional categories related to T-cell function including T-cell receptor signaling pathway, regulation of T-cell activation, and regulation of lymphocyte activation (Fig. 4A). These specific functional categories still remain significant when the set of all expressed genes in CD4+ T cells are used as background for comparison (SI Appendix, Fig. S14), highlighting the specific T-cell relevant functional pathways possibly regulated by MIR insulators. The most strikingly enriched category is the T-cell receptor signaling pathway (KEGG: hsa04660). The analysis reveals that 21 genes found in the T-cell receptor signaling pathway are located adjacent to MIR insulators on the active chromatin side (Fig. 4B and SI Appendix, Fig. S15). Among this list, there are several transmembrane receptor proteins, which mediate interactions with antigen-presenting cells, including a colocated genomic cluster of two T-cell costimulators (CD28 and ICOS) and the coinhibitor CTLA4 (Fig. 4C). The chromatin environment at this genomic cluster, along with the cell-type–specific expression patterns of these three genes, exemplifies the T-cell–specific regulatory function of the MIR-insulator encoded barrier activity (Fig. 4D and SI Appendix, Fig. S16). In CD4+ T cells, these three genes are flanked by pairs of MIR insulators that surround an open and active chromatin environment (H3K4me3 and H3K36me3) to the exclusion of repressive chromatin marks (H3K27me3) in the adjacent regions. This pattern stands in contrast to what is seen for GM12878 and K562 cells where the entire locus is marked by repressive chromatin. Accordingly, CD28, ICOS, and CTLA4 are highly expressed in CD4+ T cells compared with GM12878 and K562 cells (Fig. 4D). Similar cell-type–specific distributions of chromatin and gene expression for MIR insulators and their adjacent genomic regions are observed when the same histone marks and expression levels are compared for all 21 MIR-insulator proximal genes found in the T-cell receptor pathway (SI Appendix, Fig. S17).
Fig. 4.
T-cell–specific functions of predicted MIR insulators. (A) Results of gene ontology (GO) and pathway (KEGG) analysis of proximal genes on the active chromatin side of MIR insulators. P values (−log10 normalized) are shown for the KEGG (red) and GO biological process (orange) analyses; the gray line corresponds to P = 0.05. (B) List of 21 T-cell receptor signaling pathway genes located on the active domain side proximal to MIR insulators. (C) Portion of the T-cell receptor pathway showing membrane receptors that mediate T-cell stimulation via antigen presenting cells. (D) Expression levels and the chromatin environment across a genomic cluster of three T-cell receptor genes—CD28, CTLA4, and ICOS (blue gene models)—and their colocated MIR insulators (purple bars) are shown for CD4+ T cells, GM12878 and K562. Relative gene expression levels (high-red to low-green) are shown coincident with the gene models. Genomic distributions of three histone modifications are shown as H3K4me3 (red), H3K36me3 (orange), and H3K27me3 (blue).
We expanded the tissue-specific chromatin and expression analysis to include all MIR insulators predicted here. We first classified MIR insulators as cell-type specific based on the relative distributions of chromatin marks across MIR insulators in CD4+ T cells versus GM12878 and K562 cells. A total of 681 of 1,178 (58%) of predicted MIR insulators show skewed distributions of active versus repressive marks in CD4+ T cells, with divergent peaks on opposing sides of the MIR insulators, compared with relatively flat distributions of the same histone marks in GM12878 and K562 cells (Fig. 5 A–C). Accordingly, these tissue-specific MIR insulators have proximal genes on the active domain side that are expressed at higher levels in CD4+ T cells than the same genes in GM12878 and K562 (Fig. 5D). Furthermore, these MIR insulators separate pairs of genes, on the active versus repressive chromatin sides of the insulators, that have greater differences in their levels of expression in CD4+ T cells than seen for the same pairs of genes in GM12878 and K562 (Fig. 5E). As a comparison, we applied the same analysis on CTCF barriers and found similar fraction [947 of 1,607 (59%)] to be tissue specific. Comparisons of histone mark signals and gene expression for the subset of cell-type–specific CTCF barriers show similar patterns as seen for MIR insulators (SI Appendix, Fig. S18), consistent with the previous finding that CTCF barriers are highly tissue specific (36). The remaining 42% of MIR insulators that do not show evidence of tissue-specific function may have broader activity reflecting chromatin boundary establishment earlier in development. It is also possible that additional MIRs not detected in our bioinformatic screen, e.g., those that lack intact B-boxes or those do not bind RNA Pol III, may also serve as insulators in CD4+ T cells and/or in other tissues.
Fig. 5.
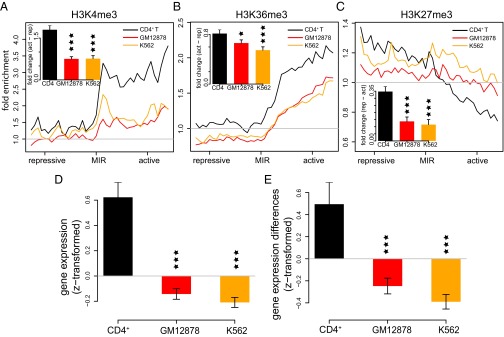
Cell-type–specific chromatin barrier activity and gene regulation by MIR insulators. ChIP-seq fold enrichment levels around tissue-specific MIR insulators are shown for (A) H3K4me3, (B) H3K36me3, and (C) H3K27me3 in CD4+ T cells (black), GM12878 cells (red), and K562 (orange) cells. Insets show the average differences (±SE) between the active versus repressive domains surrounding MIR insulators for the marks and cells. (D) Average gene expression levels (±SE) are shown for genes located in the active domain side proximal to MIR insulators. Gene expression levels are z transformed within each cell type. (E) Average (±SE) differences in the gene expression levels for genes located on the opposite sides of individual MIR insulators. Gene expression difference values are z transformed within each cell type. For all bar plots, significance of the differences between CD4+ T cells and other cells are indicated as *P < 0.05, **P < 0.01, and ***P < 0.001.
Discussion
MIRs are relatively ancient and conserved TEs, i.e., formerly selfish genetic elements, that have been coopted to provide a variety of regulatory sequences to their host genomes. Together with their conservation and regulatory capacity, the tRNA-derived sequence features of MIRs suggested to us that they might help to organize human chromatin via the provisioning of insulator elements. Therefore, we screened the human genome for putative MIR insulators and attempted to validate their activity using a combined computational and experimental approach. The results of our analysis suggest that numerous MIR sequences serve as insulators across the human genome. These predicted MIR insulators show evidence of both chromatin barrier and enhancer-blocking activity. Interestingly, whereas the chromatin barrier activity of the MIR insulators appears to be cell-type specific (Figs. 4 and 5 and SI Appendix, Fig. S13), the mechanisms underlying MIR’s enhancer-blocking activity are seemingly conserved between cell types and between species (Fig. 2). This finding may be attributed to the fact that MIR sequences in isolation possess an innate capacity to provide enhancer-blocking activity via the interaction with conserved protein factors, but in situ MIRs interact with cell-type–restricted factors to yield a more narrow and specific range of activity. Given that the EBAs were performed with minimal (<400 bp) constructs, it may be the case that synergistic binding of sites outside the MIR insulators help to provide cell-type–specific barrier activity.
The MIR insulators identified here have a distinct local chromatin environment (Fig. 3) that may yield some clues as to their mechanisms of action. For example, whereas RNA Pol II and RNA Pol III CD4+ T-cell binding profiles are highly correlated across the human genome (41), their patterns at and around MIR insulators are quite distinct. RNA Pol III occupancy levels peak right at the MIR insulators, whereas RNA Pol II levels steadily increase from the MIR insulators into the adjacent active chromatin domains. This suggests the possibility that RNA Pol III is specifically recruited to MIR insulators to help establish their activity, thus priming the adjacent chromatin for opening and transcriptional activity as reflected by the increasing RNA Pol II levels. The histone modification profiles around MIR insulators are consistent with this model. There are clear local peaks of modifications right at the MIR insulators, such as seen for H3K4me1 and H3K4me2, but these same marks of open chromatin are also maintained at relatively higher levels in the adjacent active domains. H3K4me3 shows a similar pattern, but its peak is shifted further into the active domain and it is maintained at higher levels through this domain. Thus, there may be a wave of progressive methylation of the H3K4 position starting at the MIR insulator locations and continuing with the addition of methyl groups into the active domain, similar to what we observed previously for human chromatin barriers (21).
The location of MIR insulators relative to proximal gene promoters also sheds some light on their mechanism of action. MIR insulators are located much closer to the promoters of the genes that are located on the active side of the insulator compared with the genes located on the repressive side (SI Appendix, Fig. S10). This suggests that MIR insulators are not only located in such a way to protect proximal promoters from the encroachment of repressive chromatin, but they also restrict interactions with promoters to only those enhancers that are located nearby or within genes. This scenario can be illustrated by the clustering of the colocated T-cell receptors—CD28, CTLA4, and ICOS—each of which is flanked by a pair of MIR insulators (Fig. 4D). This apparent restriction to local enhancers would seem to be at odds with the textbook definition of enhancers as regulatory elements that exert their effects over long ranges. However, recent genome-wide analyses of chromatin reveal that gene bodies are enriched for enhancer elements (43–45) and these local regulatory sequences may be largely responsible for cell-type–specific expression.
TE-derived insulators have previously been associated with CTCF binding events (13). Whereas there is weak enrichment of CTCF binding signal at MIR insulators, only 6 of 1,178 MIR insulators overlap with the CTCF barriers (SI Appendix, Fig. S12). These results raise the possibility that MIR insulators discovered here function in a CTCF-independent manner. Many questions as to the specific mechanisms underlying MIR-insulator activity remain to be answered. For example, whereas the compound insulator activity of the mouse tRNA-derived SINE B2 is related to the transcriptional activity of the element (14), it is not clear if the same can be said for MIR insulators (SI Appendix, Fig. S9). Furthermore, many of the protein factors that interact with MIR insulators remain to be elucidated. Nevertheless, the finding that numerous MIRs across the human genome can provide insulator activity raises intriguing possibilities. In particular, when their repetitive nature is considered together with their role in organizing chromatin, it suggests a possible mechanism for the establishment of cell-type–specific regulatory networks by TEs as long ago envisioned by McClintock (46) and Britten and Davidson (47).
Materials and Methods
Genomic and Functional Genomic Datasets.
The human genome reference sequence (National Center for Biotechnology Information, NCBI build 36.1, University of California Santa Cruz, UCSC version hg18) was analyzed with respect to the locations of MIR TE sequences and NCBI RefSeq gene locations using the UCSC Genome Browser “RepeatMasker” and “RefSeq Genes” tracks, respectively. ChIP-seq data (48, 49) were used to characterize the genomic locations of 38 histone modifications and one histone variant in CD4+ T cells. ChIP-seq data were used to characterize the genomic locations of RNA Pol II, CTCF (49), and RNA Pol III (41) binding sites in CD4+ T cells. ChIP-seq data from the ENCODE consortium were used to characterized the locations of three histone modifications in GM12878 and K562 cells (44, 50). Microrray data were used to characterized gene expression levels across 79 human tissues (51), including CD4+ T cells, along with GM12878 and K562 (52, 53). Microarray signal intensity values were normalized using the z transformation to compare relative expression levels across tissues and microarray platforms. RNA-seq data from CD4+ T cells (41) were used to characterize genome expression levels. ChIA–PET data from CD4+ T cells (35) were used to identify regulatory interactions between enhancers and promoters.
Bioinformatic Prediction and Validation of MIR Insulators.
Human genome MIR sequences (candidate insulators) were screened through a series of filters to identify a final set of predicted MIR-derived insulators (Fig. 1A). The final set of predicted MIR insulators (n = 1,178) contains the following set of properties: intact B-box promoter sequences, occupancy by RNA Pol III, segregation of active versus repressive chromatin domains, and segregation of expressed versus silent genomic regions. Details of the MIR-insulator prediction algorithm can be found in SI Appendix, SI Methods. The performance of the algorithm to select MIR sequences to segregate individual histone modifications and to partition active and repressive modifications were computationally tested using multidimensional statistical analysis of ChIP-seq data for the 39 CD4+ T-cell histone modifications (SI Appendix, SI Methods and Figs. S4–S6).
Predicted MIR insulators were computationally validated with respect to their chromatin barrier activity and enhancer-blocking activity based on additional functional genomics data that was not used for their prediction. For chromatin barrier activity, the relative distances (normalized by domain sizes) between MIR insulators and LAD boundaries (33) or TAD boundaries (34) were calculated and compared with the relative distances of randomly selected B-box–containing MIR sequences (Mann–Whitney tests). LAD boundaries were characterized in human fibroblast cells and TAD boundaries were characterized in embryonic stem cells and IMR90 cells. For enhancer-blocking activity, ChIA–PET interactions (≤1 Mb interactions) in human CD4+ T cells (35) were integrated into the analysis. To control for the different regional abundances of ChIA–PET interactions, only local interactions that flank MIR insulators (i.e., both anchors of the interaction are located ≤500 kb from a MIR insulator or enclosed by two adjacent MIR insulators) were considered. Those local interactions were further classified into one-side interactions (i.e., both anchors are located on only one side of the MIR insulator) and cross-interactions (i.e., the two anchors are located on opposite sides of the MIR insulator). The ratio between cross-interactions and one-side interactions was used to characterize the degree of depletion of cross-interactions, as a measurement of the enhancer-blocking activity of MIR insulators. One thousand shuffled sets of ChIA–PET interactions were generated by randomly linking anchor sites (≤1 Mb from each other) and used as control to evaluate the statistical significance of the observed cross-interaction depletion around MIR insulators (Z test).
Average sequence conservation levels (54) were calculated for the predicted MIR insulators (which were divided into B-box subregions and other parts of MIRs) and 100-bp upstream/downstream flanking sequences. Randomly selected B-box–containing MIR sequences were used as controls. Proximal genes (i.e., the top two nearest genes within 300 kb) on the active chromatin sides of MIR insulators were used for functional and pathway analysis (55). All expressed genes in CD4+ T cells were used as controls.
EBAs.
A score was calculated to quantify the difference between the levels of active and repressive chromatin marks on the opposite side of predicted MIR insulators and the top three ranked MIR sequences were then selected for experimental validation using EBAs in human and zebrafish. Human EBAs were performed as previously described (14, 56) using the pELuc vector and transient transfection HEK 293 cells. Selected MIR-insulator sequences (SI Appendix, Table S2) were cloned upstream (negative control) or between (test) enhancer and promoter sequences and enhancer-blocking activity was measured based on relative levels of luciferase expression. The 5′ HS4 insulator from the chicken beta-globin locus and the minimal insulator sequence motifs (II/III) from this same element were used as positive controls in this assay. Mutated II/III sequence motifs, incapable of binding CTCF, were used as negative controls. Three replicates were performed for each EBA.
Zebrafish EBAs were performed as previously described (57) using a Tol2 transposon-based vector and transient transfection of zebrafish embryos. Selected MIR-insulator sequences were cloned between a CNS enhancer and a promoter that drives somite expression, and enhancer-blocking activity was measured based on relative levels of somite/CNS GFP expression. The 5′ HS4 insulator from the chicken beta-globin locus was used as a positive control in this assay; an empty vector was used as a negative control. For each putative MIR-insulator sequence tested, 41–46 replicates were assayed to control for chromatin position effects. Point mutations of B-box (T45/C to G45/G) were introduced within the MIR2-insulator element using the GENEART Site-Directed Mutagenesis System (Invitrogen) and the overlapping oligonucleotides: 5′-ATAAAGTGTAGATATCCACCCTGGCCATCAGGCCC-3′ and 5′-CTGATGGCCAGGGTGGATATCTACACTTTATCACT-3′. The same EBA procedure was carried out on this mutated MIR2 insulator. Statistical analyses (median test) were calculated with IBM-SPSS v.21.
Supplementary Material
Acknowledgments
This work was supported by an Alfred P. Sloan Research Fellowship in Computational and Evolutionary Molecular Biology (BR-4839 to J.W. and I.K.J.); a Georgia Tech Integrative BioSystems Institute pilot program grant (to J.W. and I.K.J.); the Spanish Ministry of Science and Innovation (BIO2009-1297 and BIO2012-39980 to L.M., and BFU2010-14839 and CSD2007-00008 to J.L.G.-S); and by Junta de Andalucía (CVI-3488 to J.L.G.-S.). E.M. was supported by Centro de Investigación Biomédica en Red de Enfermedades Raras, Instituto de Salud Carlos III and D.S. was supported by a PhD fellowship from La Caixa program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507253112/-/DCSupplemental.
References
- 1.Gaszner M, Felsenfeld G. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7(9):703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 2.Valenzuela L, Kamakaka RT. Chromatin insulators. Annu Rev Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 3.Capelson M, Corces VG. Boundary elements and nuclear organization. Biol Cell. 2004;96(8):617–629. doi: 10.1016/j.biolcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Lunyak VV. Boundaries. Boundaries...Boundaries??? Curr Opin Cell Biol. 2008;20(3):281–287. doi: 10.1016/j.ceb.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 5.West AG, Gaszner M, Felsenfeld G. Insulators: Many functions, many mechanisms. Genes Dev. 2002;16(3):271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 6.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9(5):397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gdula DA, Gerasimova TI, Corces VG. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc Natl Acad Sci USA. 1996;93(18):9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82(4):587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 9.Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992;6(10):1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- 10.Labrador M, Corces VG. Setting the boundaries of chromatin domains and nuclear organization. Cell. 2002;111(2):151–154. doi: 10.1016/s0092-8674(02)01004-8. [DOI] [PubMed] [Google Scholar]
- 11.Román AC, et al. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011;21(3):422–432. doi: 10.1101/gr.111203.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42(7):631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt D, et al. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148(1-2):335–348. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lunyak VV, et al. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317(5835):248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- 15.Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 2001;20(3):520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125(5):859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 17.Oki M, Kamakaka RT. Barrier function at HMR. Mol Cell. 2005;19(5):707–716. doi: 10.1016/j.molcel.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 18.Scott KC, Merrett SL, Willard HF. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr Biol. 2006;16(2):119–129. doi: 10.1016/j.cub.2005.11.065. [DOI] [PubMed] [Google Scholar]
- 19.Ebersole T, et al. tRNA genes protect a reporter gene from epigenetic silencing in mouse cells. Cell Cycle. 2011;10(16):2779–2791. doi: 10.4161/cc.10.16.17092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raab JR, et al. Human tRNA genes function as chromatin insulators. EMBO J. 2012;31(2):330–350. doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Lunyak VV, Jordan IK. Genome-wide prediction and analysis of human chromatin boundary elements. Nucleic Acids Res. 2012;40(2):511–529. doi: 10.1093/nar/gkr750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7(12):e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurka J, Zietkiewicz E, Labuda D. Ubiquitous mammalian-wide interspersed repeats (MIRs) are molecular fossils from the mesozoic era. Nucleic Acids Res. 1995;23(1):170–175. doi: 10.1093/nar/23.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva JC, Shabalina SA, Harris DG, Spouge JL, Kondrashovi AS. Conserved fragments of transposable elements in intergenic regions: Evidence for widespread recruitment of MIR- and L2-derived sequences within the mouse and human genomes. Genet Res. 2003;82(1):1–18. doi: 10.1017/s0016672303006268. [DOI] [PubMed] [Google Scholar]
- 25.Mariño-Ramírez L, Jordan IK. Transposable element derived DNaseI-hypersensitive sites in the human genome. Biol Direct. 2006;1:20. doi: 10.1186/1745-6150-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piriyapongsa J, Mariño-Ramírez L, Jordan IK. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176(2):1323–1337. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huda A, Bowen NJ, Conley AB, Jordan IK. Epigenetic regulation of transposable element derived human gene promoters. Gene. 2011;475(1):39–48. doi: 10.1016/j.gene.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 28.Huda A, et al. Prediction of transposable element derived enhancers using chromatin modification profiles. PLoS One. 2011;6(11):e27513. doi: 10.1371/journal.pone.0027513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jjingo D, Huda A, Gundapuneni M, Mariño-Ramírez L, Jordan IK. Effect of the transposable element environment of human genes on gene length and expression. Genome Biol Evol. 2011;3:259–271. doi: 10.1093/gbe/evr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smit AF, Riggs AD. MIRs are classic, tRNA-derived SINEs that amplified before the mammalian radiation. Nucleic Acids Res. 1995;23(1):98–102. doi: 10.1093/nar/23.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007;23(4):158–161. doi: 10.1016/j.tig.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Jurka J, Smith T. A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci USA. 1988;85(13):4775–4778. doi: 10.1073/pnas.85.13.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453(7197):948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 34.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chepelev I, Wei G, Wangsa D, Tang Q, Zhao K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 2012;22(3):490–503. doi: 10.1038/cr.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19(1):24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tiana M, et al. A role for insulator elements in the regulation of gene expression response to hypoxia. Nucleic Acids Res. 2012;40(5):1916–1927. doi: 10.1093/nar/gkr842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin D, et al. Genome-wide CTCF distribution in vertebrates defines equivalent sites that aid the identification of disease-associated genes. Nat Struct Mol Biol. 2011;18(6):708–714. doi: 10.1038/nsmb.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleinschmidt RA, LeBlanc KE, Donze D. Autoregulation of an RNA polymerase II promoter by the RNA polymerase III transcription factor III C (TF(III)C) complex. Proc Natl Acad Sci USA. 2011;108(20):8385–8389. doi: 10.1073/pnas.1019175108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simms TA, et al. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7(12):2078–2086. doi: 10.1128/EC.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barski A, et al. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol. 2010;17(5):629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Bortle K, Corces VG. tDNA insulators and the emerging role of TFIIIC in genome organization. Transcription. 2012;3(6):277–284. doi: 10.4161/trns.21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471(7339):480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McClintock B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 47.Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46(2):111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 50.Thomas DJ, et al. ENCODE Project Consortium The ENCODE Project at UC Santa Cruz. Nucleic Acids Res. 2007;35(Database issue):D663–D667. doi: 10.1093/nar/gkl1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101(16):6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bemmo A, et al. Gene expression and isoform variation analysis using Affymetrix Exon Arrays. BMC Genomics. 2008;9:529. doi: 10.1186/1471-2164-9-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gardina PJ, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 56.Recillas-Targa F, Bell AC, Felsenfeld G. Positional enhancer-blocking activity of the chicken beta-globin insulator in transiently transfected cells. Proc Natl Acad Sci USA. 1999;96(25):14354–14359. doi: 10.1073/pnas.96.25.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bessa J, et al. Zebrafish enhancer detection (ZED) vector: A new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev Dyn. 2009;238(9):2409–2417. doi: 10.1002/dvdy.22051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.