Summary
Super-enhancers are clusters of gene-regulatory sites bound by multiple transcription factors that govern cell transcription, development, phenotype, and oncogenesis. By examining Epstein-Barr virus (EBV) transformed lymphoblastoid cell lines (LCLs), we identified four EBV oncoproteins and five EBV-activated NF-κB subunits co-occupying ~1800 enhancer sites. Of these, 187 had markedly higher and broader histone H3K27ac signals characteristic of super-enhancers, and were designated “EBV super-enhancers”. EBV super-enhancer-associated genes included the MYC and BCL2 oncogenes, enabling LCL proliferation and survival. EBV super-enhancers were enriched for B cell transcription factor motifs and had a high co-occupancy of the transcription factors STAT5 and NFAT. EBV super-enhancer-associated genes were more highly expressed than other LCL genes. Disrupting EBV super-enhancers by the bromodomain inhibitor, JQ1 or conditionally inactivating an EBV oncoprotein or NF-κB decreased MYC or BCL2 expression and arrested LCL growth. These findings provide insight into mechanisms of EBV-induced lymphoproliferation and identify potential therapeutic interventions.
Introduction
Epstein-Barr Virus (EBV), the first human tumor virus discovered 50 years ago from African Burkitt’s Lymphoma cells (Epstein et al., 1964), is causally associated with infectious mononucleosis, Burkitt’s lymphoma, Hodgkin’s lymphoma, HIV-related lymphomas, Post-Transplant Lymphoproliferative Diseases (PTLDs), nasopharyngeal carcinoma, and some gastric cancers (Longnecker, 2013; Young and Rickinson, 2004). In primary EBV infection, virus transits across the oropharyngeal epithelium to reach the B-cell compartment. EBV converts primary B-cells into activated blasts, which enable EBV to colonize the B-cell compartment. Indeed, EBV transformed lymphoblasts can be seen transiently in patients with infectious mononucleosis which is caused by primary EBV infection (Kurth et al., 2000). Although T- and NK-cell surveillance eventually contains lymphoblast proliferation, EBV latently-infected B-cells are the reservoir from which the virus establishes lifelong infection. With HIV infection, organ transplantation or primary immunodeficiency, impaired control of EBV latently-infected B cells leads to fatal lymphoproliferative diseases and lymphomas. In vitro, EBV transforms primary Resting B Lymphocytes (RBLs) to continuously proliferating Lymphoblastoid Cell Lines (LCLs). LCLs express the same viral genes as some EBV lymphomas. These viral genes include six Epstein-Barr Virus Nuclear Antigens (EBNA), three Latent Membrane Proteins (LMP), and multiple microRNAs (Longnecker, 2013; Young and Rickinson, 2004). LCLs are therefore a useful model for studying EBV mediated B-Lymphoid oncogenesis (Longnecker, 2013).
Reverse genetic studies indicate that viral oncoproteins EBNA2, EBNALP, EBNA3A, EBNA3C, and LMP1 are each required for LCL growth and survival (Longnecker, 2013). EBNAs bind to viral and cell DNA through their interactions with cell DNA binding proteins. EBNA2 and EBNALP are the first EBV genes expressed after B cell infection (Alfieri et al., 1991). EBNA2 mostly binds to DNA through the cell DNA binding protein RBPJ, and activates cell gene transcription, including MYC, the EBV cell surface receptor CD21, and CD23 (Grossman et al., 1994; Henkel et al., 1994; Kaiser et al., 1999; Wang et al., 1987; Zhao et al., 2011b). EBNA2 binds to B-cell enhancer sites ~ 428Kb and 525Kb upstream of MYC and loops to the MYC TSS to activate MYC transcription (Zhao et al., 2011b). The EBNA2 C-terminal acidic activation domain recruits basal and activation related transcription factors (TFs), including Pol II, p300/CBP, TFIID, and TFIIH (Tong et al., 1995; Wang et al., 2000). EBNALP co-activates with EBNA2 and de-represses transcription by removing NCoR and associated repressors from promoter DNA (Harada and Kieff, 1997; Portal et al., 2006; Portal et al., 2011; Portal et al., 2013). EBNA3A and EBNA3C repress p16INK4A and p14ARF expression, thereby preventing senescence and enabling continuous LCL growth (Maruo et al., 2011; Skalska et al., 2013). EBNA3A and EBNA3C also affect the expression of many host genes (Hertle et al., 2009; Zhao et al., 2011a). However, the growth inhibiting effects of EBNA3A or EBNA3C deficiency can only be rescued by restoring EBNA3A or EBNA3C expression respectively (Maruo et al., 2005; Maruo et al., 2006), indicating that they both have unique functions that are non-redundant. LMP1 constitutively activates NF-κB to promote growth and survival. NF-κB are dimeric TFs assembled from the RelA, RelB, cRel, p50 and p52 subunits. In resting B-cells, NF-κB is tethered in the cytosol by IκBα. In LCLs, LMP1 signaling triggers IκBα degradation and NF-κB homo- or hetero-dimer nuclear translocation. Inducible over expression of a non-degradable IκBα blocks NF-κB activity and causes LCL apoptosis (Cahir-McFarland et al., 2004; Cahir-McFarland et al., 2000). Conditional inactivation of EBNA2, EBNA3A, EBNA3C or NF-κB, identified cell genes regulated by these EBV oncoproteins. However, only a limited repertoire of cell genes are co-regulated by EBNAs or NF-κB (Cahir-McFarland et al., 2004; Hertle et al., 2009; Zhao et al., 2011a) (Zhao et al., 2006).
Super-enhancers are recently discovered enhancers with extraordinary high and broad ChIP-seq signals for activation-related TFs, H3K27ac modification, bromo-domain binding protein, BRD4, or mediator Med1 (Chapuy et al., 2013; Hnisz et al., 2013; Lovén et al., 2013; Parker et al., 2013; Whyte et al., 2013). Super-enhancers are principle determinants of cell identity and oncogenesis, although a super-enhancer role in host-pathogen interactions has not yet been reported. Super-enhancers are associated with genes critical for cell function, are co-occupied by multiple TFs in dense clusters, and are more sensitive to perturbation than typical enhancers (Chapuy et al., 2013; Hnisz et al., 2013; Lovén et al., 2013; Whyte et al., 2013). Super-enhancer formation can also be rapidly induced de novo upon cytokine stimulation accompanied by the decommission of parental cell super-enhancers (Brown et al., 2014). BRD4 inhibition by bromo-domain inhibitor JQ1 has significantly larger effects on super-enhancer associated gene expression than on typical enhancer associated gene expression (Hnisz et al., 2013; Lovén et al., 2013). In Multiple myeloma (Lovén et al., 2013) and Diffuse Large B Cell Lymphomas (DLBCL) (Chapuy et al., 2013), the MYC oncogene is controlled by super-enhancers.
We have now used EBNA2, EBNALP, EBNA3A, EBNA3C, NF-κB subunits RelA, RelB, cRel, p50, p52 and ENCODE GM12878 LCL TF ChIP-seq data for integrated analyses of EBV super-enhancer effects on LCL growth. We were surprised to find all 4 oncogenic EBNAs and all 5 NF-κB subunits co-occupying 187 sites that had extraordinarily high H3K27ac signals, indicative of super-enhancers. As characteristic of super-enhancers, EBV super-enhancers regulated key B-cell growth and survival genes, and super-enhancer disruption by the BRD4 inhibitor JQ1, or by EBNA2 or NF-κB inactivation, inhibited EBV super-enhancer associated gene expression and LCL growth.
Results
EBNA2 super-enhancers
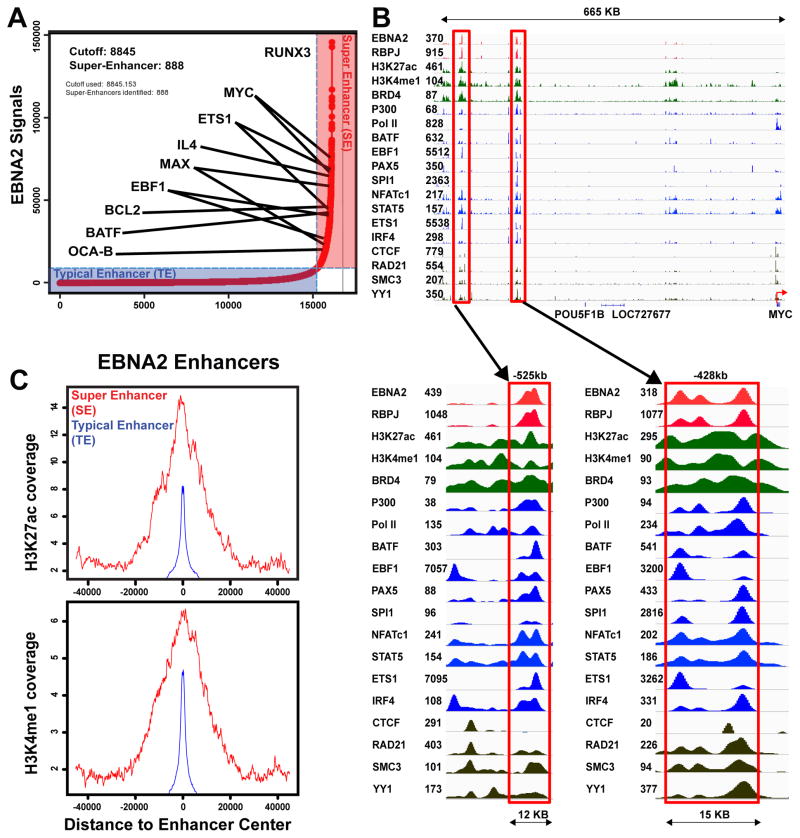
Markedly elevated ChIP-seq signals of the B-cell master TF SPI1/PU.1 distinguish B cell super-enhancers from typical enhancers (Whyte et al., 2013). Interestingly, we previously found that LCL EBNA2 sites were enriched for the SPI1 motif, and were frequently co-occupied by SPI1, suggesting possible EBNA2 incorporation into super-enhancers (Zhao et al., 2011b). EBNA2 ChIP-Seq signals were therefore tested for hallmarks of super-enhancer formation (Whyte et al., 2013). Model-based analysis of ChIP-Seq (MACS) identified 42,251 EBNA2 sites with p<10−5 (Zhang et al., 2008). Many EBNA2 sites were in broad clusters, characteristic of super-enhancers. EBNA2 sites within 12.5kb windows were then merged into 16,133 EBNA2 clusters (Whyte et al., 2013) and ranked by their EBNA2 ChIP-Seq signal. Interestingly, 888 (5.5%) of the EBNA2 sites had ChIP-seq signals 23 times higher than typical EBNA2 sites and were >4kb wide (Table S1 and Figure 1A). These sites were therefore subsequently referred to as EBNA2 super-enhancers.
Figure 1. EBNA2 Super-enhancers.
(A) Rank-order of EBNA2 ChIP-seq signals for all EBNA2 sites. Overall, 888 EBNA2 super-enhancer sites have >23 fold higher ChIP-seq signals than the average signals of ~17,000 other EBNA2 sites. These EBNA2 super-enhancer sites are annotated to their nearest cell gene. EBNA2 super-enhancer-associated cell genes particularly important for LCL growth or survival are indicated. (B) EBNA2, other TFs, and histone modification ChIP-seq signals at super-enhancers near the MYC locus are shown. Numbers near gene names indicate tag density. A red arrow indicates the MYC TSS. Magnified views of EBNA2 super-enhancers (−525 and −428kb) of MYC are shown (blue arrows and red boxes). (C) Average ChIP-seq signals for H3K27ac and H3K4me1 in 80kb windows around EBNA2 super and typical enhancers are shown. Red indicates EBNA2 super- enhancers and blue indicates typical enhancers.
As expected, most EBNA2 super-enhancers were not near transcription start sites (TSS) (Zhao et al., 2011b). We first assigned EBNA2 super-enhancers to their nearest genes (Whyte et al., 2013). Chromatin Conformation Capture (3C) followed by deep sequencing (Hi-C) captures long-range enhancer-promoter interactions and defines genome Topological Association Domains (TAD) (Lieberman-Aiden et al., 2009). We tested our EBNA2 super-enhancer associated gene assignments using high resolution GM12878 LCL Hi-C data (Selvaraj et al., 2013). ~86% of EBNA2 super-enhancers and their associated gene pairs occurred within the same Hi-C TAD, in agreement with super-enhancers and their targeted genes frequently co-occurring within CTCF/Cohesin domains (Dowen et al., 2014).
EBNA2 super-enhancers frequently localized near genes encoding relevant B cell TFs, including MYC, MAX, EBF, RUNX3, ETS1, and BATF, as well as the B cell specific co-activator OCAB (Figure 1A). Many other cell TFs including BATF, EBF, ETS1, IRF4, SPI1, NFAT, STAT5, and PAX5 co-occupied EBNA2 super-enhancer sites. RBPJ also had significant signals at these sites. Cell TFs involved in chromatin looping, such as CTCF, SMC3, and RAD21 were at adjacent sites (Figure 1B).
EBNA2 up-regulation of MYC is essential for LCL growth (Faumont et al., 2009). EBNA2 super-enhancers at ~ −525 kb and −428 kb of the MYC TSS likely induce MYC expression (Alfieri et al., 1991; Kaiser et al., 1999; Zhao et al., 2011b) (Figure 1B). The −428 kb EBNA2 super-enhancer site loops to the MYC TSS in an EBNA2 dependent manner by Fluorescence In Situ Hybridization (FISH) and 3C-qPCR (Zhao et al., 2011b). Our re-analysis of ENCODE Chromosome Conformation Capture Carbon Copy (5C) data from GM12878 LCLs also found MYC super-enhancers linked to MYC TSS. Although quantitative enrichment could not be assessed due to lack of background controls, the capture of ligation products linking these EBNA2 super-enhancers to MYC indicated that these EBNA2 MYC super-enhancers can loop to MYC to increase MYC expression. MYC heterodimerizes with MAX to activate Cyclin D2 expression, promote cell cycle entry, and enable LCL DNA replication. Two EBNA2 super-enhancer sites also localized near MAX and likely affect MAX expression (Figure 1A). EBNA2 also up-regulates RUNX3 expression and represses RUNX1 expression (Spender et al., 1999).
The 888 EBNA2 super-enhancer sites (SE in Figure 1C) had much higher H3K27ac and H3K4me1 signals than EBNA2 typical enhancer (TE) sites, indicative of a higher transcription activation state (Figure 1C).
EBV super-enhancers
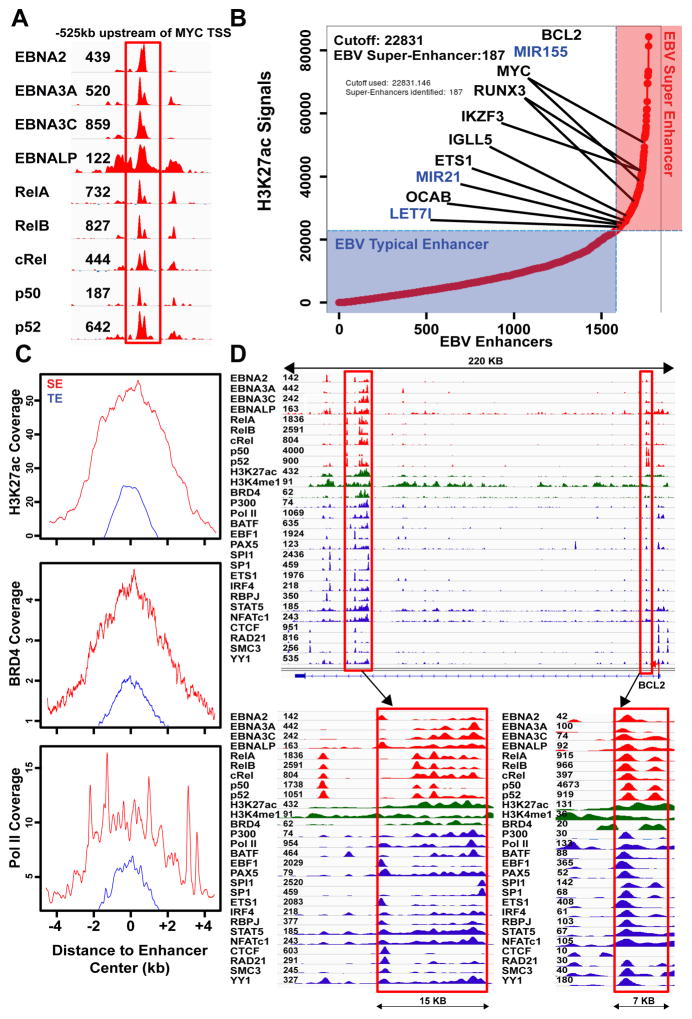
EBNA2 and EBNA3C co-localize at MYC enhancers, with RelA and EBNALP (Jiang et al., 2014; Portal et al., 2013; Zhao et al., 2014; Zhao et al., 2011b). EBNA3A, RelB, cREL, p50, and p52 binding at EBNA2 MYC super-enhancers was evaluated. Surprisingly, all oncogenic EBNAs and NF-κB subunits were at the MYC enhancer sites (Figure 2A). We therefore searched for genome-wide co-occurrences of all EBNAs and NF-κB subunits. In total, 1771 sites had significant signals for all EBNAs and NF-κB subunits, and were therefore designated as EBV enhancers.
Figure 2. EBV super-enhancers.
EBV super-enhancers are defined by high H3K27ac signals and the presence of all EBNAs and NF-κB subunits. (A) EBNA2, EBNALP, EBNA3A, EBNA3C, NF-κB subunits RELA, RELB, cRel, p50, and p52 are all significantly present at the EBNA2 super-enhancer −525kb of MYC. (B)1771 sites with significant EBV oncoproteins and NF-κB subunit binding are ranked by H3K27ac signals. 187 EBV super-enhancers with 4 fold higher H3K27ac signals than EBV typical enhancers are annotated to their nearest genes. Genes important for LCL growth and survival are indicated. (C) Anchor plots for H3K27ac, BRD4, and PolII show substantially higher and broader signals (normalized coverage) at EBV super-enhancers than at EBV typical enhancers. (D) ChIP-seq signals for virus and cell TFs and histone modifications at the BCL2 locus.
Since unusually high and broad H3K27ac signals are indicative of super- enhancers, EBV enhancers were ranked based on their H3K27ac signals. Overall, 187 EBV enhancers had >4 fold higher H3K27ac signals than the rest EBV enhancers. These were therefore designated as “ EBV super-enhancers” (Figure 2B, Table S2). In contrast to typical enhancers, which had average H3K27ac signals flanking a TF binding site without central elevation, the H3K27ac signals at EBV super-enhancer sites were broad and further elevated at the center of TF binding sites (Figure 2C).
In addition to H3K27ac, EBV super-enhancers also had much higher signals than typical enhancers for histone modifications, chromatin remodeling proteins, and basal transcription machinery, including H3K4me1 (3 fold), BRD4 (4.4 fold), Pol II (4.2 fold), BCLAF (3.6 fold), CHD1 (3.9 fold), MTA3 (3.8 fold), PML (4 fold), TAF1 (3.1 fold), and WHIP (3.9 fold) (Figure 2C, S1); proteins indicative of high super-enhancer transcription activity.
EBV super-enhancer associated genes are important for LCL growth and survival
EBV super-enhancers were assigned to their target genes by proximity. 96% of EBV super-enhancers and their regulated gene pairs resided within the same GM12878 LCL Hi-C TAD (Selvaraj et al., 2013). EBV super-enhancer associated genes included IGLL5, MYC, RUNX3, IKZF3/AIOLOS, ETS1, OCAB, and BCL2 (Figure 2B, 2D). IGLL5 encodes Igλ, which has the strongest super-enhancer in myeloma cells (Lovén et al., 2013). OCAB is also controlled by a super-enhancer in DLBCLs (Chapuy et al., 2013), whereas IKZF3 is an IKAROS family B cell TF that regulates B-cell proliferation. NF-κB induced BCL2 blocks apoptosis (Henderson et al., 1991). EBV super-enhancers were also associated with 3 miRNAs that are highly expressed in LCLs, including oncomir MIR155, MIR21, and Let-7i (Figure 2B) (Skalsky et al., 2012).
Pathway enrichment analyses for EBV super-enhancer associated genes identified enrichment for apoptosis, DNA damage response and MAPK signaling pathways (Table S3).
EBV typical enhancer associated genes included TCF3/E2A, EBF, REL, IKZF1, BATF, and IRF4, TFs important for B cell-specific transcription and B cell identity.
EBV super-enhancer enriched motifs and co-occurring cell TFs
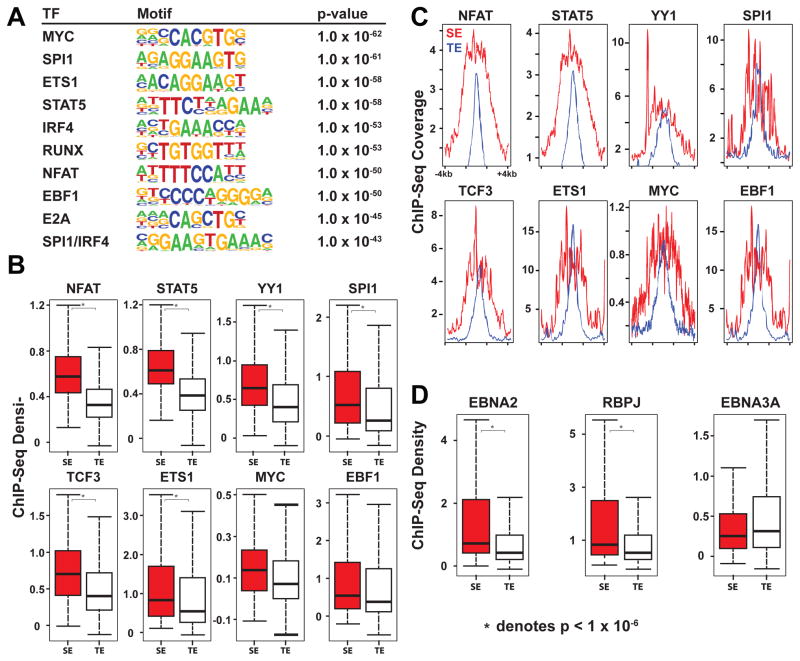
Super-enhancers are frequently enriched for cell type specific TF motifs (Whyte et al., 2013). Similarly, EBV super-enhancers were significantly enriched for B cell specific TF motifs, compared to the control, the other enhancers which were co-occupied by at least 1 EBV TF or NF-κB subunit but less than all nine. Enriched motifs included MYC, SPI1, ETS1, STAT5, IRF4, RUNX, NFAT, EBF, E2A, and SPI1/IRF4 composite site (p<10−45) (Figure 3A). These motifs were also enriched when compared with EBV typical enhancers (p<10−5). Even though many cell type specific TF motifs are enriched in super-enhancers, only a smaller number of TFs can distinguish super-enhancers from typical enhancers (Whyte et al., 2013). For example, E2A signals distinguish B-cell super-enhancers from typical enhancers (Whyte et al., 2013). In LCLs, NFAT and STAT5 signals at EBV super-enhancers were 5.1 and 3.8 fold higher than at typical enhancers (p<2×10−16) (Figure 3B). NFAT is implicated in B cell lymphomas (Pham et al., 2010), whereas STAT5 is constitutively active in LCLs (Weber-Nordt et al., 1996). STAT5 is also important for maintaining IL7 levels which are critical for B cell development and survival (Clark et al., 2014). Enrichment for these TFs in EBV super-enhancers likely contributes to super-enhancer formation and function. Notably YY1 signals at EBV super-enhancers were 2.8 fold higher than at typical enhancers (p<1×10−16) (Figure 3B). YY1 is important for long-range chromatin looping and transcription (Atchison, 2014). YY1 motifs were only moderately enriched at EBV super-enhancer sites (p<10−5) and only 11% of EBV super-enhancers had YY1 motifs. Therefore, increased YY1 binding at EBV super-enhancers was likely through interaction with other DNA binding proteins. EBV super-enhancers were also highly co-occupied by important B-cell TFs, including EBF (100%), BATF (100%), SPI1 (92%), PAX5 (99%), ETS1 (100%), and IRF4 (100%) (Figure 3B, C).
Figure 3. Transcription Factors that differentiate EBV super-enhancers from EBV typical enhancers.
(A) TF Motifs enriched at EBV super-enhancers over other EBV enhancers. Other EBV enhancers have more than 1 EBV TF or NF-κB subunit, but less than all 9. (B) Boxplots of TF ChIP-seq signals density at EBV super-enhancers and typical enhancers. Signals for NFAT, STAT5, YY1, and ETS1 at EBV super-enhancers are significantly higher than typical enhancers (Wilcoxon Rank-Sum Test p values NFATc1: p<2×10−16, STAT5: p<2×10−16, YY1: p<1×10−14, and ETS1: p<3.8×10−6, * indicates p<10−6). (C) Anchor plots of normalized TF ChIP-seq signals (coverage) around EBV super-enhancers and EBV typical enhancers. (D) Boxplots of EBNA2, RBPJ, and EBNA3A ChIP-seq signals (density) at EBV super-enhancers and typical enhancers. EBNA2 and RBPJ ChIP-seq signals at EBV super-enhancers are significantly higher than EBV typical enhancers (Wilcoxon Rank-Sum Test p values: EBNA2 p<7.3×10−12, RBPJ p<1.4×10−10, EBNA3A p<0.019). For boxplots, Middle line indicates the median. The edges indicate the first and the third quartile. The whiskers indicate minimum and maximum.
Comparison of EBNA and NF-κB signals at EBV super-enhancers and typical enhancers
ChIP-seq signals for EBNA2, EBNALP, EBNA3A, EBNA3C and NF-κB subunits at EBV super-enhancers were compared to typical enhancers. EBNA2 signals at super-enhancers were 3.6 fold higher than at typical enhancers (p<7.3×10−12) (Figure 3D), whereas, ChIP-seq signals for other EBV TFs or NF-κB subunits were far less significantly different or even similar between EBV super-enhancers and typical enhancers.
As expected, RBPJ motifs were also enriched (p<10−28) at EBV super-enhancer sites and RBPJ signals at EBV super-enhancer sites were 3.6 fold higher than at typical enhancers (p<1.4×10−10) (Figure 3D). EBNA2 can increase RBPJ DNA binding (Portal et al., 2011).
EBV super-enhancer associated genes are expressed at significantly higher levels than typical enhancer associated genes
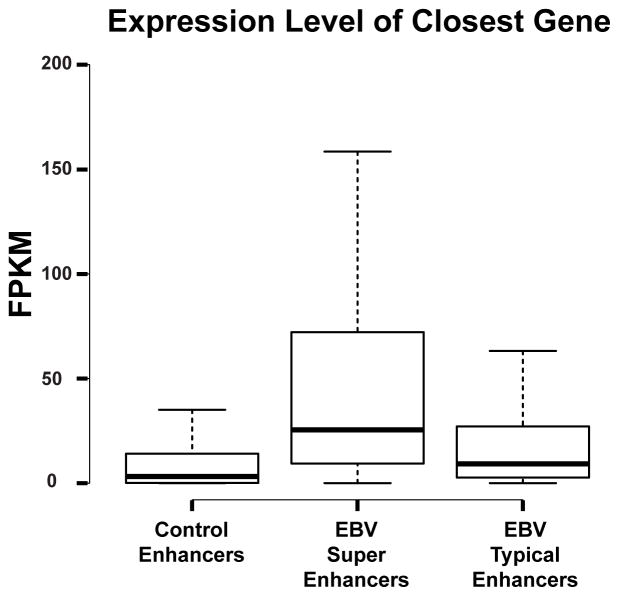
Expression level of super-enhancer associated genes are higher than typical enhancer associated genes (Whyte et al., 2013). We therefore compared expression levels of EBV super-enhancer associated genes with typical enhancer associated genes using RNA-seq data from 3 different LCLs that have minimal EBV replication (Arvey et al., 2012; Montgomery et al., 2010). Super-enhancer associated genes were expressed at significantly higher levels than typical enhancer associated genes (Wilcoxon Rank-Sum Test p< 3×10−10) (Figure 4). Furthermore, EBV super-enhancer associated gene expression levels and typical enhancer associated gene expression levels were significantly higher than other enhancer associated gene expression levels (Wilcoxon Rank-Sum Test p< 2×10−16 ).
Figure 4. EBV super-enhancer-associated genes have higher expression levels than typical enhancer associated genes.
Boxplots of LCL EBV super-enhancer associated, EBV typical enhancer associated, and other EBV enhancer associated RNA-seq gene expression levels (Fragments Per Kilobase Of Exon Per Million Fragments Mapped (FPKM)) are shown. EBV super- enhancer associated genes are expressed significantly higher than EBV typical enhancer associated genes (Wilcoxon Rank- Sum Test p<3×10−10). EBV Super-Enhancer associated genes or EBV typical enhancer associated genes are expressed significantly higher than EBV other enhancer associated genes (Wilcoxon Rank- Sum Test p<2×10−16). Middle line indicates the median. The edges indicate the first and the third quartile. The whiskers indicate minimum and maximum.
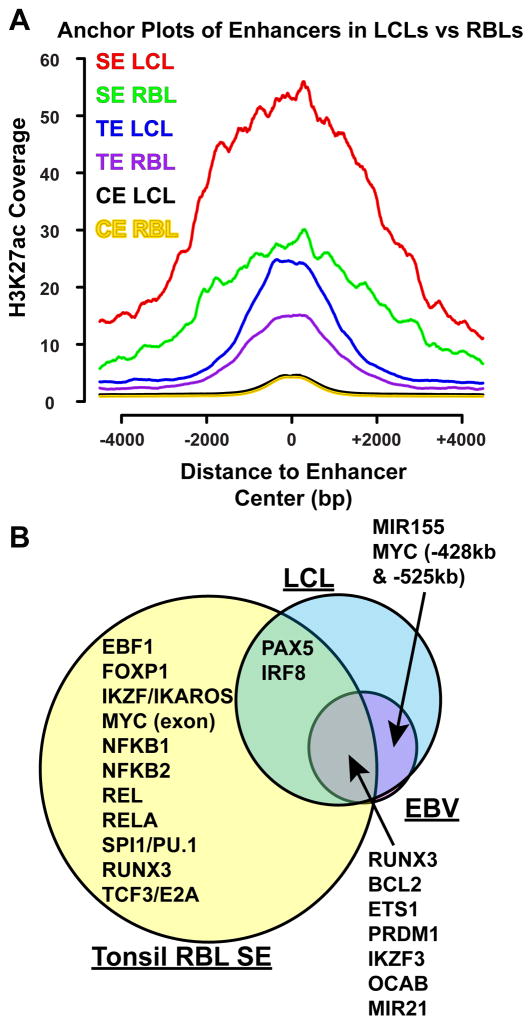
H3K27ac signals at EBV super-enhancer sites in LCLs and RBLs have similar patterns
Previous analyses of LCL and RBL H3K4me1 signals at EBNA2 sites indicated that EBNA2 sites in LCLs and RBLs have remarkably similar patterns, although LCLs in general have higher H3K4me1 signals than RBLs (Zhao et al., 2011b). We therefore analyzed tonsil RBL H3K27ac signals (Chapuy et al., 2013) at EBV super-enhancer sites. Interestingly, in RBLs, these sites also had elevated H3K27ac signals, as compared with neighboring genomic regions. However, RBL H3K27ac signals were ~50% lower than LCL signals (Figure 5A). The elevated RBL H3K27ac signals suggested B cell pioneering TF occupancy at these sites. Recruitment of p300, CBP, and PCAF histone acetyl transferases (HAT) or other chromatin remodeling proteins by EBV and EBV activated TFs such as EBNA2, EBNA3C, and NF-κB subunits may contribute to the increased H3K27ac signals (Perkins et al., 1997; Subramanian et al., 2002; Wang et al., 2000; Wu et al., 1996). Other enhancers with less than all 9 EBV TFs had much lower H3K27ac signals in both LCLs and RBLs (Figure 5A).
Figure 5. H3K27ac signals at EBV super-enhancer and typical enhancers in LCLs and RBLs.
(A) GM12878 LCL and tonsil RBL H3K27ac normalized coverage +/− 4kb of EBV super- enhancers, typical enhancers, and other enhancers are shown. (B) Overlap between RBL, LCL, and EBV super-enhancer associated genes. Genes important for B cell or LCL functions are indicated.
LCL and RBL super-enhancers
To compare LCL super-enhancers with RBL super-enhancers and identify super-enhancers in LCLs that are not co-occupied by all EBV TFs, tonsil RBL H3K27ac (Chapuy et al., 2013) and ENCODE GM12878 LCL H3K27ac ChIP-seq data were analyzed. The algorithm used to identify EBV super-enhancers identified 1756 RBL and 655 LCL super-enhancers (Figure S2A, B). RBL super-enhancers were associated with most B cell specific TFs or TFs important for B cell functions. These included EBF, IKZF1/Ikaros, TCF3/E2A, PAX5, PRDM1/BLIMP1, IKZF3, OCAB, SPI1/Pu.1, IRF8/ICSBP, RUNX1/3, and FoxP1. MYC had a super-enhancer at MYC exon in RBL (Figure 5B, Figure S2A, B). 375 (57.3%) of LCL super-enhancers also overlapped with RBL super-enhancers. These included PAX5, IRF8, RUNX3, PRDM1, IKZF3, and OCAB. LCLs lost the MYC exon super-enhancer. Instead, LCLs gained two super-enhancers >400kb upstream of MYC. LCLs also gained a super-enhancer at MIR155. The vast majority (~97%) of EBV super-enhancers were also LCL super-enhancers (Figure S3A, B, C). However, in LCLs, even though PAX5 and IRF8 were still linked to super-enhancers, they were not targeted by all EBV TFs. RBL and LCL super-enhancer associated genes were enriched for B cell functions (Table S4).
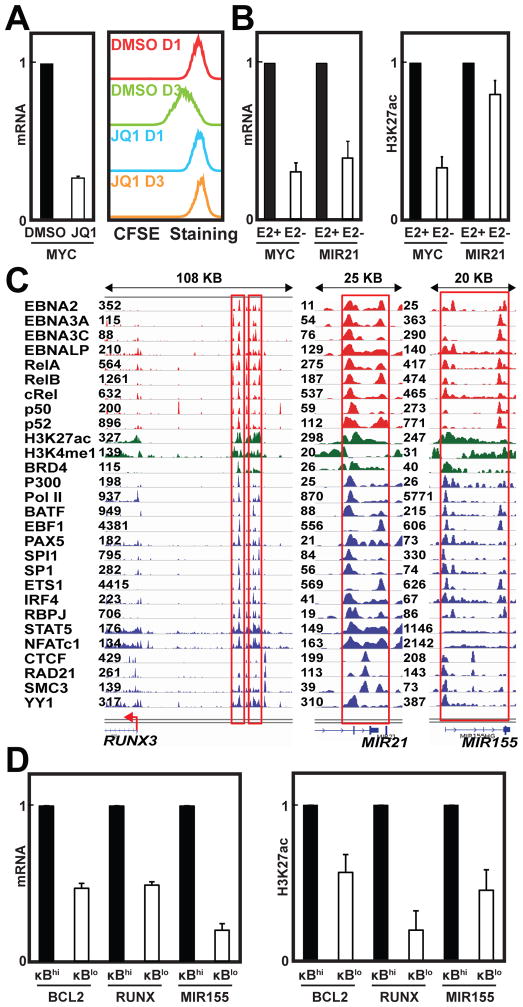
EBV super-enhancers are sensitive to perturbation
BRD4 binds to acetylated Histone lysine residues and further recruits CyclinT and CDK9 to phosphorylate PolII and activate transcription. The BET bromodomain small molecule inhibitor JQ1 specifically blocks BRD4 binding to acetylated lysine (Filippakopoulos et al., 2010) and hence disrupts DLBCL super-enhancer activity (Chapuy et al., 2013). Since BRD4 signals were evident at EBV super-enhancers and JQ1 disruption has been a super-enhancer hallmark (Figure 1B, 2D), the effect of JQ1 treatment on EBV super-enhancer activity was tested. GM12878 LCLs were treated with 500 nM JQ1 or DMSO vehicle for 24 hours. MYC expression was evaluated by quantitative RT-PCR. JQ1 treatment reduced MYC expression by more than 60% (p<0.004) (Figure 6A). JQ1 treatment also halted LCL growth, as indicated by CarboxyFluorescein diacetate, Succinimidyl Ester (CFSE) staining (Figure 6A), indicating that EBV super-enhancers are similar to cell super-enhancers in sensitivity to BRD4 inhibition.
Figure 6. Perturbation of super-enhancer constituents reduces cell growth, gene expression, and H3K27ac signals.
All Error bars represent SD. (A) Left: 24 hours post DMSO or 500nM JQ1 treatment of GM12878 LCLs, MYC mRNA levels were measured using qRT-PCR and normalized to GAPDH. MYC mRNA levels in DMSO treated cells were set to 1. Right: CFSE staining of GM12878 LCLs treated with DMSO or 500nM JQ1 for one or three days. (B) Left: Normalized MYC and MIR21 RNA levels, in conditional EBNA2 LCLs, grown under permissive (+) or non-permissive (−) conditions for EBNA2 expression. Right: H3K27ac levels at the −525kb MYC and MIR21 EBV super-enhancers, as determined by ChIP qPCR. EBNA2 (+) condition was set to 1. (C) EBV super-enhancers associate with RUNX3, MIR21, and MIR155. Super-enhancers are high-lighted by red lines with TFs signals indicated on the left. (D) Left: Normalized BCL2, RUNX3, and MIR155 mRNA levels in LCLs with high NF-κB activity (hi) versus low NF-κB activity (lo). Right: H3K27ac ChIP qPCR at BCL2, RUNX3, and MIR155 EBV super-enhancers. The NF-κB hi condition was set to 1.
LCLs transformed by a recombinant EBV that express a conditional EBNA2, grow normally under permissive conditions for EBNA2 expression (Zhao et al., 2006). When these LCLs are grown under non-permissive conditions for EBNA2, EBNA2 levels decrease and LCLs enter growth arrest (Zhao et al., 2006). Using these LCLs, we found MIR21 and MYC RNA levels were reduced by ~60–70% at 72 hours after EBNA2 inactivation (p<0.0005). Concurrently, ChIP-qPCR found MYC and MIR21 super-enhancer H3K27ac signals were also significantly reduced (p<0.05) (Figure 6B).
Likewise, all NF-κB subunits were evident at BCL2, RUNX3, and MIR155 super-enhancers (Figure 2D, 6C). Induction of a conditional IκBα dominant negative mutant in LCLs inactivates NF-κB (Cahir-McFarland et al., 2000). NF-κB inactivation resulted in a ~50–80% reduction in BCL2, RUNX3, and MIR155 gene expression (p<0.0005) and caused a ~ 40–80% reduction in H3K27ac signals at BCL2, RUNX3, and MIR155 EBV super-enhancer sites (p<0.05) (Figure 6D). Furthermore, basal promoter luciferase reporters under the control of 4 different EBV super-enhancers or 3 different EBV typical enhancers were transfected into these LCLs. Luciferase activities were determined in LCLs with wild-type or reduced NF-κB activity. Inactivation of NF-κB reduced super-enhancer reporter activity by more than 40% and typical enhancer reporter activity by less than 20% (p<0.02) (Figure S4), indicating that EBV super-enhancers were more sensitive to NF-κB inactivation than EBV typical enhancers.
Discussion
B cell specific tissue imprinting and associated TFs maintain B-cell precursors in the bone marrow. EBF expression and pioneering effects in B-cell lineage TFs, IKZF1/Ikaros, SPI1, PAX5, and other B cell specific TFs coordinately establish mature B cell identity and control gene expression (Lin et al., 2010). Subsequent antigen binding to B cell receptors and T-cell CD40 ligand-stimulation mediate RBL transcription activation.
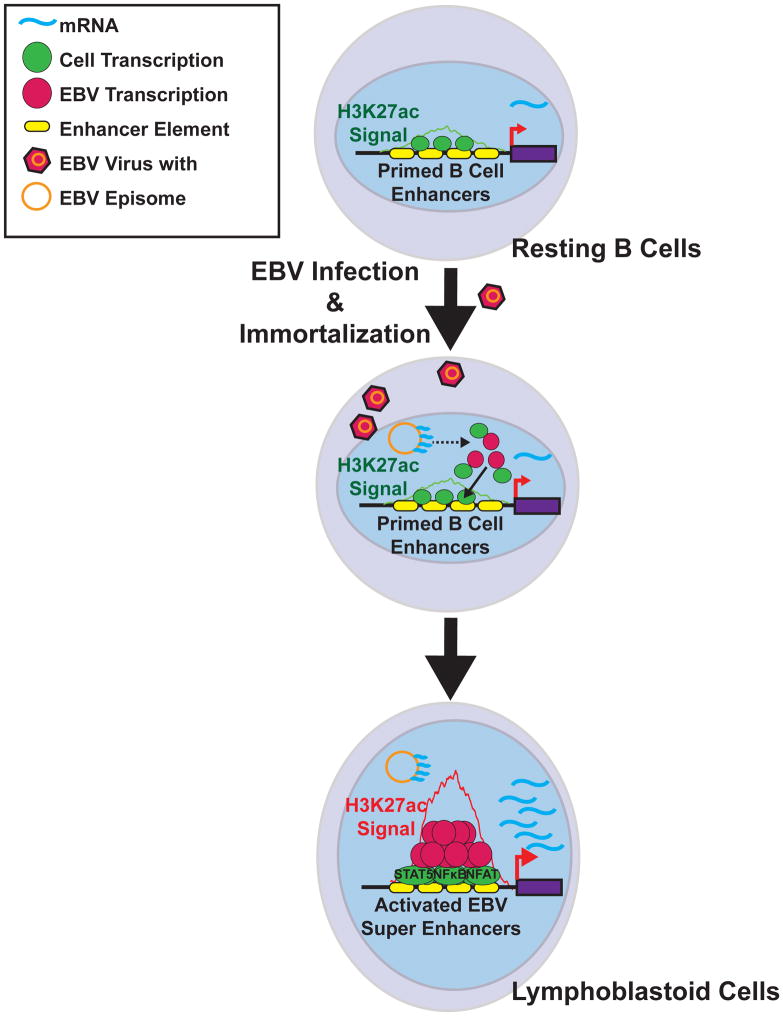
During primary EBV infection, EBV transforms B-cells into proliferating blasts, some of which ultimately differentiate into memory B-cells. Latent EBV infection of the B-cell compartment enables lifelong EBV infection. EBV infected B cells can cause lymphomas in immune suppressed individuals. This phenomenon is recapitulated in vitro, where EBV oncoprotein expression converts RBLs into LCLs. Our data suggest that EBV evolved to usurp B cell-intrinsic programs to support rapid growth and survival of latently infected B-cells (Figure 7).
Figure 7. EBV super-enhancer Model.
RBLs have broad enhancer regions with moderate H3K27ac signals. These enhancers are occupied by a limited repertoire of cell TFs to maintain chromatin accessibility. Upon EBV infection, EBNA and LMP1 oncoproteins are expressed. LMP1 activates NF-κB. All EBNAs and NF-κB subunits then co-occupy primed B cell enhancer sites, recruit additional cell TFs, and nucleate EBV super-enhancers to up-regulate transcription.
We herein identify a class of EBV super-enhancers that are comprised of EBV encoded, EBV activated, and EBV associated B-cell TFs. These EBV super-enhancers have exceptionally high signals for activation associated histone modifications, Pol II, and chromatin remodeling factors, indicative of highly active transcription. EBV super-enhancers were associated with EBF, a principal pioneering B-cell lineage factor, which increases chromatin accessibility, STAT5 and NFAT, which may nucleate EBV super- enhancer formation. These EBV super-enhancers likely exploit their high YY1 binding to loop to the TSS of affected genes, including genes critical for B cell growth and survival. EBV super-enhancers were sensitive to BRD4 perturbations as well as EBNA2 and NF-κB inactivation.
EBV efficiently transforms RBLs within a week after infection. Thereafter, infected RBLs replicate every 24 hours, in vitro (Nikitin et al.). Similarly, EBV infected cells can replicate continuously in immune deficient humans and express the same EBV genes as LCLs (Young et al., 1989). The convergence of all the EBV oncoproteins and EBV activated NF-κB subunits at EBV super-enhancers has now been dynamically demonstrated to be a key determinant of continuous LCL growth.
EBNA2 is the principal EBV super-enhancer component that up-regulates MYC. MYC overexpression is frequently the result of distal strong enhancers that loop to MYC TSS. In prostate, breast, and colon cancers, the 8q24 cancer risk variant rs6983267, which is −335kb from MYC TSS, preferentially binds TCF7L2 and loops to MYC (Pomerantz et al., 2009). Similarly, EBNA2 mediates looping from a −428kb MYC enhancer to MYC (Zhao et al., 2011b). Another EBV super-enhancer which is ~525 kb upstream of MYC is also likely to affect MYC activation. A long noncoding RNA 515kb upstream of MYC can also loop to the MYC rs6983267 enhancer and affect enhancer activity (Xiang et al., 2014). Therefore, MYC expression is complex, and likely involves multiple distinct super-enhancers.
MYC overexpression induced cell cycle entry causes apoptosis in the absence of strong pro-survival effects from BCL2 or activated tyrosine kinase growth factor receptors. EBV associated Burkitt’s lymphomas overexpress MYC as a consequence of chromosome translocations placing MYC under control of strong immunoglobin enhancers. To overcome MYC induced apoptosis, p53 is mutated in >50% of Burkitt’s lymphomas and B-cell Lymphomas in MYC transgenic mice also often have inactivated p53 or ARF (Eischen et al., 1999; Love et al., 2012). In LCLs, two EBV super-enhancers up-regulate BCL2 expression and thereby prevent MYC induced apoptosis in LCLs. Lymphomas with both MYC and BCL2 over-expression have very poor clinical outcomes (Hu et al., 2013).
EBV infection of primary B cells up-regulates MIR155, MIR21, and let-7i, which are important for LCL growth (Skalsky et al., 2012). MIR155 is essential for LCL proliferation. EBV super-enhancers, enriched with EBNA2, EBNALP, and LMP1-activated NF-κB subunits up regulate MIR155 and MIR21 expression in LCLs (Rosato et al., 2012; Yang et al., 2013). MIR21 regulates PTEN. Let-7i is associated with high grade lymphomas (Lawrie et al., 2009). Control of these critical mirRNAs by EBV super-enhancer will likely ensure LCL growth and survival.
Chromosome conformation capture carbon copy assays suggest that as few as 7% of the enhancers regulate their nearest genes, although this can vary substantially among cell types (Sanyal et al., 2012). In contrast, >90% of super-enhancers affect their nearest gene by Hi-C (Whyte et al., 2013). In support of most EBV super-enhancers also regulating the nearest promoter, we found that perturbation of EBV super-enhancers very frequently reduced expression of the nearest LCL genes and associated super-enhancer H3K27ac signals. These findings correlated the EBV super-enhancers to their nearby regulated genes.
Super-enhancers are highly accessible open chromatin regions with multiple co- occurring cell TFs. Although “accessible” loci maybe prone to artifacts (Teytelman et al., 2013), EBV super- enhancers are specifically composed of EBNA2, EBNALP, EBNA3A, EBNA3C, EBV activated NF-κB subunits, other EBV protein associated cell TFs, as well as co-factors, active chromatin regulators, BRD4 and H2AZ, and core TFs. EBV super- enhancers are highly unlikely due to ChIP-seq “artifacts” since dynamic perturbation of these super-enhancers by conditional EBNA2 or NF-κB inactivation led to down- regulation of EBV super-enhancer associated genes and cessation of cell growth. Furthermore, EBV super- enhancers are functionally relevant to LCL biology.
EBNA2 inactivation substantially decreased MYC expression, but had surprisingly little effect on BCL2 expression, whereas NF-κB inactivation had a dramatic effect on BCL2 expression and small effect on MYC expression, confirming that MYC and BCL2 are largely affected by EBNA2 and NF-κB, respectively. Increased activities from other EBV super-enhancer constituents likely to compensate the deficiency. Thus, the co-occurrence of other EBV TFs and NF-κB subunits at MYC and BCL2 may be indicative of an underlying, fail-safe transcription mechanism that assures proliferation and survival.
Like other super-enhancers (Lovén et al., 2013), EBV super-enhancers were sensitive to perturbations. JQ1 inhibition decreased super-enhancer associated transcription more than typical enhancer associated transcription (Chapuy et al., 2013; Lovén et al., 2013). As reflected in BRD4 inhibition, EBNA2 and NF-κB inactivation, EBV super-enhancers were sensitive to perturbation. These data indicate that EBV super-enhancer disruption may be effective in controlling EBV transformed cell growth.
Experimental Procedures
Sequencing Data Alignment
All ChIP-Seq reads were mapped to hg19 using Bowtie (version 0.12.9). Alignments were done with parameters: -S -t -p 1 -k 1 -m 1.
Identifying ChIP-Seq-Enriched Regions
MACS (1.4.2) was used to identify ChIP-seq TF binding sites. Default parameters were used with the exception of “to-large”, which was set due to low sequencing depth of older ChIP-seq datasets.
Motif Enrichment
HOMER (4.4) “findMotifsGenome.pl” was used to identify enriched motifs (Heinz et al., 2010) with “other enhancers” set as background.
Identification of Overlapping Binding Sites
HOMER “MergePeak” was used to identify the co-occurrence of binding sites (default parameters). If there was a overlap between the start and end coordinates of both enhancer regions, these enhancers were considered “overlapping”.
Definition of EBV Enhancers
EBV enhancers were defined by the co-localization of four EBNAs and five NF-κB subunits. Binding sites of these transcription factors were identified by MACS. “Other Enhancers” were defined by the presence of at least one, but less than all nine TFs.
Identifying EBV Super-Enhancers and EBNA2 Super-Enhancers
To identify EBV super-enhancers, all EBV Enhancers were ranked according to their total background-subtracted H3K27ac ChIP-seq signal. EBV enhancers were sorted and plotted based on H3K27ac signals in ascending order. The X axis shows H3K27ac ChIP-seq signals rank order; the Y axis shows normalized H3K27ac signals. A line was drawn from the first enhancer with lowest signal to the last enhancer with the highest signal to determine a diagonal slope. A point on the ranked plot with a tangent line identical to the diagonal slope was identified. This X axis point was set as the cut-off to distinguish EBV super-enhancers from EBV typical-enhancers. The EBV enhancers with H3K27ac signals higher than this point were assigned as EBV super-enhancers.
EBNA2 super-enhancers were identified as described above using EBNA2 ChIP-seq signals. EBNA2 binding sites within 12.5kb were stitched together as previously described (Whyte et al., 2013).
Hi-C validation of super-enhancer and their associated gene assignment
GM12878 LCL Hi-C topological association domains (generated by Bing Ren Lab) (Selvaraj et al., 2013) was used to validate the assignment of super-enhancer and associated-gene pairs. Correct assignments were scored based on the co-occurrence of a super-enhancer and its associated-gene within the same TAD.
Anchor Plots
Anchor plots show the distribution of TFs and Histone modifications ChIP-Seq signals at various binding sites, as previously described (Portal et al., 2013).
Gene Expression Analysis
“EBV super-enhancers”, “EBV typical-enhancers”, and “EBV other enhancers” associated gene expression were determined using LCL RNA-seq data. The average gene expression level (FPKM) was calculated with three LCLs (NA06985, NA07000, NA07347). RNA-seq data from these three LCLs were chosen because they had the least amount of EBV late gene expression. Boxplots were drawn using R, and the statistical significance of the difference between each two pairs of the three groups were determined using the Wilcoxon signed-rank test.
Pathway Enrichment Analysis
The enriched pathways of super-enhancer-associated genes were identified using the “Identify Pathways” function of the IntPath database (Zhou et al., 2012) and DAVID database (Huang da et al., 2008).
Reporter Assays
Four EBV super-enhancer and three typical enhancer sequences were PCR amplified and cloned into pGL3 promoter luciferase reporter vectors (Promega). 20μg of control or enhancer luciferase vectors, together with 2μg of Renilla expression vector, were electroporated (Gene Pulser II, Bio-Rad), into 7 × 106 LCLs containing an inducible IκBα mutant. Electroporated cells were split into permissive or non-permissive conditions for mutant IκBα expression and grown for 48 hour. Dual Luciferase/Renilla assays were done following manufacturer’s direction (Promega).
Supplementary Material
Acknowledgments
We thank Brian J. Abraham, Ellen Cahir-McFarland, Hongfang Wang, Shirley Liu and Tao Liu for helpful discussions. We thank Bing Ren and Anthony Schmitt for the GM12878 Hi-C TAD dataset. We thank Jay Bradner for the JQ1 compound. We thank Amy Holthaus and Amy Zheng for technical assistant. E.K. was supported by R01CA047006, R01CA170023, and R01CA085180 from the National Cancer Institute. B.G. was supported by Burroughs Wellcome Medical Scientist career award and K08CA140780. E.J. was supported by R01DE023939 from NIDCR.
Footnotes
ACCESSION NUMBERS
EBNA3A ChIP-seq data has been deposited to GEO. Accession number: GSE59181.
AUTHOR CONTRIBUTIONS
B.Z. and E.K. designed the study, B.Z. and E.K. supervised research, B.Z., S.C.S., S.J., B.W., K. B., B.E.G., and J.L. performed experiments, E.C.J., P.K., provided new reagents, H.Z. performed the computational analysis, H.Z. and S.J. prepared figures and tables, B.Z., H.Z., S.C.S., S.J., B.E.G., and E.K. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare they have no competing financial interests.
Supplemental Information includes four figures and four tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. doi: 10.1016/0042-6822(91)90893-g. [DOI] [PubMed] [Google Scholar]
- Arvey A, Tempera I, Tsai K, Chen HS, Tikhmyanova N, Klichinsky M, Leslie C, Lieberman PM. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe. 2012;12:233–245. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison ML. Function of YY1 in Long-Distance DNA Interactions. Frontiers in immunology. 2014;5:45. doi: 10.3389/fimmu.2014.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, Bair S, Newton G, Lichtman AH, Kung AL, et al. NF-kappaB Directs Dynamic Super Enhancer Formation in Inflammation and Atherogenesis. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahir-McFarland ED, Carter K, Rosenwald A, Giltnane JM, Henrickson SE, Staudt LM, Kieff E. Role of NF-kappa B in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J Virol. 2004;78:4108–4119. doi: 10.1128/JVI.78.8.4108-4119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahir-McFarland ED, Davidson DM, Schauer SL, Duong J, Kieff E. NF-kappa B inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc Natl Acad Sci U S A. 2000;97:6055–6060. doi: 10.1073/pnas.100119497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy B, McKeown Michael R, Lin Charles Y, Monti S, Roemer Margaretha GM, Qi J, Rahl Peter B, Sun Heather H, Yeda Kelly T, Doench John G, et al. Discovery and Characterization of Super-Enhancer-Associated Dependencies in Diffuse Large B Cell Lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MR, Mandal M, Ochiai K, Singh H. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nature reviews Immunology. 2014;14:69–80. doi: 10.1038/nri3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of cell identity genes occurs in insulated neighborhoods in Mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein MA, Achong BG, Barr YM. Virus Particles in Cultured Lymphoblasts from Burkitt’s Lymphoma. Lancet. 1964;1:702–703. doi: 10.1016/s0140-6736(64)91524-7. [DOI] [PubMed] [Google Scholar]
- Faumont N, Durand-Panteix S, Schlee M, Gromminger S, Schuhmacher M, Holzel M, Laux G, Mailhammer R, Rosenwald A, Staudt LM, et al. c-Myc and Rel/NF-kappaB are the two master transcriptional systems activated in the latency III program of Epstein-Barr virus-immortalized B cells. J Virol. 2009;83:5014–5027. doi: 10.1128/JVI.02264-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci U S A. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S, Kieff E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J Virol. 1997;71:6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S, Rowe M, Gregory C, Croom-Carter D, Wang F, Longnecker R, Kieff E, Rickinson A. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell. 1991;65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- Henkel T, Ling PD, Hayward SD, Peterson MG. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- Hertle ML, Popp C, Petermann S, Maier S, Kremmer E, Lang R, Mages J, Kempkes B. Differential gene expression patterns of EBV infected EBNA-3A positive and negative human B lymphocytes. PLoS Pathog. 2009;5:e1000506. doi: 10.1371/journal.ppat.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham Brian J, Lee Tong I, Lau A, Saint-André V, Sigova Alla A, Hoke Heather A, Young Richard A. Super-Enhancers in the Control of Cell Identity and Disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Xu-Monette ZY, Tzankov A, Green T, Wu L, Balasubramanyam A, Liu WM, Visco C, Li Y, Miranda RN, et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121:4021–4031. doi: 10.1182/blood-2012-10-460063. quiz 4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID gene ID conversion tool. Bioinformation. 2008;2:428–430. doi: 10.6026/97320630002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Willox B, Zhou H, Holthaus AM, Wang A, Shi TT, Maruo S, Kharchenko PV, Johannsen EC, Kieff E, et al. Epstein-Barr virus nuclear antigen 3C binds to BATF/IRF4 or SPI1/IRF4 composite sites and recruits Sin3A to repress CDKN2A. Proc Natl Acad Sci U S A. 2014;111:421–426. doi: 10.1073/pnas.1321704111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Laux G, Eick D, Jochner N, Bornkamm GW, Kempkes B. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J Virol. 1999;73:4481–4484. doi: 10.1128/jvi.73.5.4481-4484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth J, Spieker T, Wustrow J, Strickler GJ, Hansmann LM, Rajewsky K, Kuppers R. EBV-infected B cells in infectious mononucleosis: viral strategies for spreading in the B cell compartment and establishing latency. Immunity. 2000;13:485–495. doi: 10.1016/s1074-7613(00)00048-0. [DOI] [PubMed] [Google Scholar]
- Lawrie CH, Chi J, Taylor S, Tramonti D, Ballabio E, Palazzo S, Saunders NJ, Pezzella F, Boultwood J, Wainscoat JS, et al. Expression of microRNAs in diffuse large B cell lymphoma is associated with immunophenotype, survival and transformation from follicular lymphoma. Journal of Cellular and Molecular Medicine. 2009;13:1248–1260. doi: 10.1111/j.1582-4934.2008.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R, Kieff E, Cohen JI. Epstein-Barr Virus. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott, Williams and Wilkins; 2013. pp. 1898–1959. [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Hoke Heather A, Lin Charles Y, Lau A, Orlando David A, Vakoc Christopher R, Bradner James E, Lee Tong I, Young Richard A. Selective Inhibition of Tumor Oncogenes by Disruption of Super-Enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo S, Johannsen E, Illanes D, Cooper A, Zhao B, Kieff E. Epstein-Barr virus nuclear protein 3A domains essential for growth of lymphoblasts: transcriptional regulation through RBP-Jkappa/CBF1 is critical. J Virol. 2005;79:10171–10179. doi: 10.1128/JVI.79.16.10171-10179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo S, Wu Y, Ishikawa S, Kanda T, Iwakiri D, Takada K. Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc Natl Acad Sci U S A. 2006;103:19500–19505. doi: 10.1073/pnas.0604919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo S, Zhao B, Johannsen E, Kieff E, Zou J, Takada K. Epstein-Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression. Proc Natl Acad Sci U S A. 2011;108:1919–1924. doi: 10.1073/pnas.1019599108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin PA, Yan CM, Forte E, Bocedi A, Tourigny JP, White RE, Allday MJ, Patel A, Dave SS, Kim W, et al. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe. 8:510–522. doi: 10.1016/j.chom.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, Program NCS, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND, Felzien LK, Betts JC, Leung K, Beach DH, Nabel GJ. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- Pham LV, Tamayo AT, Li C, Bueso-Ramos C, Ford RJ. An epigenetic chromatin remodeling role for NFATc1 in transcriptional regulation of growth and survival genes in diffuse large B-cell lymphomas. Blood. 2010;116:3899–3906. doi: 10.1182/blood-2009-12-257378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portal D, Rosendorff A, Kieff E. Epstein-Barr nuclear antigen leader protein coactivates transcription through interaction with histone deacetylase 4. Proc Natl Acad Sci U S A. 2006;103:19278–19283. doi: 10.1073/pnas.0609320103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portal D, Zhao B, Calderwood MA, Sommermann T, Johannsen E, Kieff E. EBV nuclear antigen EBNALP dismisses transcription repressors NCoR and RBPJ from enhancers and EBNA2 increases NCoR-deficient RBPJ DNA binding. Proc Natl Acad Sci U S A. 2011;108:7808–7813. doi: 10.1073/pnas.1104991108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portal D, Zhou H, Zhao B, Kharchenko PV, Lowry E, Wong L, Quackenbush J, Holloway D, Jiang S, Lu Y, et al. Epstein-Barr virus nuclear antigen leader protein localizes to promoters and enhancers with cell transcription factors and EBNA2. Proc Natl Acad Sci U S A. 2013;110:18537–18542. doi: 10.1073/pnas.1317608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato P, Anastasiadou E, Garg N, Lenze D, Boccellato F, Vincenti S, Severa M, Coccia EM, Bigi R, Cirone M, et al. Differential regulation of miR-21 and miR-146a by Epstein-Barr virus-encoded EBNA2. Leukemia. 2012;26:2343–2352. doi: 10.1038/leu.2012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj S, JRD, Bansal V, Ren B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nature biotechnology. 2013;31:1111–1118. doi: 10.1038/nbt.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalska L, White RE, Parker GA, Sinclair AJ, Paschos K, Allday MJ. Induction of p16(INK4a) is the major barrier to proliferation when Epstein-Barr virus (EBV) transforms primary B cells into lymphoblastoid cell lines. PLoS Pathog. 2013;9:e1003187. doi: 10.1371/journal.ppat.1003187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA, et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spender LC, Cannell EJ, Hollyoake M, Wensing B, Gawn JM, Brimmell M, Packham G, Farrell PJ. Control of cell cycle entry and apoptosis in B lymphocytes infected by Epstein-Barr virus. J Virol. 1999;73:4678–4688. doi: 10.1128/jvi.73.6.4678-4688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian C, Hasan S, Rowe M, Hottiger M, Orre R, Robertson ES. Epstein-Barr virus nuclear antigen 3C and prothymosin alpha interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J Virol. 2002;76:4699–4708. doi: 10.1128/JVI.76.10.4699-4708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman L, Thurtle DM, Rine J, van Oudenaarden A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci U S A. 2013;110:18602–18607. doi: 10.1073/pnas.1316064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Drapkin R, Reinberg D, Kieff E. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc Natl Acad Sci U S A. 1995;92:3259–3263. doi: 10.1073/pnas.92.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Gregory CD, Rowe M, Rickinson AB, Wang D, Birkenbach M, Kikutani H, Kishimoto T, Kieff E. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc Natl Acad Sci U S A. 1987;84:3452–3456. doi: 10.1073/pnas.84.10.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Grossman SR, Kieff E. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc Natl Acad Sci U S A. 2000;97:430–435. doi: 10.1073/pnas.97.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber-Nordt RM, Egen C, Wehinger J, Ludwig W, Gouilleux-Gruart V, Mertelsmann R, Finke J. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood. 1996;88:809–816. [PubMed] [Google Scholar]
- Whyte Warren A, Orlando David A, Hnisz D, Abraham Brian J, Lin Charles Y, Kagey Michael H, Rahl Peter B, Lee Tong I, Young Richard A. Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DY, Kalpana GV, Goff SP, Schubach WH. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J Virol. 1996;70:6020–6028. doi: 10.1128/jvi.70.9.6020-6028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang JF, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell research. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GD, Huang TJ, Peng LX, Yang CF, Liu RY, Huang HB, Chu QQ, Yang HJ, Huang JL, Zhu ZY, et al. Epstein-Barr Virus_Encoded LMP1 Upregulates MicroRNA-21 to Promote the Resistance of Nasopharyngeal Carcinoma Cells to Cisplatin-Induced Apoptosis by Suppressing PDCD4 and Fas-L. PLoS ONE. 2013;8:e78355. doi: 10.1371/journal.pone.0078355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L, Alfieri C, Hennessy K, Evans H, O’Hara C, Anderson KC, Ritz J, Shapiro RS, Rickinson A, Kieff E, et al. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N Engl J Med. 1989;321:1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Barrera LA, Ersing I, Willox B, Schmidt SC, Greenfeld H, Zhou H, Mollo SB, Shi TT, Takasaki K, et al. The NF-kappaB genomic landscape in lymphoblastoid B cells. Cell reports. 2014;8:1595–1606. doi: 10.1016/j.celrep.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Mar JC, Maruo S, Lee S, Gewurz BE, Johannsen E, Holton K, Rubio R, Takada K, Quackenbush J, et al. Epstein-Barr virus nuclear antigen 3C regulated genes in lymphoblastoid cell lines. Proc Natl Acad Sci U S A. 2011a;108:337–342. doi: 10.1073/pnas.1017419108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Maruo S, Cooper A, MRC, Johannsen E, Kieff E, Cahir-McFarland E. RNAs induced by Epstein-Barr virus nuclear antigen 2 in lymphoblastoid cell lines. Proc Natl Acad Sci U S A. 2006;103:1900–1905. doi: 10.1073/pnas.0510612103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Zou J, Wang H, Johannsen E, Peng CW, Quackenbush J, Mar JC, Morton CC, Freedman ML, Blacklow SC, et al. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci U S A. 2011b;108:14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Jin J, Zhang H, Yi B, Wozniak M, Wong L. IntPath--an integrated pathway gene relationship database for model organisms and important pathogens. BMC systems biology. 2012;6(Suppl 2):S2. doi: 10.1186/1752-0509-6-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.