Abstract
The transcription factor Pax6 is comprised of the paired domain (PD) and homeodomain (HD). In the developing forebrain, Pax6 is expressed in ventricular zone precursor cells and in specific subpopulations of neurons; absence of Pax6 results in disrupted cell proliferation and cell fate specification. Pax6 also regulates the entire lens developmental program. To reconstruct Pax6-dependent gene regulatory networks (GRNs), ChIP-seq studies were performed using forebrain and lens chromatin from mice. A total of 3514 (forebrain) and 3723 (lens) Pax6-containing peaks were identified, with ∼70% of them found in both tissues and thereafter called ‘common’ peaks. Analysis of Pax6-bound peaks identified motifs that closely resemble Pax6-PD, Pax6-PD/HD and Pax6-HD established binding sequences. Mapping of H3K4me1, H3K4me3, H3K27ac, H3K27me3 and RNA polymerase II revealed distinct types of tissue-specific enhancers bound by Pax6. Pax6 directly regulates cortical neurogenesis through activation (e.g. Dmrta1 and Ngn2) and repression (e.g. Ascl1, Fezf2, and Gsx2) of transcription factors. In lens, Pax6 directly regulates cell cycle exit via components of FGF (Fgfr2, Prox1 and Ccnd1) and Wnt (Dkk3, Wnt7a, Lrp6, Bcl9l, and Ccnd1) signaling pathways. Collectively, these studies provide genome-wide analysis of Pax6-dependent GRNs in lens and forebrain and establish novel roles of Pax6 in organogenesis.
INTRODUCTION
Cellular differentiation is driven by a combinatorial action of lineage-specific DNA-binding transcription factors and extracellular signaling that results in activation and repression of specific batteries of genes, accompanied by dramatic changes in their chromatin structure. These transcription factors are molecular determinants of cell type specification and differentiation. To control cell fate decisions during development, a group of ‘top hierarchy’ transcription factors, including BSAP/Pax5, Gata1, Gata2, MyoD, Pax6, PU.1, Runx2, Sox9 and a few others (often called ‘master’ regulators), control expression of ‘secondary’ transcription factors to establish specific cell fates. The same transcription factors often control the expression of terminal differentiation products, such as globins (regulated by Gata1 in erythrocytes) and crystallins (regulated by Pax6 in lens). Interestingly, a number of lineage-specific transcription factors fulfill a dual role in activating specific genes in one cell type while repressing their expression in a different cell type. Consequently, reconstruction of gene regulatory networks (GRNs) in multiple cell types or tissues that are regulated by a common transcription factor can help elucidate specific molecular mechanisms that govern gene activation and/or repression during differentiation.
The lineage-specific and evolutionarily conserved transcription factor Pax6 exerts a multitude of critical roles in brain and eye development. In the brain, Pax6 functions in early stages of cortical neurogenesis (1,2) and in differentiation of specialized types of neurons such as glutamatergic projection neurons (2,3) as well as dopaminergic neurons in the olfactory bulb (4). In the eye, Pax6 is essential for the formation of lens progenitor cells that form the lens placode (5,6). In parallel, Pax6 both controls formation of retinal progenitor cells and is essential for their multipotency within the optic vesicle neuroepithelium (7,8). Pax6 is also essential in both formation of retinal pigmented epithelium (RPE) (9) and corneal epithelium (10). Finally, Pax6 regulates many subsequent stages of lens morphogenesis including the expression of crystallin genes (5). However, little is known about the molecular mechanisms of Pax6 regulation in vivo as it is unclear how Pax6 is engaged in these diverse and tissue-specific cellular differentiation processes. Consequently, the structure and organization of Pax6-dependent gene regulatory networks (GRNs) in different cell types and throughout differentiation remain to be established. Use of genome-wide ChIP-seq platforms in conjunction with differential RNA profiling now opens new opportunities to reconstruct Pax6-dependent GRNs to unravel mechanisms of Pax6-mediated cellular differentiation.
Studies of chromatin and gene transcription have lead to a general model in which DNA-binding transcription factors recruit a plethora of chromatin remodeling complexes to dynamically regulate local chromatin structure (11,12). Genome-wide studies have shown that promoter regions are marked by H3K4me3 post-translational histone modification (PTM) and RNA polymerase II and that distal enhancers are marked by abundant H3K4me1 and H3K27ac core histone modifications (13). Interestingly, certain enhancers (class II) even contain repressive H3K27me3 modifications (14). In addition, some enhancers are also marked by RNA polymerase II and eRNA (enhancer RNA) transcripts (15,16). Embryonic stem (ES) cell differentiation has provided insight into the chromatin structure at the ‘ground’ state (17,18) as well as information on transitions during the differentiation of specific cell types such as neuronal progenitors (19). It has been shown that many developmental regulatory genes posses a ‘bivalent domain’ structure comprised of a broad H3K27me3 and a narrow H3K4me3 region at their promoters (17). Ezh1/2, a catalytic subunit of the Polycomb group repressive complex (PRC), methylates H3K27 to H3K27me3 (18). Upon receiving differentiation signals, the bivalent domain promoters adopt ‘open’ chromatin structure through removal of PRCs (18). These differentiation processes are driven by top-hierarchy factors, e.g. Pax6, which is expressed in both early neural and lens progenitors. In both systems, Pax6 regulates cell type specification and total numbers of individual committed progenitors, including radial glial and lens placode cells (20–22). Identification of genes directly regulated by Pax6 is required to explain how Pax6 mediates these ‘global’ roles. We have shown earlier that Pax6 directs multiple positive feed-forward loops both in lens (23) and RPE cells (9) to control expression of proteins encoded by terminally differentiated genes. Whether this specific molecular mechanism is more broadly used among DNA-binding factors during cortical development and differentiated lens remains to be determined.
Here we conducted comparative genome-wide study of Pax6 binding in mouse E12.5 embryonic forebrain and newborn lens chromatin. We examined binding with Pax6-dependent gene expression to identify those genes directly regulated by Pax6. Novel Pax6-dependent GRNs that contribute to the function of Pax6 were identified in both tissues. Analysis of RNA polymerase II, H3K4me1, H3K4me3, H3K27ac and H3K27me3 in ES cell, forebrain and lens chromatin was used to annotate both Pax6-bound and Pax6-free tissue-specific enhancers. Collectively, these studies revealed three types of in vivo consensus Pax6 DNA-binding sites (PD, PD/HD and HD), determined the proportion of lens-specific promoters and enhancers occupied by Pax6, and established novel GRNs that regulate cortical neurogenesis and lens differentiation.
MATERIALS AND METHODS
Antibodies
The antibodies recognizing H3K4me1 (Abcam, ab8895), H3K4me3 (Abcam ab8580), H3K27ac (Millipore, #07–360), H3K27me3 (Millipore, #07–449), Pax6 (Millipore, ab2237), and RNA polymerase II (Abcam ab817) were used in ChIP-seq and qChIP assays.
Chromatin immunoprecipitation and library preparation
Forebrain (E12.5) and newborn (P1) lenses of CD-1 mice (Charles River Laboratories) were dissected as described earlier (24). Dissected tissues were kept at −80°C. Tissue (100 mg) was freshly crosslinked with 1% formaldehyde for 15 min at room temperature. Tissue was then homogenized in a Dounce glass homogenizer (pestle B) with 10 ml of lysis buffer A (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100, 1× protease inhibitors). Lysate was centrifuged at 2000 rpm for 10 min and supernatant was discarded. The pellet was resuspended in 3 ml of lysis buffer B (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.2% SDS) and sonicated (settings: 40% amplitude, 15 s on 45 s off, 10 min in total) in a dry ice/ethanol bath. Lysate was collected by centrifugation (12 000 x g) for 15 min and the supernatant was collected for immunoprecipitation. Sonicated lysate was clarified by centrifugation at 4°C for 15 min. Chromatin was diluted in buffer C (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, 1× protease inhibitors) and preclarified with Protein G Dynabeads (Invitrogen). Each immunoprecipitation was performed with 5–10 μg antibodies. After overnight incubation at 4°C, 50 μl of Dynabeads were added and incubation was continued for 1 h. Beads were sequentially washed two times in buffer B or C, two times in wash buffer (0.1% SDS, 1% Triton X-100, 1mM EDTA, 20 mM Tris-HCl [pH 8.1], 500 mM NaCl), two times in LiCl buffer (100 mM Tris-HCl [pH 8.1], 500 mM LiCl, 1% NP-40, 1% Na deoxycholate), and finally once in Tris-EDTA. Antibody-bound chromatin complexes were eluted by incubating for 15 min at 25°C in 250 μl of elution buffer (50 mM Tris-HCl [pH 8.1] and 1% SDS). Incubating samples at 65°C overnight reversed crosslinking. Samples were digested with proteinase K for 1 h at 56°C and phenol-chloroform extracted. The DNA pellet was dissolved in water and used for downstream qPCR or library preparation.
ChIP-seq analysis, initial assignment of peak regions and peak visualization
Sequencing of Pax6 and histone ChIP-seq experiments was performed on Illumina HiSeq 2500 and Genome Analyzer IIx instruments. ChIP-seq reads were processed by the Einstein WASP analysis pipeline (25) and aligned to the mouse genome (GRCm37/mm9) using Bowtie (26), with the option ‘– tryhard’ used to find valid alignments for low quality reads. For Pax6-binding and histone modifications (H3K4me3 and H3K27ac) with sharp enriched regions, peaks were called using the MACS2 program (27) with default settings. For histone modifications with broadly enriched regions (H3K4me1 and H3K27me3), peaks were called using the SICER program (28) with default settings. Where multiple ChIP-seq replicates were available, reads from all replicates were pooled for final peak calling. We also filtered out peaks that mapped to the modENCODE blacklisted genomic regions (29). The IGV browser (30) was used for data visualization. The program MAnorm (31) was used to find tissue-specific Pax6 peaks, with 0.01 as p-value cutoff (for unique peaks) and default for the rest of the settings. The data were deposited into GEO under an accession number GSE66961. ES cell ChIP-seq data were downloaded from the GEO (GSM1000099, GSM1000089, GSM881352, GSM769008, GSM918749) (32,33).
Unbiased analysis of enriched DNA motifs in Pax6-bound regions
MEME 4.9.0 (34) and MEME-ChIP (35) suites were used to find de novo enriched motifs within 200 bp sequences centered on the summits of the Pax6 peaks. The enrichment of known motifs from JASPAR (36) and de novo identified motifs from MEME, as well as previously identified Pax6 motifs (23) was determined by using MAST (37).
Identification of genes with Pax6 binding and data visualization
The association of Pax6-peaks to genes was carried out by in-house scripts using the RefSeq (38) annotation, downloaded in February 2013 from the UCSC Genome Browser (39). This annotation contained 31 864 transcripts from 23 397 genes, including both protein-coding and non-coding genes. The script assigned peaks to genes in a step-wise manner: to promoter regions (−2 kb to +2 kb of transcriptional start sites, TSSs), to exons, to introns, or to distal regulatory regions (−50 kb of transcription starts to +50 kb of transcription ends). Only the gene with the closest TSS was selected when mapping peaks to either promoter regions or distal regions. A peak could be mapped to multiple genes, but only if it was located to exons or introns shared by multiple genes or if is equidistant from the TSS of multiple genes.
The heatmaps to visualize ChIP-seq data were generated by the seqMiner program (40), which calculated ChIP-seq read density and resulted in an array that consisted of the maximal number of overlapping ChIP-seq reads (extended to 200-bp) in 50-bp bins from −5 kb to +5 kb of the Pax6 peak summits. The enrichment of the sequencing-depth-normalized reads over those of input experiments for each cell type was calculated. The final matrix of enrichment values was used to generate profile plots in the R package.
Reanalysis of Pax6-dependent gene expression
For forebrain differential expression analysis, we utilized Pax6 null mutant versus WT microarray expression data from GSE38703 (22). We averaged probes that were assigned to the same gene across replicates to get an average ratio for the forebrain expression. We selected a ratio of 1.5 for a gene as a cutoff for Pax6-dependent expression. For lens, we used lens Pax6 conditional knockout versus WT microarray expression data and differential gene identification from (41).
Secondary validations by qChIP and qRT-PCR
Secondary validations of selected genomic regions by qChIP were conducted using independent chromatin material using the standard curve normalization as described earlier (23,42). Differentially expressed transcripts between E9.5 Pax6−/− conditionally inactivated and wild type lens placode (24) were examined by qRT-PCR as described earlier (24). In addition,E12.5 Pax6−/− forebrain was generated by crossing Pax6lacZ/+ mice (43). Transcripts that were differentially expressed between E12.5 wild type and Pax6−/− forebrain were examined by qRT-PCR as described earlier (24). The primers are listed in Supplemental Table S1.
RNA-seq sequencing, analysis and data processing
Mouse tissues were freshly dissected and homogenized in Trizol reagent, and total RNA was extracted using Trizol. The purified RNAs were treated with DNaseI and RNA integrity was checked by bioanalyzer. Sequencing of RNA-seq experiments was performed on an Illumina HiSeq 2500 instrument. The reads were processed by the Einstein WASP analysis pipeline (25) and aligned to the mouse genome (GRCm37/mm9) using TopHat (v2) (44). Genes studied in the analysis are from Illumina iGenomes gene annotation with UCSC data source mm9 (ftp://igenome:G3nom3s4u@ussd-ftp.illumina.com/Mus_musculus/UCSC/mm9/Mus_musculus_UCSC_mm9.tar.gz). The Cuffdiff tool in the Cufflinks package (v2) (45) was used to determine RPKM (Reads Per Kb exons per Million mapped reads), with default geometric mean normalization and the options of correcting for sequence bias and multiple hits.
RESULTS
In vivo identification of Pax6-binding regions in forebrain and lens chromatin within known and novel genes by ChIP-seq
In contrast to earlier studies of Pax6-binding in vivo, which used promoter ChIP-chip in forebrain, lens and pancreas chromatin (24,46), ChIP-seq analysis provides whole genome data with much improved signal to noise ratio (47). In E12.5 forebrain and newborn lens chromatin we found comparable numbers (3514 and 3723, respectively) of Pax6 ‘peaks’ (Figure 1A). Among those peaks, ∼2500 (i.e. ∼70%) were shared between the two tissues. To illustrate the specificity of tissue-specific Pax6 peaks, we examined ChIP-seq enrichment signals (by number of normalized reads) at peak summits. We found that both forebrain- and lens-specific peaks exhibited higher enriched ChIP-seq signals compared to the shared peaks and only background signals were observed in the other tissue (Supplementary Figure S1A). Partition of Pax6 peaks based on their locations within the gene body (exons and intron), in promoters, or in distal or intergenic regions revealed that the promoters, intron and distal regions contained the majority of Pax6 binding, while intergenic (‘far’ distal) regions had less representation, relative to the genome-wide expectation (Figure 1B).
Figure 1.
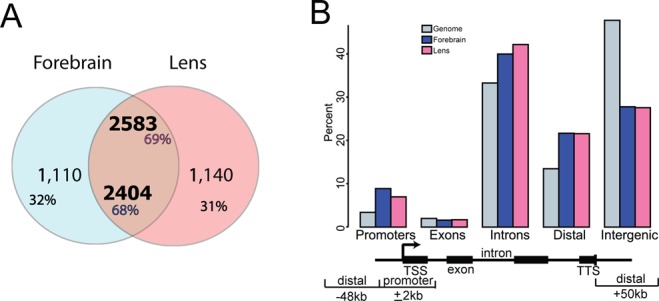
Identification, initial characterization and genomic distribution of Pax6 peaks in embryonic forebrain and newborn lens chromatin. (A) Venn diagram showing a large overlap of Pax6 peaks in lens and forebrain chromatin. (B) Genomic location of Pax6 peaks. A bar diagram indicates the percentages of Pax6-binding sites in the promoter (±2 kb relative to TSS), gene body (exons and introns), distal regions (±50 kb relative to TSS/TES) and intergenic regions.
A representative panel of six loci occupied by Pax6, including Pax6, Cryaa, Crybb3, Fezf2, Mab21l1 and Ngn2, is shown to illustrate Pax6-bound peaks at the individual gene level (Figure 2). Pax6 is known to autoregulate its own expression via an ectodermal enhancer (EE) found in the 5′-flanking region of the P0 promoter (48–50), which we identified here in the forebrain chromatin. A common novel peak region was found between exons 7 and 8 in both forebrain and lens chromatin (Figure 2). In addition, three novel peaks were found at positions −16, −14.5 and −11 kb upstream of the P1 promoter only in lens chromatin. These findings suggest that Pax6 autoregulation employs additional enhancers that may also be required for initiation of Pax6 expression in the surface ectoderm prior the EE (48–50). Cryaa, encoding the αA-crystallin, is a highly expressed lens-specific gene regulated by Pax6 (42). As expected, Pax6-binding was found in the distal (∼8 kb) DCR1 enhancer in lens (42) but not in forebrain chromatin (Figure 2). Crybb3 is another crystallin gene; however its transcriptional control is unknown. We found here that Pax6 binds to its proximal 5′-flanking promoter in lens but not in forebrain chromatin (Figure 2). Fezf2 is a zinc-finger transcription factor that directs differentiation of multipotent proneural cells in the subventricular zone (SVZ) of the prospective cerebral cortex (51). Pax6 binds to its 3′-distal region of Fezf2 in forebrain but not in lens chromatin (Figure 2). Mab21l1 is an important Pax6-dependent regulatory gene (24,52) that protects cells in the invaginating lens placode from apoptosis by an unknown mechanism (52). Pax6 binds to Mab21l1 promoter and distal 5′-region (−31 kb) in both lens and forebrain chromatin (Figure 2). Neurogenin 2 (Ngn2) is a bHLH transcription factor directly regulated by Pax6 in dorsal forebrain (53). Two Pax6-binding regions were found −7.2 and −0.4 kb from the transcriptional start-site (TSS) in forebrain but not in lens chromatin (Figure 2). Taken together, the present ChIP-seq studies provide new and useful insights to understand the molecular basis of forebrain and lens-enriched gene expression and serve as ‘reference’ for analysis of Pax6 at different developmental stages in mouse embryos or during ES-cell differentiation towards specialized neuronal cells.
Figure 2.
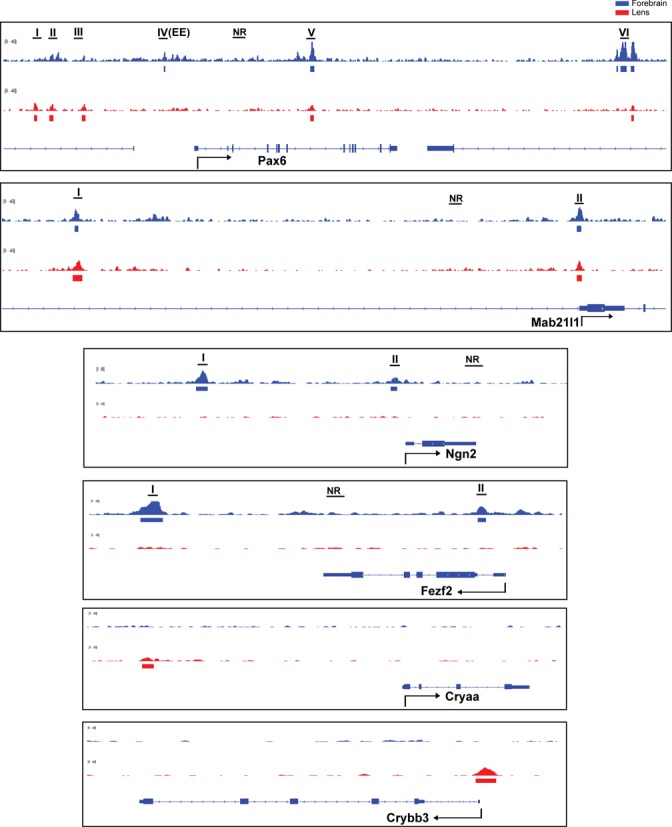
Pax6 binding in forebrain and lens chromatin at Pax6, Mab21l1, Fezf2, Ngn2, Cryaa, and Crybb3 loci. A representative panel of six loci occupied by Pax6: Pax6, and Mab21l1 (common peaks), Fezf2, and Ngn2 (forebrain-specific binding), and Cryaa and Crybb3 (lens-specific peaks). The Pax6 peaks (labeled by roman numbers) were independently validated by qChIPs as shown in Figure 5A. The previously identified Pax6 ectodermal enhancer (EE) corresponds to Pax6 peak IV. The non-bound regions (NR) examined by qChIPs are also indicated.
Identification and characterization of in vivo Pax6 DNA-binding sites
To identify de novo DNA sequence motifs enriched in the Pax6 peaks, we used the MEME suite (as described in Material and Methods). The motif analysis of Pax6 peaks was conducted separately for the forebrain and lens data and revealed multiple motifs (Figure 3). The top motif (E-value 3.2e−389) found in forebrain chromatin is a 15 bp sequence that resembles in vitro defined Pax6-binding site recognized by the paired domain (PD) (54); this motif is enriched at the peak summits (Figure 3A). The top motif (E-value 9.8e−326) found in lens chromatin is nearly identical (Figure 3A). Interestingly, the 3′-region of the bipartite PD binding site, though not optimal for PD binding in in vitro assays (55,56), corresponds to the de novo sites identified here. Taken together, the de novo identified common motifs were found in over 3000 individual peaks immunoprecipitated by antibody specific to C-terminal region of Pax6 and establishes the in vivo Pax6 consensus binding sequence.
Figure 3.
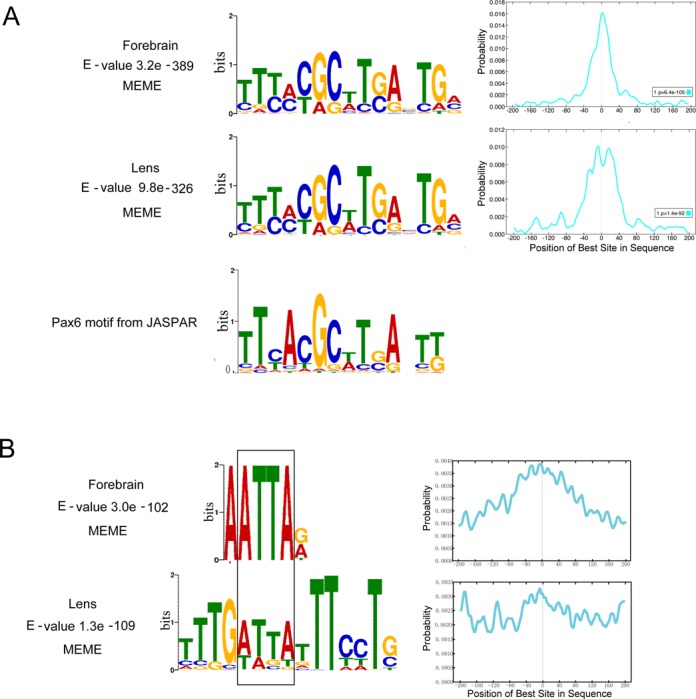
De novo enriched motifs within the Pax6 peaks. (A) A comparison between top motifs identified in lens and forebrain using all peaks (centered ± 100 bp from the peak summits) with a consensus Pax6 binding site determined by in vitro SELEX studies (JASPAR database (36)). The graphs show distribution of the de novo identified motifs within the 200 bp region examined. (B) A comparison between the second most frequent motifs and Pax6-binding sequences and identification of the common 5′-ATTA-3′ motif (boxed).
The second most frequent motif (E-value 3.0 e−102) found in forebrain chromatin is a 6 bp sequence with a highly conserved ATTA motif and a preferred occurrence around the peak summit regions (Figure 3B). The second most frequent motif (E-value 1.3e−109) found in lens chromatin is a 15 bp sequence that resembles the AT-rich binding sites of forkhead proteins (Figure 3B); nevertheless, a closer inspection revealed a distinct 5′-ATTA-3′ motif known to be recognized by many homeodomain-containing transcription factors (57), including Pax6 (23,58). Out of more than 70 Pax6-binding sites none examined by in vitro protein-DNA studies contain just the ATTA motif, such as the AT-rich sequence (called G1 element) that binds Pax6 via its PD but not HD in the rat glucagon promoter (59).
To further characterize DNA sequences recognized by Pax6, we conducted additional analyses. An appreciable number of Pax6 promoters and enhancers are comprised of multiple adjacent Pax6-binding sites (23,53,59,60). Thus, we tested if some Pax6 peaks contained multiple Pax6-recognition sites. In both forebrain and lens chromatin, we indeed found a large but comparable number of Pax6 peaks (16.5% and 15.4%, respectively) containing multiple (n = 2–4) instances of the predicted PD-motif (Table 1). Earlier studies identified 10 Pax6-binding sites where the ATTA HD-recognition motif is immediately located to the 5′ end of the PD binding sequences, forming the Pax6 HD-PD recognition site (23,61). Here, we detected the co-existence of the Pax6 HD-PD motifs in 31.4% and 26.6% of the PD motif contained Pax6 peaks in forebrain and lens, respectively (Supplementary Figure S2A, B). Finally, Pax3 and Pax7 proteins have similar molecular structures and conserved DNA binding domains as Pax6 (62,63). ChIP-seq analysis of Pax3- and Pax7-binding in myoblast chromatin identified similar consensus sequences for both these proteins; nevertheless the HD sequences were more abundant than the PD motif (64). Specifically, the prevailing motif of Pax3/7 is comprised of two ATTA palindromes separated by two base pairs, 5′-TAATTGATTA-3′, suggesting a cooperative binding mediated by two Pax3/7 HDs (Supplementary Figure S2C). The 10 bp Pax3/Pax7 PD motif (Supplementary Figure S2C) (64) is comprised of nucleotides that are recognized by the β-turn and PAI-domain, as judged from crystallographic data of Pax6 (65). In contrast, Pax6 PD-binding sites involve all four DNA-binding modules of Pax6, including β-turn, PAI-domain HTH, linker and RED-domain HTH. Previous DNase I footprinting studies have shown that Pax6 proteins occupy between 15 and 20 bp of DNA (54,60,65). From these data we propose that in vivo DNA-binding preferences of Pax6 are both quantitatively and qualitatively different from the structurally similar Pax3 and Pax7 proteins.
Table 1. Distribution of Pax6 peaks with multiple (n ≤ 4) PD or HD motifs in forebrain and lens chromatin.
| Tissue | % of peaks (n) with multiple paired domain (PD) motifs | % of peaks (n) with multiple homeodomain (HD) motifs | % of peaks with PD and HD motifs | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | 4 | 3 | 2 | 1 | 0* | 4 | 3 | 2 | 1 | 0* | N/A |
| Forebrain | 0.1 | 1.6 | 14.8 | 51.3 | 32.3 | 0 | 0.5 | 5.1 | 27.4 | 66.9 | 23 |
| Lens | 0.1 | 1.1 | 14.2 | 52.5 | 32.1 | 0 | 0.1 | 2.0 | 19.2 | 78.6 | 27 |
*) There are at least two possibilities to explain the finding that no Pax6 PD, or HD motifs were identified in a certain number of peaks. First, there are Pax6-binding sites with lower level of sequence conservation (24). Second, some peaks are the result of promoter-enhancer looping and the actual binding site is not present in the DNA that was immunoprecipitated from the crosslinked chromatin.
Not applicable, N/A.
Pax6-dependent and direct gene regulation during forebrain and lens development
To identify genes directly regulated by Pax6 in forebrain and lens, we linked Pax6 peaks to their adjacent genes (see Materials and Methods), and studied the expression of Pax6-bound genes upon Pax6 gene expression reduction. A number of earlier studies examined differential gene expression in mouse E12 (66), E12.5 (22), E14 (67) and E15 (66) wild type and Pax6−/− cortices. In lens, transcriptome analysis of E9.5 and E10.5 lens placodes (mutated prospective lens ectoderm) (68,69) or E14.5 lens (41) were conducted following conditional inactivation of Pax6. By comparing differentially expressed genes obtained with microdissected E12.5 cortices (22) and E14.5 lens (41) with current Pax6-bound genes, we found 90 and 191 genes were directly regulated by Pax6 in forebrain and lens, respectively (Figure 4). The overlap was statistically significant (binomial test; forebrain: P = 5.4E-9, lens: P = 1.9E-14). In forebrain, Pax6 repressed a slight majority of Pax6-regulated genes (47/90, 52%). In contrast, Pax6 activated a clear majority of lens genes identified here (150/191, 79%). A group of 13 genes, including Aldh1a3/Raldh3, Ccnd1, Dusp6, Epbh1, Foxp2, Lmo1, Mab21l1, Mrps28, Pax6, Slc39a11, Spsb4, Tenm2 and Tspan7, is directly regulated by Pax6 in both tissues as each has altered expression. Direct forebrain Pax6 target genes code for proteins of varying function; nevertheless the three largest functional classes are DNA-binding transcription factors, cell surface receptors including cell adhesion molecules and their ligands, and intracellular signaling molecules (Figure 4). In lens, the top five functional categories include DNA-binding transcription factors, cell surface receptors and their ligands, intracellular signaling molecules, cytoskeletal proteins, and extracellular matrix (ECM) proteins. Notably, many of these proteins function in Wnt signaling and cell polarity control. We found that direct Pax6 targets in both tissues were slightly enriched in comparison to all Pax6-bound targets in each tissue for HD-motif containing Pax6 peaks (∼5% increase), but only forebrain direct targets had a preference for PD motif containing peaks (∼10% increase)
Figure 4.
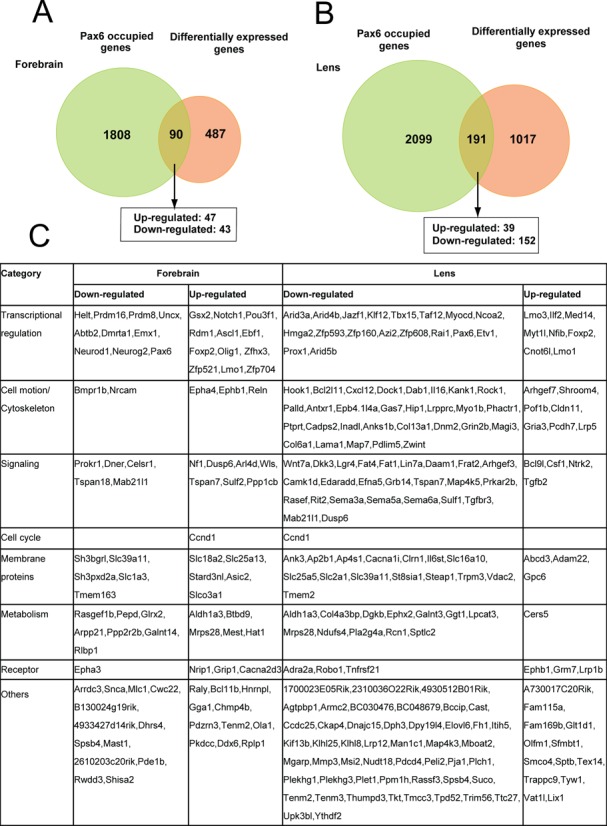
Identification of directly regulated Pax6 genes in forebrain and lens. (A) Venn diagram of genes identified by Pax6 ChIP-seq in E12.5 forebrain compared to differentially expressed genes in E12.5 wild type and Pax6 null embryos (22). (B) Venn diagram of genes identified by Pax6 ChIP-seq in newborn lens compared to differentially expressed genes in E14.5 wild type and conditionally inactivated Pax6 in lens (41). Note that 12 genes were differentially expressed and bound by Pax6 in both tissues. The direct target genes are shown within larger functional groups. Genes up-regulated (down-regulated) by Pax6 are shown in red (blue), respectively.
Validation of differential gene expression and Pax6-binding
Among the 90 genes identified in forebrain (191 in lens), several of them have been reported previously as Pax6 targets (22,24), but the majority of them are novel Pax6 direct targets. To independently demonstrate their dependence on Pax6, we conducted analysis of mRNAs and qChIPs in both tissues of interest. Our qChIP studies validated Pax6-binding regions in Ascl1, Ccnd1, Dmrta1, Emx1, Emx2, Ephb1, Fezf2, Ngn2, and Pax6 in E12.5 forebrain, and Fgfr2, Mab21l1, Pax6, and Prox1 in lens chromatin (Figure 5A). In addition, studies of transcripts encoding Dmrta1, Ngn2, Pax6, and Prox1 showed that these genes were down-regulated in mutated forebrains while expression of Ascl1, Emx2, Ephb1, Fezf2, and Gsx2 were up-regulated (Figure 5B). Interestingly, while Bcl11b, Ccnd1, Foxp2, and Mab21l1 had Pax6-binding, there was no change in their expression in forebrain (Figure 5B, data not shown). Finally, down-regulation of Pax6, Prox1 and c-Maf was found between wild type E9.5 lens placodes and Pax6 depleted surface ectoderm (Figure 5B). In sum, these RNA data validate earlier gene-expression studies in both forebrain (46,70,71) and lens (24,68).
Figure 5.
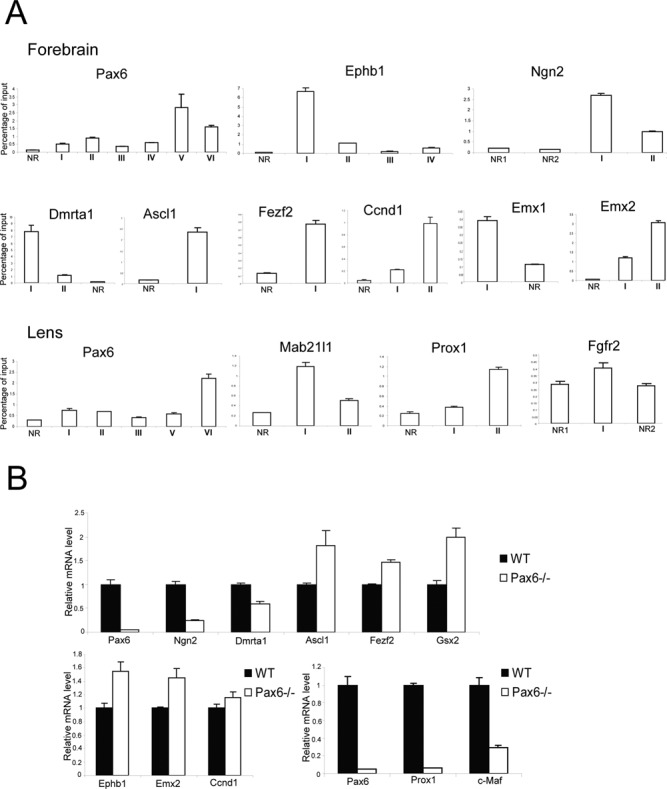
Validation of Pax6 directly regulated genes in forebrain and lens by qChIPs and qRT-PCR. (A) ChIP analysis of Pax6, Ephb1, Ngn2, Dmrta1, Ascl1, Fezf2, Ccnd1, Emx1, and Emx2 in E12.5 forebrain, and Pax6, Mab21l1, Prox1 and Fgfr2 in newborn lens chromatin. Position of the peaks and non-bound regions (NRs) is shown in Figure 1B and Supplementary Figure S4. The NRs were arbitrarily selected in nearby regions to the Pax6-bound peaks within the same loci. (B) RT-PCR analysis of Pax6, Ngn2, Dmrta1, Ascl1, Fezf2, Gsx2, Ephb1, Emx2 and Ccnd1 in Pax6−/- E12.5 forebrain, and Pax6, Prox1, and c-Maf in conditionally deleted E9.5 prospective lens ectoderm.
Distribution of Pax6 sites within promoters and enhancers and the regulatory effects
To better understand partition of chromatin in forebrain and lens related to the presence of Pax6, we determined localization of RNA polymerase II and four core histone PTMs, including H3K4me1, H3K4me3, H3K27ac and H3K27me3. These PTMs and their specific combinations are known to mark active, poised and inactive promoters and enhancers (13). We also included previously published PTM data from mouse ES cells (32,33) as a ‘ground state’ chromatin for embryonic development. We computed ChIP-seq read densities in ±5 kb regions of the 3514 Pax6 sites in forebrain and 3723 in lens chromatin (Figure 1A) and clustered the data by k-means clustering using the seqMINER software (40), generating heatmaps as shown in Figure 6A, B. The results revealed eight clusters of Pax6 peaks in the forebrain and seven in the lens each with unique combination of chromatin marks. Clusters I to III from the ESC-forebrain comparison and clusters I and II from the ESC-lens comparison were marked by H3K4me3 in forebrain or lens, as well as in ES cells, and thus represented Pax6-bound gene promoters. Indeed, about 50% (229/459 in forebrain and 185/373 in lens) of these Pax6 peaks are within Refseq annotated promoters. In contrast, clusters IV (III) to VIII in forebrain (VII in lens) tended to colocalize with various combinations of H3K4me1 and H3K27ac, suggesting that these regions were Pax6-bound enhancers. While 22.7% of forebrain Pax6 binding sites had high levels of H3K27me3 proximal to the peak, there were almost no Pax6 peaks with H3K27me3 in lens, an interesting finding that may be due to the highly differentiated status of lens cells (see below).
Figure 6.
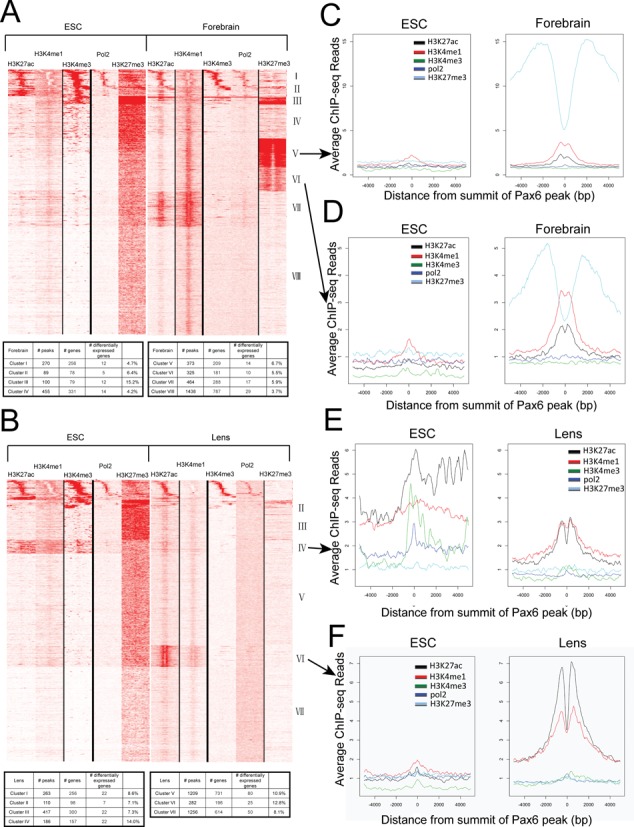
Heatmaps showing the co-occurrence of Pax6, core histone PTMs and RNA polymerase II. (A) Heatmap of maximal read coverage in 50 bp bins from −5 kb to +5 kb of the peak summits at forebrain Pax6 peaks (n = 3514). ChIP-seq data from H3K4me1, H3K4me3, H3K27ac, H3K27me3 and RNA polymerase II in ES cell and forebrain chromatin shown as labeled. The numbers of Pax6 peaks from clusters I to VIII are shown in table format. (B) Heatmap of maximal read coverage in 50 bp bins from −5 kb to +5 kb of the peak summits at lens Pax6 peaks (n = 3723). ChIP-seq data from H3K4me1, H3K4me3, H3K27ac, H3K27me3 and RNA polymerase II in ES cell and lens chromatin shown as labeled. The numbers of Pax6 peaks from clusters I to VII are shown in table format. (C–F) Profiles of H3K4me1, H3K4me3, H3K27ac, H3K27me3 and RNA polymerase II at forebrain Pax6 peaks in ES cell, forebrain and lens chromatin at individual clusters. Y-axis shows the read density per 50 bp averaged over Pax6-bound peaks in each tissue from −5 kb to +5 kb of the peak summits. Data were normalized to a read depth of 10 million mapped reads. Formation of forebrain-specific class II enhancers compared to their status in ES cells in clusters V and VI marked by H3K27me3 in both cell types (panels C,D). Activation of poised and inactive enhancers in ES cells in lens identified by clusters IV and VI (panels E,F), respectively.
At the genome-wide level, we have also examined genomic regions enriched with the four PTMs and RNA polymerase II ChIP-seq signals. To interpret these data, we took the advantage of experimentally validated correlations between specific PTMs and individual promoter/enhancer activity (13,14). The results are given in Table 2. Of the ∼ 32 000 genes in the mm9 mouse annotation, we observed that 13 734 and 14 880 of the annotated promoters were putatively active (marked by the presence of RNA polymerase II and H3K4me3) in forebrain and lens, respectively, of which 324 and 324 were bound by Pax6 in forebrain and lens. Even with the most generous promoter-Pax6 association, Pax6 bound no more than 1.4% of promoters and 2.4% of active promoters.
Table 2. Genome-wide combinations of enhancer-specific chromatin marks and co-localization with RNA polymerase II and Pax6.
| Chromatin modification/Pol II/Pax6 | Forebrain | Lens | ||||
|---|---|---|---|---|---|---|
| Peak Number | Total Base Pairs | Enhancer [%] | Peak Number | Total Base Pairs | Enhancer [%] | |
| H3K4me1 | 40,774 | 145,564,626 | 20,811 | 74,059,189 | ||
| H3K27ac | 26,863 | 19,594,746 | 20,053 | 11,848,208 | ||
| H3K27me3 | 25,474 | 185,092,127 | 31,034 | 123,586,366 | ||
| H3K4me1 AND H3K27ac ( = Enhancer, Enh) | 13852 | 100.0% | 4998 | 100.0% | ||
| H3K4me1 AND H3K27ac AND Pol II | 967 | 7.0% | 564 | 11.3% | ||
| H3K4me1 AND H3K27ac AND H3K27me3 (Enhancer Class II) | 3194 | 23.1% | 516 | 10.3% | ||
| H3K4me1 AND H3K27ac AND Pax6 | 949 | 6.9% | 625 | 12.5% | ||
| H3K4me1 AND H3K27ac AND Pol II AND Pax6 | 49 | 0.4% | 21 | 0.4% | ||
| H3K4me1 AND H3K27ac AND H3K27me3 AND Pax6 (Pax6 - Enhancer Class II) | 342 | 2.5% | 46 | 0.9% | ||
| H3K4me1 OR H3K27ac (‘poised’ enh) | 47627 | 100.0% | 32900 | 100.0% | ||
| H3K4m1 OR H3K27ac AND H3K27me3 (bivalent poised enhancers) | 10804 | 22.7% | 3484 | 10.6% | ||
| H3K4m1 OR H3K27ac AND Pol II | 1586 | 3.3% | 3185 | 9.7% | ||
| H3K4me1 OR H3K27ac AND Pax6 | 2160 | 4.5% | 1566 | 4.8% | ||
With regard to transcriptional enhancers, we found a greater number of active enhancers in forebrain (13 852) than in lens (4998), marked by high abundance of both H3K4me1 and H3K27ac. Among them, a total of 949 (6.9%) and 625 (12.5%) overlapped with Pax6 binding regions in forebrain and lens, respectively (Table 2). Recent genome-wide studies have identified two additional subtypes of enhancers: one marked by RNA polymerase II (transcribing eRNA) (15,16) and the other with abundant H3K27me3 (class II) (14). In forebrain, we found 49 (5.2%) eRNA and 342 class II (36.0%) Pax6-bound enhancers, which both target neuronal differentiation genes. In lens, we found reduced numbers of only 21 (3.4%) eRNA and 46 class II (7.4%) enhancers, which did not show significant enrichment for any gene ontology terms. Taken together, while Pax6 binds to ∼1/40 of the active promoters, it binds ∼1/10 (6.9–12.5%) of all active enhancers (Table 2). The identification of an appreciable number of enhancers marked by H3K27me3, especially in forebrain chromatin, is interesting as H3K27 residues are substrates for competing (i.e. H3K27ac versus H3K27me3) PTMs.
To address how chromatin environment may affect Pax6 regulating its targets, we examined which genes located in specific clusters were more likely to respond to Pax6 binding, by differential expression data in Pax6 conditionally inactivated or null tissues. In forebrain, we found that the clusters with the largest fractions of genes exhibiting differential expression were clusters III and V, 15.2% and 6.7%, respectively (Figure 6A). Interestingly, these clusters had increased H3K27me3 enrichment in forebrain. Unlike in forebrain, the clusters with the largest fraction of genes with differential expression upon deletion of Pax6 in lens were those with active enhancer marks, clusters IV-VI, where 10.9%-14.0% of the genes were differentially expressed (Figure 6B). These results suggest that chromatin environment may have a different effect on the regulation of Pax6 targets in forebrain and lens.
Comparing ES cell, forebrain and lens chromatin, we found that a subset of Pax6-bound regions was enriched with H3K27me3 and H3K4me1/H3K27ac in forebrain (clusters V and VI, Figure 6A). This enrichment was not observed in ES cell chromatin nor was it detected in lens Pax6-bound regions, suggesting that those Pax6-bound regions may represent class II enhancers (14), repressed at neural progenitors but poised for later stages of neuronal differentiation or brain development. The lack of H3K27me3 marking Pax6-bound regions in lens is also interesting; however, it may be due to the reduced amounts of H3K27me3 in differentiated lens cells, as immunofluorescence analysis of H3K27me3 during lens development revealed that this modification is significantly reduced in newborn compared to the embryonic lens (Supplementary Figure S4). In contrast, in lens, the enhancers identified in cluster VI were formed from initially relatively highly abundant H3K27ac-enriched regions, the poised enhancers (Figure 6E). Finally, lens cluster VI reflected the transition from repressed (in ESC, marked by H3K27me3) to activated enhancers, marked by abundant H3K4me1 and H3K27ac histone PTMs (Figure 6F).
Pax6, enhancers and transcription
To link the chromatin signatures of Pax6-bound regions with transcription, we conducted RNA-seq to establish E12.5 forebrain and newborn lens transcriptomes (separating lens epithelium and lens fiber cells). We first examined the genes with significant expression differences in forebrain and lens (Figure 7A), identifying 155 genes that were expressed 20x higher in forebrain than in both lens epithelium and fiber cells, and 98 genes expressed 20x greater in lens. As expected, these genes are enriched with functions critical for either forebrain development or lens development (Figure 7A, bottom), such as Emx1/2, Dlx1/2, Nkx2.1 and Neurog2 for forebrain, and Lim2, Aldh1a1 and 13 crystallin genes for lens. While 1.6% of the mouse genes were bound by Pax6 only in forebrain, 20 (12.9%) of the 155 forebrain high-expressing genes had Pax6 binding only in forebrain. Conversely, 18 (18.4%) of the 98 lens high-expressing genes had Pax6 binding only in lens, whereas only 2% of the mouse genes had such a pattern. These genes included Emx1/2, Neurog2 and Igfbpl1 for forebrain and Cryaa, Crybb3 and Iggfbp7 for lens.
Figure 7.
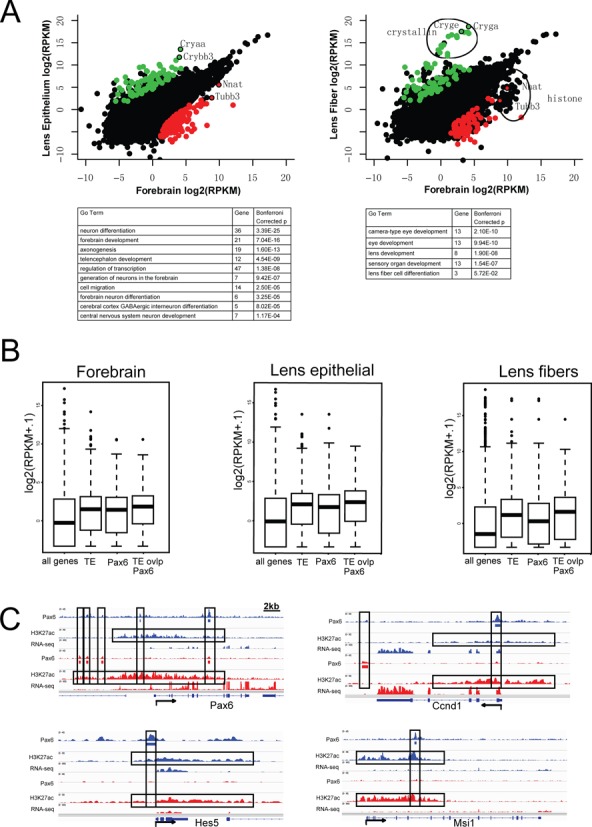
Pax6-binding, RNA abundance and extended H3K27ac regions in Ccnd1, Hes5, Msi1 and Pax6 loci. (A) A comparison of genes highly expressed in forebrain (red dots) or lens (green dots) determined by RNA-seq and their enriched gene ontology (GO) functions. (B) Comparison of transcripts levels (box plots) linked to ‘Pax6-occupied’ and ‘Pax6-free enhancers’. TE, typical enhancer (i.e. marked by H3K27ac). (C) Examples of extended regions marked by abundant H3K27ac identified in Ccnd1, Hes5, Msi1, and Pax6 loci.
At a more global level, we found that in both tissues the Pax6-bound genes in combination with the presence of H3K4me3/RNA polymerase II (cluster I) tended to be highly expressed compared to all genes bound by Pax6. Similarly, genes bound by Pax6 in combination with strong H3K4me1/H3K27ac and without H3K27me3 (clusters VII in forebrain and VI in lens) also tended to be highly expressed. Finally, genes bound by Pax6 and H3K27me3 (clusters V and VI in forebrain) were more likely to be lowly expressed (data not shown).
Finally, given the recent interest in large chromatin domains with extended active histone modifications in mammalian development (72,73), we examined whether our data sets contain extended and broad regions with enriched H3K27ac as previously described (72). We identified a total of 308 and 301 of such extended enhancer domains in the mouse forebrain and lens, respectively. Interestingly, ∼1/3 of these regions (97 in forebrain; 112 in lens) also contained Pax6 binding (see four examples in Figure 7C), significantly more than expected by chance (binomial P-value < 4.7E-59). These regions are significantly enriched with genes important for forebrain and lens development and functions. Their tissue-specific functions and the role of Pax6 in the formation of these large enhancer domains, however, need further investigation.
Pax6-dependent GRNs and cortical neurogenesis
The onset of cortical neurogenesis is concomitant with the transition from neuroepithelial stem cells into radial glial (RG) stem cells followed by direct or indirect neurogenesis (1,2). At the molecular levels, multiple cell signaling pathways have been implicated in this transformation. A sparse group of transcription factors, mostly from HD and bHLH families, have been shown to function in RG cells and in the generation of basal progenitors. Expression domains of these factors vary between dorsal and ventral progenitors including at the dorso-ventral boundary established at E12.5. The present data show that Pax6 directly activates dorsally expressed transcription factors, including Dmrta1, Emx1, Helt and Ngn2, while it directly represses expression of ventral factors, such as Ascl1/Mash1, and Gsx2, in the boundary region between the dorsal (pallium) and ventral (subpallium) regions of the telencephalon. In addition, Pax6 appears to repress expression of Fezf2, expressed dorsally. We integrated these data into previously established regulatory interactions between Pax6/Ngn2 and Pax6/Gsx2 identified through earlier analyses of corresponding mouse mutants (53,74) and Pax6 ChIP-chip studies (22). The expanded GRNs that control early stages of cortical neurogenesis demonstrate that Pax6 is engaged in multiple positive and negative feed-forward loop subcircuits comprised of DNA-binding transcription factors (Figure 8).
Figure 8.
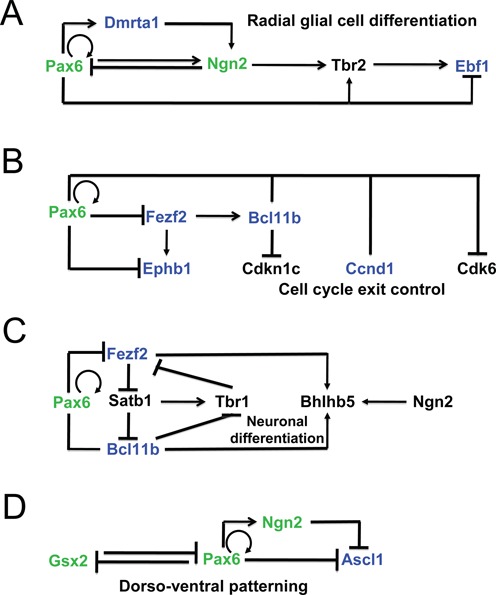
Pax6-dependent gene regulatory networks and analysis of enhancers of key regulatory genes in forebrain. (A) GRN of early stages of cortical neurogenesis. (B) GRN of cell cycle exit control in radial glial cells. (C) Dual roles of Pax6 in a subcircuit that indirectly controls expression of Tbr1 and Bhlhb5. (D) Direct repression of Ascl1 and Grx2 in the boundary region. Pax6 direct targets identified (validated) here are shown in blue (green), respectively. Pax6-binding in both forebrain and lens, and distribution of histone PTMs and RNA polymerase II is shown in Supplementary Figure S5.
In dorsal telencephalon, two consecutive feed-forward loops include transcription factors Pax6, Dmrta1, Ngn2 and Tbr2 and operate in the radial glial cells (Figure 8A). Within the network comprised of DNA-binding factors Pax6 and Fezf2, and ephrin B1 (Ephb1, membrane protein and ligand of Eph-related receptor tyrosine kinases), positive and negative feed forward loops form a novel regulatory circuit (Figure 8B) that links Pax6 activity with the regulation of cell cycle exit (via Ccnd1 and Cdk6) and axon guidance (via Ephb1). In addition, the transcription factors Fezf2 and Bcl11b/Ctip2 also control projection identities via a complex network, including Tbr1, Bhlhb5 and Satb2 transcription factors (Figure 8C) (75). In the border region, our data now show that Pax6 directly represses HD factor Ascl1 (Figure 8D), a key regulator of neurogenesis in the ventral telencephalon (76). Earlier studies have shown that Pax6 also represses expression of Gsx2 (70,71), a HD-containing multifunctional regulator of self-renewal and differentiation of radial glial cells. Among the remaining transcription factors regulated by Pax6, including Emx1 and Helt/Hesl/Mgn (activated group) and Foxp2, Lmo1 and Zfhx3 (repressed group), only genes directly regulated by Foxp2 (77) have been identified. We conclude that Pax6 is a pleiotropic direct regulator of neurogenesis (via regulation of multiple bHLH factors such as Ngn2 and Helt/Hesl/Mgn), cell cycle regulation (via Ccnd1 and Cdk6) of radial glial cells, and axonal guidance (via Ephb1).
Pax6-dependent GRNs and embryonic lens development
The present data show that Pax6 directly regulates Prox1 and Mab21l1 (Figure 9A). Prox1 regulates expression of various β/γ-crystallins (78,79). Pax6 also regulates expression of Foxe3 (80). Interestingly, expression of Foxe3 is attenuated in the severely disrupted lens placode of Mab21l1−/- embryos (52). Taken together, Pax6 directly regulates the expression of an increasing number of regulatory genes, including c-Maf, Foxe3 and Prox1, within the crystallin-specific GRN.
Figure 9.
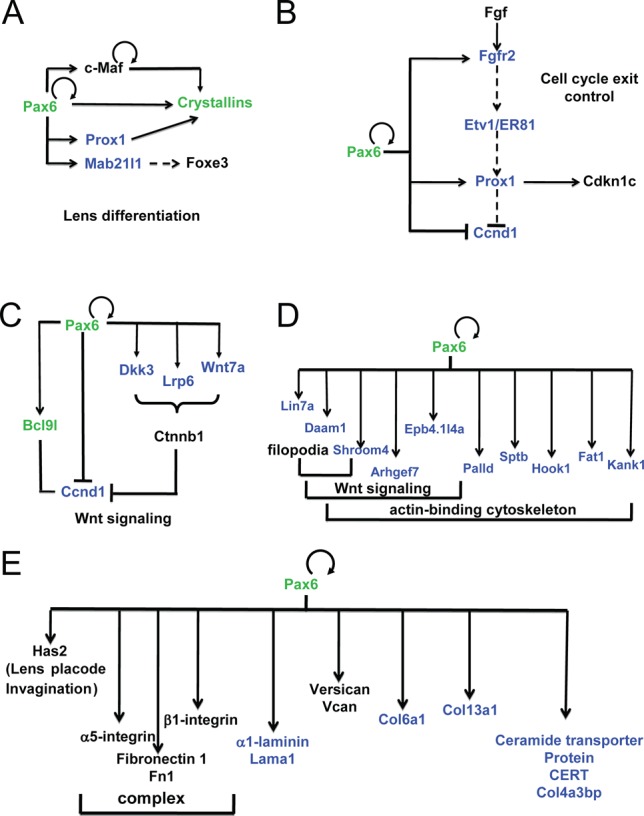
Pax6-dependent gene regulatory networks and analysis of enhancers of key regulatory genes in lens. (A) GRN of crystallin gene expression. (B) GRN of cell cycle exit control via FGF signaling in lens cells. (C) Pax6 and Wnt signaling in lens differentiation. (D) Pax6 directly regulates genes implicated in filopodia, Wnt signaling and cytoskeleton. (E) GRN of extracellular matrix (ECM) gene expression. Pax6 direct targets identified (validated) here are shown in blue (green), respectively. Pax6-binding in both forebrain and lens, and distribution of histone PTMs and RNA polymerase II is shown in Supplementary Figure S6.
It has been shown earlier that cell cycle exit of lens precursor cells in the posterior part of the lens vesicle is regulated by a combination of BMP and FGF signaling (81). In vivo, the inactivation of transcription factors Pax6 (82), Gata3 (83), Pitx3 (84), Prox1 (79), and Rbpj (85,86) (a nuclear target of Notch signaling) disrupted cell cycle exit. Reduced expression of Etv1/ER81 was found when FGF receptors were inactivated in the differentiating lens (87). Depletion of Prox1 in the lens vesicle inhibited expression of negative regulators of Cdks, including Cdkn1b/p27Kip1 and Cdkn1c/p57Kip2 (79). The present data show that Pax6 binds regulatory regions of Fgfr2, Prox1 and cyclin D1 (Ccnd1). As all of these genes have altered expression following somatic loss of Pax6 from the lens, we infer that these genes are directly regulated by Pax6 in lens cells (Figure 9B).
Our data also show that Pax6 directly regulates expression of multiple ‘early’ components of Wnt signaling in lens, including Bcl9l, Ccnd1, Dkk3 (88), Lrp6 and Wnt7a (Figure 8C, D) consistent with recent data showing that zinc finger-containing protein Bcl9l regulates lens Wnt signaling independent of β-catenin (89). Pax6 directly regulates expression of 10 genes that can be classified within a broader group of ‘actin-binding, cytoskeleton, filopodia and Wnt signaling’ (90), including Arhgef7/Pak3bp, Bcl2l11/Bim, Daam1, Hook1, Kank1, Lin7a, Lix1, Palld, Rock1 and Shroom4 (Figure 9D). Finally, Pax6 also directly regulates expression of four novel genes related to extracellular matrix (ECM), including laminin1 (Lama1), collagens (Col6a1 and Col13a1), and collagen binding proteins (Col4a3bp) (Figure 9E). Taken together, the present data expand our knowledge about the full range and spectrum of lens morphogenesis that directly requires Pax6.
DISCUSSION
Genome-wide identification of Pax6-directly regulated genes in forebrain and lens is required to gain new insights into molecular mechanisms that govern formation of both tissues. Reconstruction of Pax6-dependent GRNs revealed repetitive utilization of three common regulatory mechanisms, including feed-forward loops, top to bottom pathway control, and use of multiple types of active chromatin assembled at the distal enhancer regions. In this study, we mapped Pax6-binding sites throughout the entire embryonic forebrain and newborn lens chromatin including a set of key histone PTMs. Specific combinations of these PTMs, coupled with data on RNA polymerase II localization, are associated with promoters and enhancers. Earlier studies to determine genes regulated by Pax6/Eyeless at genome level were based on the initial prediction of putative Pax6 binding through the use of a consensus-binding site for Pax6 PD within evolutionary conserved non-coding regions (71,91). More recently, unbiased identifications of Pax6 regions were facilitated through the use of a ChIP-chip platform (24,46); however, the outputs were limited to approximately 10 kb promoter regions printed on the arrays. In contrast, the data generated here show that the majority of Pax6-binding occurs at distal regulatory regions and revealed distinct types of putative enhancers bound by Pax6. Importantly, our data show that 7–13% of all active enhancers are occupied by Pax6, a range that indicates the selectivity of Pax6 binding as well as extensive utilization of Pax6 within these enhancers. Furthermore, we showed that Pax6 binding in the context of H3K4me1/H3K27ac was highly enriched at targets that were differentially regulated upon Pax6 gene targeting.
Mammalian cortical development is directly connected to cell fate decisions of radial glial cells. Neurons develop from radial glial cells by two related processes known as direct and indirect neurogenesis (2). Radial glial cells can undergo two types of cell divisions in the ventricular zone, including self-renewal by symmetric cell division and asymmetric cell division. The outcome of the asymmetric division results is the generation of another radial glial cell while the other cell differentiates toward its neuronal fate (1,2). Genetic studies of Pax6 have shown that Pax6 is involved in the decisions of progenitors to self-renew or differentiate (92,93). In addition, Pax6 is essential for patterning and regionalization of the neocortex as well as for differentiation of special types of neurons (94). It has been shown earlier that Pax6 directly regulates multiple genes that control radial glial cell proliferation through Hmga2, Fab7, Cdk4, and Cdk6 (22,46). The present data add cyclin D1/Ccnd1 into this network (Figure 8B) though additional studies are needed to probe this mechanism at the protein level. Additionally, the present data demonstrate direct regulation of Ccnd1 by Pax6 in forebrain while lens studies raise the possibility that Pax6 also directly regulates Hmga2 in radial glial cells as well as in retinal progenitor cells (95). Our data also show that Pax6 directly regulates Bcl11b (Figure 8B), which is known to regulate expression of Cdkn1c/p57Kip2 (96).
Regarding dorso-ventral patterning of the telencephalon, loss of Pax6 leads radial glial progenitors to convert from having dorsal characteristics to having ventral characteristics (21). The present data (Figure 8A, C, D) have identified novel regulatory mechanisms, including direct repression of Ascl1, Bcl11b and Ebf1 by Pax6. The novel genes directly activated by Pax6 include Dmrta1, and expand the number of genes encoding transcription factors that regulate neurogenesis in conjunction with Pax6, such as Ngn2, Foxp2 and Etv1/ER81 (71,97). Expression of Dmrta1 is also regulated by Pax6 in rat telencephalon (98). Notably, the novel Pax6-dependent GRNs identified here are comprised of both positive and negative feed-forward loops, a universal feature of Pax6 networks we identified earlier in lens (23) and RPE (9). It is possible that evolution of such GRN subcircuits controls both robustness of the system and enables integration of upstream signal transduction pathways to target multiple (two or three) transcription factors to generate local specificity of the signaling. Since Ascl1 and other pro-neural genes are also expressed in retinal progenitor cells and repressed by Pax6 (99), some of the regulatory mechanisms identified here can also function during retinal development.
Like in radial glial cells, Pax6 is known to regulate cell cycle exit during lens fiber cell differentiation (82). A combination of BMP and FGF signaling has been shown to regulate this process (81); however, the intrinsic nuclear factors and their target genes involved in cell cycle exit regulation remain poorly characterized. The present data establish a molecular mechanism where Pax6, through direct regulation of Fgfr2, Prox1 and Ccnd1 (Figure 9), regulates this process in the lens. Additional studies by ChIP-seq are needed to integrate Gata3, Pitx3 and Prox1 as well as signal regulated transcription factors (e.g. c-Jun, Etv1/ER81, Etv5/ERM and Smads) into the emerging GRNs established here.
The direct regulation of Prox1 by Pax6, coupled with earlier studies of β- and γ-crystallin gene expression by Prox1 (78,79,100), identifies a parallel positive feed-forward loop with the Pax6/c-Maf/crystallins (23) that together control the crystallin gene transcriptional network (Figure 9). Wnt signaling is an important pathway implicated at multiple stages of lens development, though it is obvious that its individual components may vary depending on the precise developmental context. The present data show that Pax6 directly regulates expression of multiple components of Wnt signaling in lens, including Bcl9l, Ccnd1, Dkk3, Lrp6 and Wnt7a, in differentiating lens fibers (Figure 9C). Pax6 also regulates a number of genes that play roles in cytoskeletal organization (90) with some of them also regulated by Wnt signaling (Figure 9D) as well as ECM proteins, including α1-laminin (Figure 9E). It remains to be established how these proteins regulate lens fiber cell morphology, including its primary cilium located at apical surface of the elongating fiber cell (101).
It is noteworthy that the fraction of genes directly regulated by Pax6 seems quite low compared to the number of Pax6-bound genes identified here. It is likely that many of the Pax6 binding sites are actually functional but at different stages of embryonic development such as the lens placode and vesicle formation between E9 and E11.5 of mouse embryonic development, perhaps under different sets of development cues. Although the RNA expression data can be collected from quite a small number of cells, the limiting step in these analyses is recovery of the immunoprecipitated DNA, which is determined by the antibody-antigen interactions and other quantitative parameters such as the fact that even in a single cell, RNAs are present in hundreds or thousands of copies while a specific genomic fragment such as an enhancer is present just as two copies per diploid genome. Thus, to understand which genes are regulated by Pax6 at developmental stages for which only a few hundreds of cells are available, such as the lens placode, a number of technical challenges remains to be addressed.
The current data shed new light on in vivo ‘consensus’ Pax6-binding site. Three types of DNA sequences, combined with earlier data on protein-DNA contacts between individual DNA-binding subdomains of Pax6 (54–56,66,102), implicate three partially overlapping DNA-binding mechanisms, including PD alone, a combination of PD/HD, and HD alone. In addition, analysis of all Pax6-bound peaks identified here suggests that approximately 1/6 of these regions contain multiple Pax6-binding sites. The knowledge of in vivo Pax6 sites will aid studies to understand Pax6 roles in ‘small’ tissues such as iris and ciliary body epithelium (103), olfactory placode (104) and embryonic pituitary gland (105) that are difficult to obtain in quantities sufficient for chromatin studies.
Taken together, our data serve as an entry point to decipher GRNs that control self-renewal and differentiation of radial glial cells, that, when combined with data on Ascl1, Ngn2, Tbr1, Tbr2 and other DNA-binding regulatory proteins (e.g. Hes1 and Hes5), will generate complete wiring of the network underlying early stages of cortical neurogenesis. In lens, there is a relatively short list of top hierarchy (Pax6, Six3 and Sox2) and ‘core layer’ (e.g. AP-2α, c-Maf, Gata3, Hsf4, Pitx3, Prox1 and Sox1) transcription factors that need similar level of attention with the respect of identification of their direct targets as we showed here for Pax6. The repetitive use of feed-forward regulatory mechanisms and control of batteries of genes that encode products of terminal differentiation may also involve direct protein-protein interactions and/or functional synergism between Pax6 and its regulated transcription factors, as shown for Pax6 and c-Maf in crystallin gene expression (106), as well as some evolutionary advantage that originates from this type of genetic ‘wiring’.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We are grateful Dr Magdalena Götz for critical comments on the manuscript. We thank Drs John Greally and Shahina Maqbool for their extensive advice, the Einstein Genomics and Epigenomics Shared Facilities, and high performance computing core for their services. We thank Mrs Jie Zhao for her expert help with mice colonies. Data in this paper are from a thesis submitted in partial fulfillment of the requirements for the Degree of Doctor of Philosophy in the Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University (J.S.).
Authors contributions. J.S. and S.R., contributed equally, and shared first authorship. J.S. carried out ChIP-seq studies, contributed to the bioinformatic analysis and manuscript preparation. S.R. conducted the data and bioinformatics analysis and contributed to manuscript preparation. Q.X. and R.A.P. contributed to the differential gene expression analysis. D.Z. conceived the study and contributed to the ChIP-seq bioinformatics analysis and manuscript preparation. A.C. conceived the study, and contributed to data analysis and drafted the manuscript. All authors read and approved the final manuscript.
FUNDING
NIH [R01 EY012200 to A.C., R21 MH099452 to D.Z.]; Research to Prevent Blindness, Inc. to the Department of Ophthalmology and Visual Sciences. Research to R.A.P. is supported by the Israel Science Foundation [228/14], the Israel Ministry of Science [36494], the Ziegler Foundation, and the US-Israel Binational Science Foundation [2013016]. Funding for open access charge: NIH [EY012200].
Conflict of interest statement. None declared.
REFERENCES
- 1.Georgala P.A., Carr C.B., Price D.J. The role of Pax6 in forebrain development. Dev. Neurobiol. 2011;71:690–709. doi: 10.1002/dneu.20895. [DOI] [PubMed] [Google Scholar]
- 2.Martynoga B., Drechsel D., Guillemot F. Molecular control of neurogenesis: a view from the mammalian cerebral cortex. Cold Spring Harbor Persp. Biol. 2012;4:a008359. doi: 10.1101/cshperspect.a008359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikoletopoulou V., Plachta N., Allen N.D., Pinto L., Gotz M., Barde Y.A. Neurotrophin receptor-mediated death of misspecified neurons generated from embryonic stem cells lacking Pax6. Cell Stem Cell. 2007;1:529–540. doi: 10.1016/j.stem.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Weinandy F., Ninkovic J., Gotz M. Restrictions in time and space–new insights into generation of specific neuronal subtypes in the adult mammalian brain. Eur. J. Neurosci. 2011;33:1045–1054. doi: 10.1111/j.1460-9568.2011.07602.x. [DOI] [PubMed] [Google Scholar]
- 5.Cvekl A., Ashery-Padan R. The cellular and molecular mechanisms of vertebrate lens development. Development. 2014;141:4432–4447. doi: 10.1242/dev.107953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graw J. The genetic and molecular basis of congenital eye defects. Nature reviews. Genetics. 2003;4:876–888. doi: 10.1038/nrg1202. [DOI] [PubMed] [Google Scholar]
- 7.Marquardt T., Gruss P. Generating neuronal diversity in the retina: one for nearly all. Trends Neurosci. 2002;25:32–38. doi: 10.1016/s0166-2236(00)02028-2. [DOI] [PubMed] [Google Scholar]
- 8.Canto-Soler M.V., Adler R. Optic cup and lens development requires Pax6 expression in the early optic vesicle during a narrow time window. Dev. Biol. 2006;294:119–132. doi: 10.1016/j.ydbio.2006.02.033. [DOI] [PubMed] [Google Scholar]
- 9.Raviv S., Bharti K., Rencus-Lazar S., Cohen-Tayar Y., Schyr R., Evantal N., Meshorer E., Zilberberg A., Idelson M., Reubinoff B., et al. PAX6 regulates melanogenesis in the retinal pigmented epithelium through feed-forward regulatory interactions with MITF. PLoS Genet. 2014;10:e1004360. doi: 10.1371/journal.pgen.1004360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolosin J.M., Budak M.T., Akinci M.A. Ocular surface epithelial and stem cell development. Int. J. Dev. Biol. 2004;48:981–991. doi: 10.1387/ijdb.041876jw. [DOI] [PubMed] [Google Scholar]
- 11.Cvekl A., Mitton K.P. Epigenetic regulatory mechanisms in vertebrate eye development and disease. Heredity. 2010;105:135–151. doi: 10.1038/hdy.2010.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho L., Crabtree G.R. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M., et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S.A., Flynn R.A., Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim T.K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S., et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mousavi K., Zare H., Dell'orso S., Grontved L., Gutierrez-Cruz G., Derfoul A., Hager G.L., Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 18.Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeo G.W., Coufal N., Aigner S., Winner B., Scolnick J.A., Marchetto M.C., Muotri A.R., Carson C., Gage F.H. Multiple layers of molecular controls modulate self-renewal and neuronal lineage specification of embryonic stem cells. Hum. Mol. Genet. 2008;17:R67–R75. doi: 10.1093/hmg/ddn065. [DOI] [PubMed] [Google Scholar]
- 20.van Raamsdonk C.D., Tilghman S.M. Dosage requirement and allelic expression of PAX6 during lens placode formation. Development. 2000;127:5439–5448. doi: 10.1242/dev.127.24.5439. [DOI] [PubMed] [Google Scholar]
- 21.Quinn J.C., Molinek M., Martynoga B.S., Zaki P.A., Faedo A., Bulfone A., Hevner R.F., West J.D., Price D.J. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev. Biol. 2007;302:50–65. doi: 10.1016/j.ydbio.2006.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mi D., Carr C.B., Georgala P.A., Huang Y.T., Manuel M.N., Jeanes E., Niisato E., Sansom S.N., Livesey F.J., Theil T., et al. Pax6 exerts regional control of cortical progenitor proliferation via direct repression of Cdk6 and hypophosphorylation of pRb. Neuron. 2013;78:269–284. doi: 10.1016/j.neuron.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Q., Cvekl A. The orchestration of mammalian tissue morphogenesis through a series of coherent feed-forward loops. J. Biol. Chem. 2011;286:43259–43271. doi: 10.1074/jbc.M111.264580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Q., Yang Y., Huang J., Ninkovic J., Walcher T., Wolf L., Vitenzon A., Zheng D., Gotz M., Beebe D.C., et al. Pax6 interactions with chromatin and identification of its novel direct target genes in lens and forebrain. PloS One. 2013;8:e54507. doi: 10.1371/journal.pone.0054507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLellan A.S., Dubin R.A., Jing Q., Broin P.O., Moskowitz D., Suzuki M., Calder R.B., Hargitai J., Golden A., Greally J.M. The Wasp System: an open source environment for managing and analyzing genomic data. Genomics. 2012;100:345–351. doi: 10.1016/j.ygeno.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shao Z., Zhang Y., Yuan G.C., Orkin S.H., Waxman D.J. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 2012;13:R16. doi: 10.1186/gb-2012-13-3-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamatoyannopoulos J.A., Snyder M., Hardison R., Ren B., Gingeras T., Gilbert D.M., Groudine M., Bender M., Kaul R., Canfield T., et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13:418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao S., Xie D., Cao X., Yu P., Xing X., Chen C.C., Musselman M., Xie M., West F.D., Lewin H.A., et al. Comparative epigenomic annotation of regulatory DNA. Cell. 2012;149:1381–1392. doi: 10.1016/j.cell.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 1994;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machanick P., Bailey T.L. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryne J.C., Valen E., Tang M.H., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic acids research. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey T.L., Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 38.Pruitt K.D., Tatusova T., Brown G.R., Maglott D.R. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer L.R., Zweig A.S., Hinrichs A.S., Karolchik D., Kuhn R.M., Wong M., Sloan C.A., Rosenbloom K.R., Roe G., Rhead B., et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye T., Krebs A.R., Choukrallah M.A., Keime C., Plewniak F., Davidson I., Tora L. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39:e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaham O., Gueta K., Mor E., Oren-Giladi P., Grinberg D., Xie Q., Cvekl A., Shomron N., Davis N., Keydar-Prizant M., et al. Pax6 regulates gene expression in the vertebrate lens through miR-204. PLoS Genet. 2013;9:e1003357. doi: 10.1371/journal.pgen.1003357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y., Stopka T., Golestaneh N., Wang Y., Wu K., Li A., Chauhan B.K., Gao C.Y., Cveklova K., Duncan M.K., et al. Regulation of αA-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. EMBO J. 2006;25:2107–2118. doi: 10.1038/sj.emboj.7601114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.St-Onge L., Sosa-Pineda B., Chowdhury K., Mansouri A., Gruss P. Pax6 is required for differentiation of glucagon-producing α-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- 44.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts A., Pimentel H., Trapnell C., Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 46.Sansom S.N., Griffiths D.S., Faedo A., Kleinjan D.J., Ruan Y., Smith J., van Heyningen V., Rubenstein J.L., Livesey F.J. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P., et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams S.C., Altmann C.R., Chow R.L., Hemmati-Brivanlou A., Lang R.A. A highly conserved lens transcriptional control element from the Pax-6 gene. Mech. Dev. 1998;73:225–229. doi: 10.1016/s0925-4773(98)00057-4. [DOI] [PubMed] [Google Scholar]
- 49.Kammandel B., Chowdhury K., Stoykova A., Aparicio S., Brenner S., Gruss P. Distinct cis-essential modules direct the time-space pattern of the Pax6 gene activity. Dev. Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- 50.Dimanlig P.V., Faber S.C., Auerbach W., Makarenkova H.P., Lang R.A. The upstream ectoderm enhancer in Pax6 has an important role in lens induction. Development. 2001;128:4415–4424. doi: 10.1242/dev.128.22.4415. [DOI] [PubMed] [Google Scholar]
- 51.Zuccotti A., Le Magueresse C., Chen M., Neitz A., Monyer H. The transcription factor Fezf2 directs the differentiation of neural stem cells in the subventricular zone toward a cortical phenotype. Proc. Natl. Acad. Sci. USA. 2014;111:10726–10731. doi: 10.1073/pnas.1320290111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada R., Mizutani-Koseki Y., Hasegawa T., Osumi N., Koseki H., Takahashi N. Cell-autonomous involvement of Mab21l1 is essential for lens placode development. Development. 2003;130:1759–1770. doi: 10.1242/dev.00399. [DOI] [PubMed] [Google Scholar]
- 53.Scardigli R., Baumer N., Gruss P., Guillemot F., Le Roux I. Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development. 2003;130:3269–3281. doi: 10.1242/dev.00539. [DOI] [PubMed] [Google Scholar]
- 54.Epstein J., Cai J., Glaser T., Jepeal L., Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J. Biol. Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- 55.Epstein J.A., Glaser T., Cai J., Jepeal L., Walton D.S., Maas R.L. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 56.Duncan M.K., Kozmik Z., Cveklova K., Piatigorsky J., Cvekl A. Overexpression of PAX6(5a) in lens fiber cells results in cataract and upregulation of (alpha)5(beta)1 integrin expression. J. Cell Sci. 2000;113:3173–3185. doi: 10.1242/jcs.113.18.3173. [DOI] [PubMed] [Google Scholar]
- 57.Berger M.F., Badis G., Gehrke A.R., Talukder S., Philippakis A.A., Pena-Castillo L., Alleyne T.M., Mnaimneh S., Botvinnik O.B., Chan E.T., et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Czerny T., Busslinger M. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Mol. Cell. Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ritz-Laser B., Estreicher A., Klages N., Saule S., Philippe J. Pax-6 and Cdx-2/3 interact to activate glucagon gene expression on the G1 control element. J. Biol. Chem. 1999;274:4124–4132. doi: 10.1074/jbc.274.7.4124. [DOI] [PubMed] [Google Scholar]
- 60.Yang Y., Cvekl A. Tissue-specific regulation of the mouse αA-crystallin gene in lens via recruitment of Pax6 and c-Maf to its promoter. J. Mol. Biol. 2005;351:453–469. doi: 10.1016/j.jmb.2005.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie Q., Ung D., Khafizov K., Fiser A., Cvekl A. Gene regulation by PAX6: structural-functional correlations of missense mutants and transcriptional control of Trpm3/miR-204. Mol. Vis. 2014;20:270–282. [PMC free article] [PubMed] [Google Scholar]
- 62.Chi N., Epstein J.A. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- 63.Blake J.A., Ziman M.R. Pax genes: regulators of lineage specification and progenitor cell maintenance. Development. 2014;141:737–751. doi: 10.1242/dev.091785. [DOI] [PubMed] [Google Scholar]
- 64.Soleimani V.D., Punch V.G., Kawabe Y., Jones A.E., Palidwor G.A., Porter C.J., Cross J.W., Carvajal J.J., Kockx C.E., van I.W.F., et al. Transcriptional dominance of Pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Dev. Cell. 2012;22:1208–1220. doi: 10.1016/j.devcel.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu H.E., Rould M.A., Xu W., Epstein J.A., Maas R.L., Pabo C.O. Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev. 1999;13:1263–1275. doi: 10.1101/gad.13.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holm P.C., Mader M.T., Haubst N., Wizenmann A., Sigvardsson M., Gotz M. Loss- and gain-of-function analyses reveal targets of Pax6 in the developing mouse telencephalon. Mol. Cell. Neurosci. 2007;34:99–119. doi: 10.1016/j.mcn.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 67.Walcher T., Xie Q., Sun J., Irmler M., Beckers J., Ozturk T., Niessing D., Stoykova A., Cvekl A., Ninkovic J., et al. Functional dissection of the paired domain of Pax6 reveals molecular mechanisms of coordinating neurogenesis and proliferation. Development. 2013;140:1123–1136. doi: 10.1242/dev.082875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang J., Rajagopal R., Liu Y., Dattilo L.K., Shaham O., Ashery-Padan R., Beebe D.C. The mechanism of lens placode formation: a case of matrix-mediated morphogenesis. Dev. Biol. 2011;355:32–42. doi: 10.1016/j.ydbio.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolf L., Harrison W., Huang J., Xie Q., Xiao N., Sun J., Kong L., Lachke S.A., Kuracha M.R., Govindarajan V., et al. Histone posttranslational modifications and cell fate determination: lens induction requires the lysine acetyltransferases CBP and p300. Nucleic Acids Res. 2013;41:10199–10214. doi: 10.1093/nar/gkt824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cocas L.A., Georgala P.A., Mangin J.M., Clegg J.M., Kessaris N., Haydar T.F., Gallo V., Price D.J., Corbin J.G. Pax6 is required at the telencephalic pallial-subpallial boundary for the generation of neuronal diversity in the postnatal limbic system. J. Neurosci. 2011;31:5313–5324. doi: 10.1523/JNEUROSCI.3867-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coutinho P., Pavlou S., Bhatia S., Chalmers K.J., Kleinjan D.A., van Heyningen V. Discovery and assessment of conserved Pax6 target genes and enhancers. Genome Res. 2011;21:1349–1359. doi: 10.1101/gr.124115.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pott S., Lieb J.D. What are super-enhancers? Nature Genet. 2014;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 74.Toresson H., Potter S.S., Campbell K. Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development. 2000;127:4361–4371. doi: 10.1242/dev.127.20.4361. [DOI] [PubMed] [Google Scholar]
- 75.Srinivasan K., Leone D.P., Bateson R.K., Dobreva G., Kohwi Y., Kohwi-Shigematsu T., Grosschedl R., McConnell S.K. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proc. Natl. Acad. Sci. USA. 2012;109:19071–19078. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Castro D.S., Martynoga B., Parras C., Ramesh V., Pacary E., Johnston C., Drechsel D., Lebel-Potter M., Garcia L.G., Hunt C., et al. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25:930–945. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsui D., Vessey J.P., Tomita H., Kaplan D.R., Miller F.D. FoxP2 regulates neurogenesis during embryonic cortical development. Journal Neurosci. 2013;33:244–258. doi: 10.1523/JNEUROSCI.1665-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cui W., Tomarev S.I., Piatigorsky J., Chepelinsky A.B., Duncan M.K. Mafs, Prox1, and Pax6 can regulate chicken βB1-crystallin gene expression. J. Biol. Chem. 2004;279:11088–11095. doi: 10.1074/jbc.M312414200. [DOI] [PubMed] [Google Scholar]
- 79.Wigle J.T., Chowdhury K., Gruss P., Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nature Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- 80.Blixt A., Landgren H., Johansson B.R., Carlsson P. Foxe3 is required for morphogenesis and differentiation of the anterior segment of the eye and is sensitive to Pax6 gene dosage. Dev. Biol. 2007;302:218–229. doi: 10.1016/j.ydbio.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 81.Jarrin M., Pandit T., Gunhaga L. A balance of FGF and BMP signals regulates cell cycle exit and Equarin expression in lens cells. Mol. Biol. Cell. 2012;23:3266–3274. doi: 10.1091/mbc.E12-01-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shaham O., Smith A.N., Robinson M.L., Taketo M.M., Lang R.A., Ashery-Padan R. Pax6 is essential for lens fiber cell differentiation. Development. 2009;136:2567–2578. doi: 10.1242/dev.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maeda A., Moriguchi T., Hamada M., Kusakabe M., Fujioka Y., Nakano T., Yoh K., Lim K.C., Engel J.D., Takahashi S. Transcription factor GATA-3 is essential for lens development. Dev. Dyn. 2009;238:2280–2291. doi: 10.1002/dvdy.22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Medina-Martinez O., Shah R., Jamrich M. Pitx3 controls multiple aspects of lens development. Dev. Dyn. 2009;238:2193–2201. doi: 10.1002/dvdy.21924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jia J., Lin M., Zhang L., York J.P., Zhang P. The Notch signaling pathway controls the size of the ocular lens by directly suppressing p57Kip2 expression. Mol. Cell. Biol. 2007;27:7236–7247. doi: 10.1128/MCB.00780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rowan S., Conley K.W., Le T.T., Donner A.L., Maas R.L., Brown N.L. Notch signaling regulates growth and differentiation in the mammalian lens. Dev. Biol. 2008;321:111–122. doi: 10.1016/j.ydbio.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao H., Yang T., Madakashira B.P., Thiels C.A., Bechtle C.A., Garcia C.M., Zhang H., Yu K., Ornitz D.M., Beebe D.C., et al. Fibroblast growth factor receptor signaling is essential for lens fiber cell differentiation. Dev. Biol. 2008;318:276–288. doi: 10.1016/j.ydbio.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Forsdahl S., Kiselev Y., Hogseth R., Mjelle J.E., Mikkola I. Pax6 regulates the expression of Dkk3 in murine and human cell lines, and altered responses to Wnt signaling are shown in Flpln-3T3 cells stably expressing either the Pax6 or the Pax6(5a) isoform. PLoS One. 2014;9:e102559. doi: 10.1371/journal.pone.0102559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cantu C., Zimmerli D., Hausmann G., Valenta T., Moor A., Aguet M., Basler K. Pax6-dependent, but β-catenin-independent, function of Bcl9 proteins in mouse lens development. Genes Dev. 2014;28:1879–1884. doi: 10.1101/gad.246140.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chauhan B., Plageman T., Lou M., Lang R. Epithelial morphogenesis: the mouse eye as a model system. Curr. Top. Dev. Biol. 2015;111:375–399. doi: 10.1016/bs.ctdb.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ostrin E.J., Li Y., Hoffman K., Liu J., Wang K., Zhang L., Mardon G., Chen R. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006;16:466–476. doi: 10.1101/gr.4673006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heins N., Malatesta P., Cecconi F., Nakafuku M., Tucker K.L., Hack M.A., Chapouton P., Barde Y.A., Gotz M. Glial cells generate neurons: the role of the transcription factor Pax6. Nature Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- 93.Bibel M., Richter J., Schrenk K., Tucker K.L., Staiger V., Korte M., Goetz M., Barde Y.A. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nature Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- 94.Gotz M., Stoykova A., Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 95.Parameswaran S., Xia X., Hegde G., Ahmad I. Hmga2 regulates self-renewal of retinal progenitors. Development. 2014;141:4087–4097. doi: 10.1242/dev.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Topark-Ngarm A., Golonzhka O., Peterson V.J., Barrett B. Jr, Martinez B., Crofoot K., Filtz T.M., Leid M. CTIP2 associates with the NuRD complex on the promoter of p57KIP2, a newly identified CTIP2 target gene. J. Biol. Chem. 2006;281:32272–32283. doi: 10.1074/jbc.M602776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tuoc T.C., Stoykova A. Er81 is a downstream target of Pax6 in cortical progenitors. BMC Dev. Biol. 2008;8:23. doi: 10.1186/1471-213X-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kikkawa T., Obayashi T., Takahashi M., Fukuzaki-Dohi U., Numayama-Tsuruta K., Osumi N. Dmrta1 regulates proneural gene expression downstream of Pax6 in the mammalian telencephalon. Genes Cells. 2013;18:636–649. doi: 10.1111/gtc.12061. [DOI] [PubMed] [Google Scholar]
- 99.Philips G.T., Stair C.N., Young Lee H., Wroblewski E., Berberoglu M.A., Brown N.L., Mastick G.S. Precocious retinal neurons: Pax6 controls timing of differentiation and determination of cell type. Dev. Biol. 2005;279:308–321. doi: 10.1016/j.ydbio.2004.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen X., Taube J.R., Simirskii V.I., Patel T.P., Duncan M.K. Dual roles for Prox1 in the regulation of the chicken βB1-crystallin promoter. Invest. Ophthalmol. Vis. Sci. 2008;49:1542–1552. doi: 10.1167/iovs.07-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sugiyama Y., Stump R.J., Nguyen A., Wen L., Chen Y., Wang Y., Murdoch J.N., Lovicu F.J., McAvoy J.W. Secreted frizzled-related protein disrupts PCP in eye lens fiber cells that have polarised primary cilia. Dev. Biol. 2010;338:193–201. doi: 10.1016/j.ydbio.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jun S., Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- 103.Davis N., Yoffe C., Raviv S., Antes R., Berger J., Holzmann S., Stoykova A., Overbeek P.A., Tamm E.R., Ashery-Padan R. Pax6 dosage requirements in iris and ciliary body differentiation. Dev. Biol. 2009;333:132–142. doi: 10.1016/j.ydbio.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 104.Donner A.L., Episkopou V., Maas R.L. Sox2 and Pou2f1 interact to control lens and olfactory placode development. Dev. Biol. 2007;303:784–799. doi: 10.1016/j.ydbio.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kioussi C., O'Connell S., St-Onge L., Treier M., Gleiberman A.S., Gruss P., Rosenfeld M.G. Pax6 is essential for establishing ventral-dorsal cell boundaries in pituitary gland development. Proc. Natl. Acad. Sci. USA. 1999;96:14378–14382. doi: 10.1073/pnas.96.25.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chauhan B.K., Yang Y., Cveklova K., Cvekl A. Functional interactions between alternatively spliced forms of Pax6 in crystallin gene regulation and in haploinsufficiency. Nucleic Acids Res. 2004;32:1696–1709. doi: 10.1093/nar/gkh334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


