Abstract
Small cell lung carcinoma (SCLC) often features the up-regulation of the Sonic Hedgehog (Shh) pathway leading to activation of Gli transcription factors. SCLC cells secrete bombesin (BBS)-like neuropeptides (BLPs) that act as autocrine growth factors. Here, we show that SCLC tumour samples feature co-expression of Shh and BBS-cognate receptor (Gastrin-Releasing Peptide Receptor: GRPR). We also demonstrate that BBS activates Gli in SCLC cells, which is crucial for BBS-mediated SCLC proliferation because cyclopamine, an inhibitor of the Shh pathway, hampered the BBS-mediated effects. BBS binding to GRPR stimulated Gli through its downstream Gαq and Gα12/13 GTPases, and consistently, other Gαq and Gα13 coupled receptors (such as muscarinic receptor, m1, and thrombin receptor, PAR-1) and constitutively active GαqQL and Gα12/13QL mutants stimulated Gli. By using cells null for Gαq and Gα12/13, we demonstrate that these G proteins are strictly necessary for Gli activation by BBS. Moreover, by using constitutively active Rho small G-protein (Rho QL) as well as its inhibitor, C3 toxin, we show that Rho mediates GPCR-, Gαq- and Gα12/13-dependent Gli stimulation. At the molecular level, BBS caused a significant increase in Shh gene transcription and protein secretion that was dependent on BBS-induced GPCR/Gαq-12/13/Rho mediated activation of NFκB, which can stimulate a NFκB response element in the Shh gene promoter. Our data identify a novel molecular network acting in SCLC linking autocrine BBS and Shh circuitries, and suggest Shh inhibitors as novel therapeutic strategies against this aggressive cancer type.
Keywords: SCLC, neuropeptide, sonic hedgehog, G-protein
INTRODUCTION
Lung cancer is one of the leading cause of cancer-related death in the Western countries.1 Small cell lung carcinoma (SCLC) is a very aggressive, highly metastasizing and lethal lung cancer type and it accounts for about 15% of all lung cancers.1 SCLC is regarded as a neuroendocrine tumour because of its ability to synthesize various hormone peptides that can act as paracrine and autocrine growth factors.2
The vertebrate family of hedgehog extracellular ligands is represented by three members: Desert (Dhh), Indian (Ihh) and Sonic (Shh) hedgehog. Shh is the best-characterized family member and it is involved in a wide variety of developmental events. In humans, Shh gene is located on chromosome 7 (7q36) and its heterozygous mutations result in holoprosencephaly.3 Shh signals are transduced, at the cell surface, by two transmembrane proteins, Patched (Ptch) and Smoothened (Smo). Ptch is a 12-pass membrane protein that binds directly to Shh ligands and inhibits Smo function. Smo is a 7-pass transmembrane protein required to transduce the Shh signal. Shh/Ptch binding leads to the de-repression of Smo. In turn, Smoregulates canonical (Gli dependent) signaling, resulting in activation of the Gli transcription factor, and non canonical (Gli independent) signaling leading to activation of heterotrimetic Gi and small GTPase RhoA proteins. Non canonical Shh signaling is also activated independently of Smo, through Ptch regulation of cell survival.45
Shh binding to Ptch phenocopies the genetic loss of Ptch.6 In the absence of Ptch, Smo constitutively activates Shh pathway.7 Shh acts both as a short-range and a long-range diffusible morphogen and regulates epithelial-mesenchymal interactions in many organs.8 It is expressed in the embryonic lung epithelium while Ptch is found in mesenchymal cells, this system representing a model for epithelial-mesenchymal interaction in lung development.9
Gain-of-function of Shh pathway is associated with many different tumours.10 The fact that upregulation of Shh signalling occurs in familial cancer, such as basal cell carcinoma (BCC) and medulloblastoma, indicates that it is sufficient to initiate tumour formation.11 Recent findings demonstrate that deregulation of Shh signalling is also required for tumour maintenance, as neoplastic cells depend on Shh activity for survival and proliferation.10 Based on the evidence that Shh pathway is an important regulator of lung development12 and that several tumors of neuroendocrine origin overexpress molecules of the Shh pathway, Watkins and colleagues have found that SCLCs are dependent on Shh pathway for tumour development and maintenance.13 Moreover, genetically engineered mice require Shh signaling for SCLC initiation, progression, and tumor maintenance.14
SCLC derived cell lines require conditioned medium to grow in culture, suggesting that they need autocrine/paracrine growth factors to proliferate.15, 16 These growth factors have been identified as different neuropeptides. The prototypical neuropeptide secreted by SCLC cells is Gastrin-Releasing Peptide (GRP), the mammalian homologue of the amphibian bombesin (BBS).2, 17 GRP has a carboxyl-terminal peptide similar to that of BBS, and binds to the same receptor as BBS. BBS/GRP receptor (GRPR) is a 7- transmembrane-spanning protein that couples to heterotrimeric G proteins of the Gαq and Gα12/13 families. Since most of the SCLC cells express not only GRP/BBS but also its cognate GRPR, GRP/BBS has been recognized as autocrine growth factor for SCLC cells as it sustains cell proliferation, survival, angiogenesis, local invasion and distant metastases.16, 18 The presence of this receptor/ligand pair is regarded as a marker of tumor aggressiveness and poor prognosis and its inhibition represents an attractive target for SCLC therapy.2, 19 Here we demonstrate a functional interaction between Shh and BBS pathways in SCLC.
RESULTS
BBS promotes proliferation of SCLC cells through Shh pathway
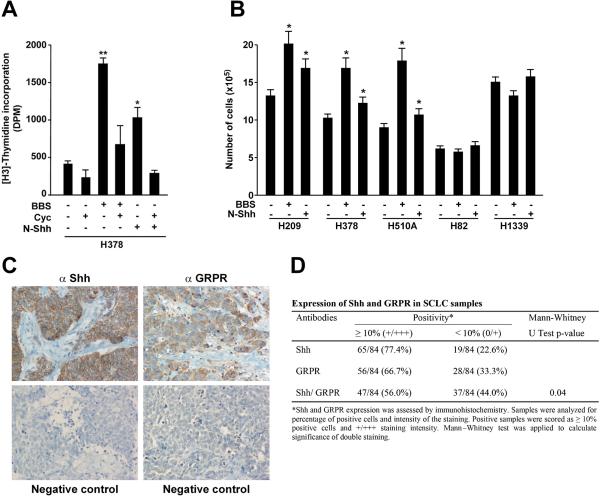
BBS functions as autocrine growth factor in SCLC cells cell growth.2 In order to test a relationship between the BBS and the Shh pathways, we measured DNA synthesis rate of H378 SCLC cells. As shown in figure 1A, BBS treatment increased DNA synthesis; interestingly, this proliferative effect was sharply reduced when the cells were treated with cyclopamine, a specific inhibitor of the Smo receptor6 (Fig. 1A). Soluble Shh (pSec-N-Shh) (Supplementary Informations, Fig. S1) had a proliferative effect on H378 cells that was abolished by cyclopamine, which hence served as an experimental control (Fig. 1A). To confirm that other SCLC cells also responded to BBS and Shh stimulation, we cultured a panel of different SCLC cell lines for 72 hours in the presence of BBS or Shh ligand and counted total number of cells. Figure 1B shows that BBS and Shh increased number of H209, H378 and H510A cells. Interestingly, other SCLC cells known to be negative for bombesin-like immunoreactivity, H82 and H133916, 18-20 did not show the same response (Fig. 1B).
Figure 1. BBS induced proliferation of SCLC cells is inhibited by cyclopamine.
A) H378 cells were pre-treated with cyclopamine or vehicle for 48 hours, serum starved (1:1, complete medium: serum free medium) and stimulated over night with 100nM BBS. DNA synthesis was measured upon 6 hours of [H3]-thymidine treatment (1μCi/ml). The results represent the average of three independent experiments +/- SD. *p<0.05, **p<0.01. B) A panel of SCLC cell lines positive (H209, H378, H510A) and negative (H82, H1339) for bombesin-like immunoreactivity were plated and cultured for 72 hours in the presenceof BBS or Shh before evaluating the number of cells. The results represent the average of three independent experiments +/- SE. *p<0.05. C, D) Co-expression of Shh and GRPR in SCLCs. Representative immunohistochemistry of a SCLC sample stained with anti Shh and anti GRPR antibodies. Negative controls for each antibody are also shown. The data set is reported in D. E) Several mass populations of H209 cells transfected with three different shRNA plasmids targeting Shh as well as scrambled control were generated and silencing of the protein was tested by Western blotting where tubulin was used for normalization. F) H209 stably silenced for Shh and scrambled control were plated and cultured for 72 hours in the presence of BBS or Shh before evaluating the number of cells. The results represent the averageof three independent experiments +/- SD. *p<0.05, **p<0.01. G) The ability of scrambled and Shh silenced cells to invade Matrigel was tested by embedding the cells in 3D Matrigel and monitoring their ability to form large colonies in the presence of BBS or Shh. Representative phase contrast micrographs are shown. H, I) H209 stably transfected with scrambled and Shh targeting shRNA were plated in soft agar and colonies were counted after two weeks under BBS stimulation. Representative phase contrast micrographs are shown in H. The average results of three independent experiments are presented in I. Significance was calculated by using a paired, two-tailed Student t test. **p= <0.01.
Co-expression of Shh-Gli1 and GRPR in human SCLCs
To verify in vivo the correlation between the Shh and the BBS pathways, we performed immunohistochemical analysis on a set of 84 human SCLC samples with Shh and GRPR specific antibodies. Representative stainings are shown in figure 1C, and the dataset is reported in figure 1D. Interestingly, the majority of the SCLC samples examined were positive for Shh (77.4%) and GRPR (66.7%). Overall, 56% of the cases showed co-expression of Shh and GRPR (p=0.04).
Silencing of Shh in SCLC cells abolishes proliferation, invasion and colony formation in response to BBS
To investigate whether the Shh pathway participates in the effects of BBS on SCLC cells, we stably silenced Shh in H209 cells by using three shRNA targeting plasmids and generating different mass culture populations (sh Shh MP1, MP2, MP3). Figure 1E shows the level of Shh protein knock-down, using tubulin as loading control. When testing the proliferative rate of these newly generated cells, we observed that Shh knock-down cells grew more slowly than H209 cells stably transfected with scrambled shRNA; moreover they did not respond to BBS stimulation (Fig. 1F). Nonetheless, treatment with Shh ligand was able to rescue the knock-down effect and to increase number of cells (Fig. 1F). To evaluate the effect of Shh silencing on other biological properties of H209 cells, we tested the ability of Shh knockdown cells to perform invasive growth in Matrigel and to form colonies in soft agar. As shown in figure 1G, when embedded in 3D Matrigel the shRNA scrambled cells were able to proliferate and generate large colonies in response to BBS and, to a lesser extent, in response to Shh ligand. On the contrary, Shh silenced cells did not proliferate. Once again, stimulation with Shh ligand reverted the effect of silenced Shh (Fig. 1G). Similarly, when testing the ability of the cells to form colonies in soft agar, we observed that Shh knock-down cells generated only few small colonies; on the contrary, cells stably transfected with scrambled shRNA generated large colonies whose number and size was further increased in response to BBS (p<0.01)(Fig 1H, I).
BBS stimulation activates the Shh-Gli1 pathway
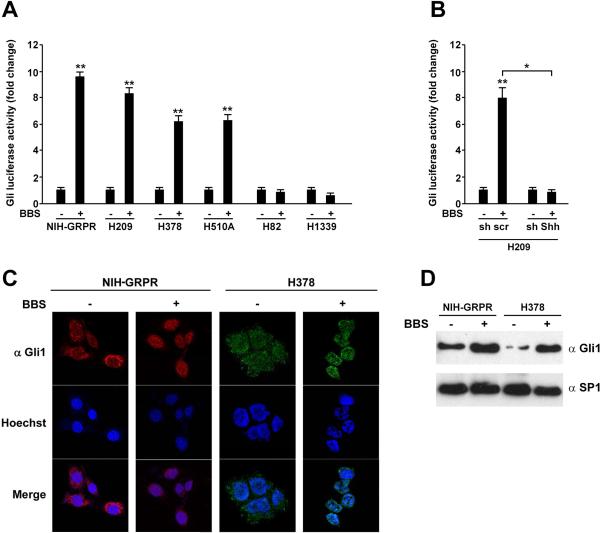
The transcriptional output of the Shh signaling is mediated by the Gli transcription factors. To investigate the possibility of a direct interaction between the Shh and the BBS pathways, we engineered a reporter plasmid (Gli-Luc), where the luciferase gene was under the control of 8 Gli1-responsive elements (Supplementary Informations, Fig. S1A) and produced mutant (Ptch insensitive) Smo (SmoA1) and soluble N-Shh (Supplementary Information, Fig. S1A, B). We stably transfected GRPR in NIH3T3 cells and used for further experiments these NIH-GRPR expressing cells together with H209, H378, and H510A SCLC cells that express BBS and GRPR endogenously.18 In NIH3T3-GRPR, H209, H378, and H510A cells, but not in H82 and H1339, BBS stimulation significantly increased Gli-luciferase activity (Fig. 2A). This activation was abolished when Shh was silenced in H209 cells, demonstrating once again that Shh mediates the effect of BBS in SCLC cells (Fig. 2B). Moreover, Gli-Luc activation was paralleled by accumulation of Gli1 protein into the nucleus, as tested by immunofluorescence in both, NIH-GRPR and H378 SCLC cells (Fig. 2C); in basal conditions, upon serum deprivation, Gli1 was mainly located into the cytosol, while it redistributed to the nucleus upon stimulation with 100 nM of BBS (Fig. 2C). Same results were also obtained by Western blotting of sub-cellular nuclear fractions where SP1 was used as a loading control (Fig. 2D).
Figure 2. BBS stimulation activates Gli in SCLC cells.
A) NIH3T3 cells expressing GRPR receptor (NIH-GRPR) and a panel of SCLC cells were plated in 6 well plates and transfected with 300 ng of pGL3-Gli-Luc reporter or the empty pGL3 together with 50 ng of Renilla plasmid. After 24 hours, the cells were serum starved and analysed for luciferase activity upon stimulation with bombesin (BBS) (100nM) or vehicle. B) H209 cells stably transfected with sh scrambled or sh Shh were transfected with pGL3-Gli-Luc reporter or the empty pGL3 together with Renilla. After 24 hours, the cells were serum starved and analysed for luciferase activity upon stimulation with BBS or vehicle. The results represent the average of three independent experiments +/- SD. **p<0.05, **p<0.01. C) NIH-GRPR and H378 cells were plated on coverslips and stained by immunofluorescence with Gli1 antibody. Cells were visualized with Hoechst nuclear dye. Gli1, Hoechst and merged pictures were taken and representative images are shown. D) NIH-GRPR and H378 cells were subjected to protein extraction and nuclear fractionation; the levels of endogenous Gli expression were tested by Western blotting after 12 hours of BBS stimulation or vehicle (-). Sp1 was used as nuclear normalization marker.
G protein αq and α13 promote activation of the Shh-Gli1 pathway
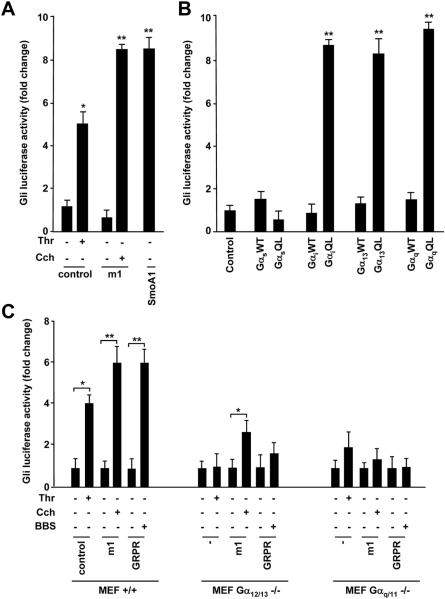
GRPR signals through heterotrimeric guanine nucleotide-binding protein Gαq and Gα12/13. Evidence that BBS activated the Gli-mediated transcription, prompted us to investigate whether this activation was mediated by Gαq and Gα12/13. We first checked if other GPCR coupled to Gαq or Gα12/13 activated Gli-Luc. We used parental NIH3T3 cells endogenously expressing thrombin (Thr) receptor PAR-1, coupled to Gα12/13, and transfected NIH3T3 cells exogenously expressing muscarinic receptor (m1), coupled to Gαq. Stimulation with both Thr (ligand for PAR-1) or carbachol, Cch, (a cholinergic agonist ligand for m1) activated Gli-Luc (Fig. 3A); SmoA1 was used as positive control (Fig. 3A).
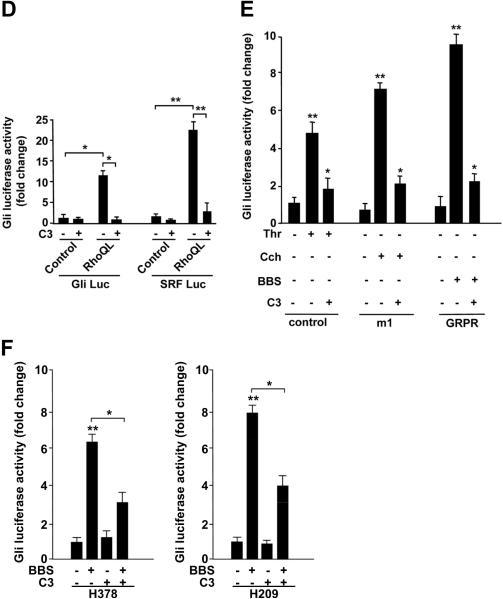
Figure 3. Activation of Gli-Luc by GPCRs and large G proteins.
A) Empty vector (control), m1 and SmoA1 receptors were transiently transfected in NIH3T3 cells together with Gli-Luc reporter and Renilla. After 24 hours, cells were serum starved and stimulated with carbachol (Cch) (1mM) or thrombin (Thr) (5U/ml) and Gli luciferase activity was tested. SmoA1 was used as positive control. B) Gli-Luc activity was measured upon transfection of α subunits of different G proteins in NIH3T3 cells. For each family (Gαq, Gα13, Gαi, Gαs) wild type (WT) and constitutively activated (QL) mutants were used. C) Wild type, Gαq/11 and Gα12/13 knock-out MEFs were transiently transfected with empty vector (control), m1 or GRPR receptors together with Gli-Luc reporter and Renilla. Gli luciferase activity was measured upon Thr, Cch and BBS stimulation. D) Empty vector or active Rho mutant (RhoQL) were transfected in NIH-GRPR cells together with either Gli-Luc or SRF-Luc reporters. Luciferase activity was measured in basal conditions and upon transfection of C3 toxin. E) NIH3T3 cells were transfected with empty vector (control), m1 or GRPR receptors together with Gli-Luc reporter and Renilla. The effect of C3 toxin on Gli luciferase activity was measured upon Thr, Cch or BBS stimulation. F) The effect of C3 toxin transfection on Gli-Luc activity was measured in H378 and H209 cells, in basal conditions as well as upon BBS stimulation. The results are the average of three independent experiments +/ SD. *p< 0.05, **p< 0.01.
To check if Gαq and Gα13 were able to directly activate Gli, we transiently transfected NIH3T3 cells with wild type (WT) or activated (QL) GTPase-deficient forms of Gα13, Gαq, Gαs, or Gαi. Constitutively active Gα13 and Gαq induced Gli-Luc response, to a similar extent as GαiQL, a known mediator of the signaling downstream of Smo receptor21 (Fig. 3B). Differently, GαsQL was unable to activate Gli (Fig. 3B).
To clarify the role of Gαq, and Gα13 in mediating BBS-dependent Gli1 response, we used mouse embryonic fibroblasts (MEFs) knock-out for Gαq/11 or Gα12/1322Supplementary Informations, Fig. S2). In these MEFs, we evaluated the activation of Gli-Luc upon transient transfection with empty vector, m1 or GRPR receptors followed by stimulation with Thr, Cch and BBS, respectively. As shown in figure 3C, Thr was ableto induce a Gli-Luc response in wild type (+/+) MEFs while the effect was lost in Gα12/13 knockout (-/-) fibroblasts and was reduced in Gαq/11 -/-MEFs, suggesting that Thr can signal to Gli mainly through Gα12/13. On the other hand, Cch was not able to elicit a Gli-Luc response in Gαq/11-/- cells and its function was reduced in Gα12/13 -/- MEFs (Fig. 3C). Interestingly, the effect of BBS on Gli-luciferase activation was lost in both knockout MEFs, indicating that, although Gαq/11 or Gα12/13 subunits can signal to the nucleus independently, they may be both required for the full stimulation of Gli by BBS (Fig. 3C).
BBS mediated Gli activation is dependent on Rho and is sensitive to C3 toxin
Monomeric Rho GTPase is the main effector of Gαq and Gα13 proteins and BBS leads to rapid Rho activation.23, 24 Moreover, Rho has been involved in Hh signaling.4, 5 To address the possibility that Rho could mediate the BBS activation of Gli,we transfected a constitutively active form of Rho (Rho QL) in NIH3T3-GRPR cells. Rho QL resulted in the stimulation of the Gli-Luc reporter, when compared to the SRF-Luc reporter, a bona fide Rho target (Fig. 3D). Moreover the Clostridium botulinum C3 toxin, which specifically ADP-ribosylates at residue 41 and inhibits Rho25, blocked Gli activation in response to BBS, Thr or Cch and in response to Rho QL in NIH3T3 cells (Fig. 3D, E), as well as in response to BBS in H378 and H209 cells (Fig. 3F). All together our results support that the small GTPase Rho plays an important role in regulating signaling to Gli in response to BBS.
Activation of a Shh autocrine loop in cells stimulated with BBS
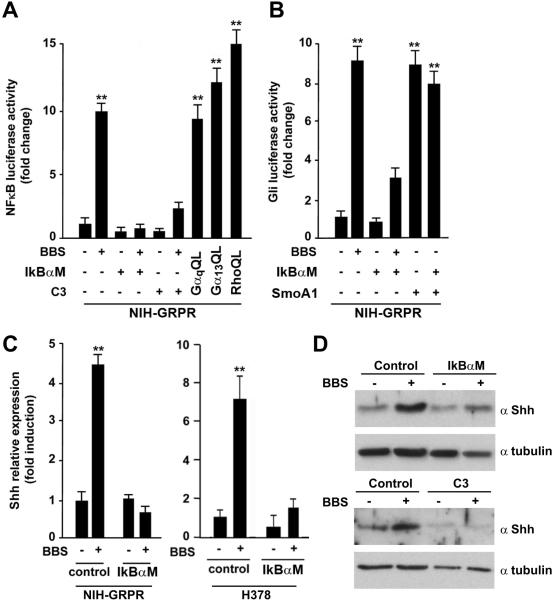
Previous studies have shown that several GPCRs (including GRPR) activate Nuclear Factor κB (NFκB) through different mechanism, initiated by heterotrimeric G- proteins and converging on IκB phosphorylation and nuclear translocation of NFκB (reviewed in26). Moreover, Rho is able to activate NFκB.27 Thus, we tested whether in SCLC cells BBS signals to NFκB through Rho. Figure 4A shows that in NIH-GRPR cells BBS potently activated NFκB Luc, and that a specific inhibitor of NFκB (IκBαM) completely abolished this activation. Interestingly, C3 toxin also impaired BBS-mediated NFκB activation (Fig. 4A). To study the role of NFκB in regulating the BBS mediated signal to Gli, we transiently transfected IκBαM in NIH-GRPR cells and observed a reduced activation of Gli Luc upon BBS treatment (Fig. 4B). IκBαM did not affect the activation of Gli Luc mediated by active Smo receptor (Smo A1), hence serving as a specificity control (Fig 4B). A recent study has characterized Shh as a direct target of NFκB; NFκB binds to the Shh promoter and triggers its transcription.28 We therefore tested whether BBS would directly stimulate Shh transcription through NFκB activation. BBS stimulation significantly increased the mRNA levels of Shh in NIH-GRPR and H378 SCLC cells, and this effect was lost in the presence of IkBαM (Fig 4C). A similar result was observed by Shh Western blot in NIH-GRPR cells transfected with IkBαM or Rho inhibitor C3 (Fig. 4D). Of direct relevance to SCLC biology, H378 and H209 SCLC cells stimulated with BBS showed a remarkable increase in Shh protein levels aligned with its impact in Shh gene expression (Supplementary Informations, Fig. S3). All together, these results suggest that BBS activates Shh production through a Gαq/Gα12/13-Rho-NFκB mediated pathway, and that in turn this likely causes Gli activation.
Figure 4. BBS induces an autocrine loop producing Shh ligand, through activation of NFkB transcription factor.
A) NIH-GRPR cells were treated with BBS or vehicle in the presence of transiently transfected NFkB (IkBαM) or Rho (C3) inhibitor and activation of NFkB-Luc was measured. Luciferase activity was tested also upon transfection of activated (QL) α subunits of Gαq and Gα13, as well as upon RhoQL transfection. B) NIH-GRPR cells were treated with BBS or vehicle in the presence of transiently transfected IkBαM or C3 and activation of Gli-Luc was measured. As control, activated Smo (SmoA1) was transfected alone or together with IkBαM. C) Production of Shh in response to BBS was tested by RNA extraction and quantitative RT-PCR upon transfection of control plasmid or IkBαM in NIHGRPR and H378 cells. The results represent the average of three independent experiments +/-SD. *p<0.05, **p<0.01. D) Production of Shh in response to BBS was tested by Western blotting with a specific Shh antibody in NIH-GRPR cells upon IkBαM or C3 transfection. Tubulin was used for normalization.
The Smo antagonist cyclopamine synergizes with Bortezomib and Cisplatin in SCLC cells
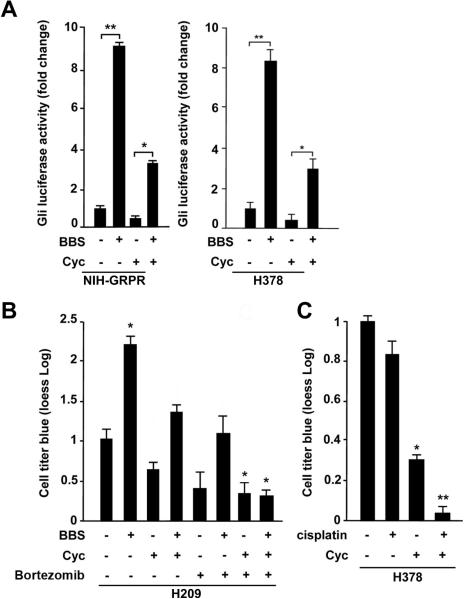
To test the dependency of the BBS effect on the Smo receptor, we performed Gli-Luc assay in the presence of cyclopamine (Cyc), an inhibitor of Smo.6 Our results indicate that Smo block resulted in substantial inhibition of BBS dependent Gli-Luc activation in NIH-GRPR cells as well as in H378 SCLC cells (Fig. 5A). Moreover, parallel pharmacological inhibition of Smo (with cyclopamine) and use of Bortezomib, a proteasome inhibitor able to block NfkB,29 abolished BBS-mediated proliferative effect in H209 cells, suggesting that Shh and NFκB pathways are both involved in the response of SCLC cells to BBS (Fig. 5B). On the contrary, NFκB Luc activation by BBS was not affected by cyclopamine (Supplementary Informations, Fig. S4), thus indicating that the activation of NFκB is independent on Smo. As a control, IkBαM reduced cell proliferation in response to BBS in H209 without affecting their survival, albeit C3 toxin abolished the BBS effect on cell proliferation and also limited their survival under the same experimental conditions (Supplementary Informations, Fig S5).
Figure 5. Cyclopamine inhibits the effect of BBS and cooperates with Bortezomib and cisplatin.
A) NIH-GRPR and H378 cells were pretreated with cyclopamine for 24 hours, serum starved and stimulated with BBS over-night to measure activity of transiently transfected Gli-Luc. B) Combination of cyclopamine and Bortezomib significantly reduces BBS stimulation of H209 cell viability as measured by cell titre blue dye reduction. C) Combination of cyclopamine and cisplatin significantly reduces cell viability of H378 SCLC cells. The results represent the average of three independent experiments +/- SD. *p<0.05, **p<0.01.
SCLCs are poorly responsive to conventional therapies, and they quite often develop resistance to routinely used chemotherapeutic agents such as cisplatin.30 We treated H378 cells with cisplatin in the presence or absence of cyclopamine. Figure 5C shows that, while as expected cisplatin treatment had only modest effect in reducing cell viability,30 cyclopamine instead induced a significant reduction of living cells. More interestingly, the combination of cisplatin with cyclopamine resulted in a dramatic decrease of cell viability (Fig. 5C).
DISCUSSION
Signaling pathways that play a crucial role during development may also be implicated in tumorigenesis.31 The Hedgehog pathway represents one important example of this concept. Loss-of-function mutations in the gene encoding the inhibitory receptor of the pathway, Patched (Ptch), are responsible for an inherited cancer predisposition syndrome known as Gorlin or naevoid basal cell carcinoma syndrome (NBCCS).11 Mammalian sonic hedgehog, Shh, a secreted ligand, mediates epithelial-mesenchymal interactions during lung development by regulating signals involved in branching morphogenesis12 and the loss of Shh results in severe lung defects in mice.32
Small-cell lung cancer (SCLC) is a very aggressive tumour with neuroendocrine features.2 Autocrine growth factors secreted from SCLC cells belonging to the bombesin (BBS) like peptide (BLP) family, mainly gastrin-releasing peptide (GRP) and its cognate receptor (GRPR), promote growth and invasion.33 Significant SCLC growth inhibition in vitro as well as in vivo has been achieved by different strategies blocking either BBS or GRPR.34 Shh signalling is activated in SCLCs, where it promotes both tumour development and maintenance.13 However, no mutations in Shh pathway components have been found in SCLCs13 and therefore the mechanisms of Shh activation in SCLC remain unclear.14
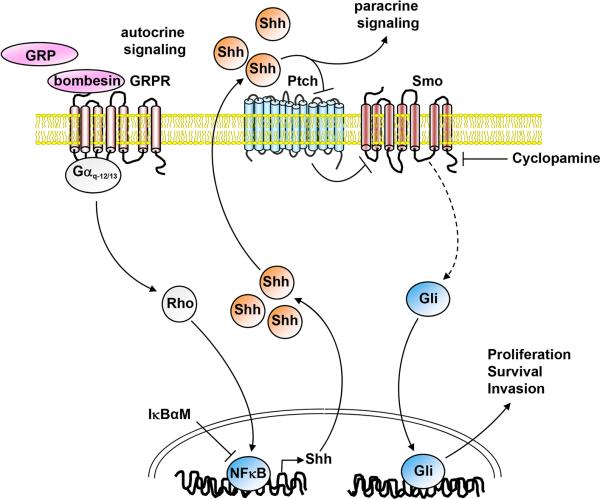
Our data show that BBS induces proliferation of SCLC cells and this effect is blocked by cyclopamine, an inhibitor of Smo. Moreover, we demonstrate that BBS activates the Gli-Lucreporter through G protein-coupled receptor (GPCR) mediated activation of Gαq and Gα12/13 proteins and through downstream activation of Rho. BBS mediates also NFkB activation through the same G proteins, and our results are aligned with the proposed model that NFkB may in turn bind to the Shh promoter directly and stimulate Shh transcription, thus generating an autocrine signaling loop initiated by BBS that results in Shh ligand production (Fig. 6).
Figure 6. Schematic representation of the mechanism of action of bombesin on the Shh pathway in SCLC cells.
Bombesin activates GRPR receptor that is linked to Gαq and Gα12/13 proteins. Gαq and Gα12/13 signaling involves Rho, that stimulates a variety of transcription factors, such as NFkB. Activation of NFkB increases production of Shh, which in turn activates Gli through Ptch and Smo membrane receptors on the same cell (autocrine signaling) or neighbouring cells (paracrine signaling).
Our findings shed light on the mechanism underlying Shh pathway activation in SCLC. The expression of BBS/GRPR-Shh pairs in many other cancer types, such as glioblastoma,35 gastrointestinal tract tumors,36 pancreatic cancers,37 head and neck squamous cellcarcinomas38 and neuroblastomas39 and the observation that the Shh pathway is increased in these malignancies opens the possibility that the mechanism described here can be more general, and maybe operational also in other cancer types. Furthermore, the existence of an autocrine loop starting from BBS and leading to Shh production can also be important in initiating paracrine signaling between cancer and stroma cells; accordingly, several reports suggest that the paracrine Shh production by tumor cells is able to activate Gli signaling in stromal cells and viceversa.40, 41
More than 50 compounds have been isolated thus far that act as inhibitors of the Hh pathway; at least five of them are in clinical trials (Cur-61414, IPI-926, GDC-0449, XL139, LDE-225).42 A recent study suggested that inhibition of the Shh pathway in mouse models cooperates with chemotherapy to delay SCLC recurrence.14 In this framework, our data may open new therapeutic approaches for this aggressive disease, as it may be possible to target the autocrine activation of GRPR, and/or disrupt their downstream pathways resulting in the local Shh secretion, as part of future combined anti-cancer therapies for SCLC.
MATERIALS AND METHODS
Cell culture
NIH3T3 and HEK293T (293T) cells were maintained in DMEM, 10% calf serum or fetal bovine serum, respectively (Gibco, Invitrogen, Groningen, The Netherlands). SCLC cells H378, H82 (ATCC, Manassas, VA, USA), H510A, H1339 (DSMZ, Braunschweig, Germany), H209 (a kind gift from Dr. A. Budillon)43were grown at 37°C in RPMI 1640, 10% heat inactivated FBS. Mouse embryonic fibroblasts from wild-type, Gαq-11 and Gα12-13 knock-out animals, a kind gift from Dr. S. Offermanns,22 were kept in DMEM supplemented with 10% FBS, with 1 mM sodium pyruvate and non-essential amino acids. Cells were transfected by Lipofectamine Plus (Invitrogen, Groningen, The Netherlands) (NIH3T3, 293T and MEFs), or Fugene 6 (Roche, Mannheim, Germany) (SCLC cells). Bombesin (BBS) was used at 100 nM (Sigma Chemical Company, St. Louis, MO, USA); Cyclopamine (cyc) (Calbiochem, La Jolla, CA, USA) was used at 10 μM; Bortezomib was used at 100 nM (Millenium, Cambridge, MA, USA); Cisplatin was used at 400 nM concentration (Pfizer, New York, NY, USA).
DNA constructs
pCDNA3 GRPR was purchased from Guthrie cDNA Resource Center (Rolla, MO, USA). pCEFL Smo and pCEFL Gli1 were obtained by cloning the corresponding full length cDNA PCR products into the pCEFL vector by the Gateway technology (Invitrogen, Groningen, The Netherlands). The oncogenic SmoA1 (Trp537Leu) mutant was obtained by site-directed mutagenesis (Stratagene, Cedar Creek, TX, USA) of the pCEFLSmo plasmid. Plasmids expressing HA-tagged-m1 receptor and AU5-tagged wild type and constitutively activated (QL) Rho, as well as expression plasmids for wild type and active forms of Gαq, Gαi, Gαs, Gα13 are described elsewhere.23,44 pEF-C3 was a kind gift of Dr. R. Treisman. pSRF-luc and pNFκB Luc were purchased from Stratagene (La Jolla, CA, USA). pCDNA3 IkBαM mutant was provided by Dr.A. Leonardi (University of Naples, Italy).45 pRenilla Luc was purchased from Promega (Madison, WI, USA). pGL3-Gli-Luc was obtained by PCR cloning eight copies of the Gli DNA binding site into the pGL3 enhancer vector from Promega (Madison, WI, USA). Shh silencing was obtained by stably transfecting three shRNA expression plasmids (TG 309454: GI337809-10-11) or scrambled control as instructed by the manifacturer (Origene, Rockville, MD, USA). Briefly, 1μg of shRNA expression plasmid was transfected with 3 μl of Fugene 6 reagent in a 6-well plate. 48 hours after transfection puromycin selection (1μg/ml) was added to the growth medium and mass populations were isolated.
Production of Secreted Shh
- PCR primers were designed for the active N-terminal portion of the human Shh. PCR product was cloned into the BamHI/NotI site of the pSecTag2B plasmid (Invitrogen, Groningen, The Netherlands). This construct was transfected into 293T cells using Lipofectamine Plus (Invitrogen, Groningen, The Netherlands). At 80% of confluence, cells were serum starved. Medium containing secreted N-terminal-Shh (N-Shh) was collected at 1 and 2 days post transfection. Medium collected from parallel transfections with the empty pSecTag2B vector was used as control.
Western blotting analysis and immunofluorescence
Protein extractions and immunoblotting experiments were performed according to standard procedures. Briefly, cells were harvested in lysis buffer (50mM HEPES, pH 7,5, 150mM NaCl, 10% glycerol, 1% Triton X-100, 1mM EGTA, 1.5mM MgCl2, 10mM NaF, 10mM sodium pyrophosphate, 1mM Na3VO4, 10mg of aprotinin/ml, 10 mg of leupeptin/ml) and clarified by centrifugation at 10.000xg. Antibodies for Western blotting: rabbit polyclonal Shh, rabbit polyclonal Sp1 and rabbit polyclonal Gαq and Gα12/13 (Santa Cruz Biotechnology, Santa Cruz, CA, USA); rabbit polyclonal Gli1 (Zymed, San Francisco, CA, USA); mouse monoclonal anti-α-tubulin (Sigma-Aldrich, St Louis, MO, USA). For nuclear cell extracts, cells were lysed with the NEPER (nuclear and cytoplasmic extraction reagents, Pierce Biotechnology, Rockford, IL, USA). For indirect immunofluorescence, NIH3T3 and H378 cells were serum starved, stimulated with BBS (100nM) for 24 hours, and then seeded on glass coverslips (H378 cells were cytospinned before seeding). 1:100 anti-Gli1 and subsequently FITC-conjugated donkey rabbit secondary antibodies were used (Jackson Immunoresearch, West Grove, PA, USA). Coverslips were then inverted and mounted onto glass slides with Vectashield containing 4',6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA) and analysed by fluorescence microscopy (Axiovert 2, Zeiss; equipped with a 40X lens) interfaced with the image analyzer software KS300 (Carl Zeiss, Thornwood, NY, USA).
Reporter assays
NIH3T3 or SCLC cells were transiently transfected with Gli-Luc or NFκB-Luc reporters together with pRL-TK (encoding the Renilla luciferase) in triplicate, as instructed by the manifacturer (Promega, Madison, WI, USA). Luciferase activity was measured 48 h after transfection by using the Dual-Luciferase Reporter System (Promega, Madison, WI, USA). The total amount of plasmid DNA was adjusted with pCDNAIII-β-gal.Light emission was quantitated using a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA, USA). Data were presented as luciferase activity present in each sample, and the values plotted were the average +/- SD of triplicate samples from representative experiments. Experiments were repeatedthree times.
RT-PCR
Total RNA was harvested by using the Rneasy Kit (Qiagen, Valencia, CA, USA). CDNA synthesis was done with Superscript II (40 U/l, Invitrogen, Groningen, The Netherlands). Real-time PCR was performed using the Biorad-iCycler IQ detector system and Biorad-iCycler IQ SYBR Green mix (Biorad, Munich, Germany). The PCR primers (Sigma Chemical Company, St. Louis, MO, USA) were: Shh 5'-CAGCGACTTCCTCACTTTCC-3' (forward) and 5'-GGAGCGGTTAGGGCTACTCT-3' (reverse); GAPDH: 5'-ACAGTCAGCCGCATCTTCTT-3' (forward) and 5'-ACGACCAAATCCGTTGACTC-3' (reverse). Fluorescent threshold values were measured in triplicate, and fold changes were calculated by the formula 2-(sample 1 ΔCt - sample 2 ΔCt), where ΔCt is the difference between the amplification fluorescence thresholds of the mRNA of interest and the GAPDH mRNA.
Proliferation assays
Thymidine incorporation: Proliferation of H378 cells was measured as amount of DNA synthesis. Cells were pretreated with cyclopamine for 48 hours and grown over night in low (2.5%) serum medium +/- BBS stimulation. [H3]-thymidine was added to a final concentration of 1μCi/ml and cells were incubated for 6 hours at 37°. Cells were then washed twice in cold PBS, incubated 10 minutes at 4° with 6% trichloroacetic acid (TCA),washed twice in water and lysed is 0.5 N NaOH for 1 hour at 37°. Thymidine incorporation was measured with a scintillation counter, and values were reported as DPM (disintegrations per minute). Cell count: The cell count was performed using a TC10 Automated Cell Counter (BioRad, Hercules, CA, USA) according to the manufacturer's instructions. Cell Titre Blue assay: Cells were plated at 1,000 cells/well in a volume of 35 μl of DMEM supplemented with 2.5% FBS. CellTiter-Blue reagent (Promega, Madison, WI, USA) (10 μl) diluted (1:1) in culture medium was added to each well. Plates were incubated at 37 °C, in 5% CO2 for 6 hours and then transferred to room temperature for overnight incubation, at dark. Cell viability was measured by dye reduction with an EnVision Multilabel plate reader at an excitation wavelength of 530 nm and emission wavelength of 590 nm (Perkin Elmer, Waltham, MA, USA). The raw data were smoothed using loess-correction and subsequently normalized to the median of the negative controls and log2-transformed (loess-log normalization).
Soft agar colony formation assay and growth in 3D matrigel
Cells (20.000) were plated in 60mm dishes and incubated at 37°C, 5% CO2 for colony formation. After 15 days, colonies were fixed with 10% (v/v) methanol for 15 min and stained with crystal violet for 30 min for colony visualisation. Each count was performed in triplicate.
For outgrowth in Matrigel, 10.000 cells were mixed with 160 μl of Matrigel (BD Biosciences, San Jose, CA, USA) and plated in 35 mm dishes containing a glass coverslip. After hardening, cells were overlaid with 2 mL growth medium and incubated at 37 degrees for up to 8 days.
Immunohistochemistry
Archival frozen lung tissue samples from 84 patients affected by SCLCs were retrieved from the files of the Pathological Anatomy, Department of Surgery of the University of Pisa uponinformed consent. All tumor samples were formalin-fixed and paraffin-embedded for microscopic examination. The most representative paraffin block of each tumor was selected for analysis. Histologic diagnosis and pathologic features were obtained by 2 pathologists (G. Alì and G. Fontanini) according to the World Health Organization (WHO) 2004 histologic criteria.46 Immunohistochemical analysis (IHC) was performed on 3-μm tissue sections. After deparaffinization through serial xylene baths, sections were rehydrated through a graded series of ethanol and water baths (5 minutes each). After a deionized water wash (3 minutes), the sections were heated to 98°C for 40 minutes to unmask target antigens, then cooled at room temperature and washed with phosphate-buffered saline (PBS) for 3 minutes. After treatment with peroxide block, sections were washed again in PBS and incubated with power block reagent (Biogenex Laboratories, San Ramon, CA, USA) for 30 minutes. Rabbit polyclonal anti-Gli, rabbit polyclonal anti-GRPR and rabbit polyclonal anti-Shh were from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and were used overnight at 4°C at 1:50, 1:20 and 1:30 dilution respectively. After washing with PBS, immunoreactivity was obtained by using the Super Sensitive Polymer-HRP Detection System (Biogenex, Fremont, CA, USA), following manufacturer's instructions. After rinsing with PBS for 3 minutes, sections were incubated 20 minutes at room temperature with Super Enhancer Reagent, followed by incubation with Poly-HRP reagent. The reaction was developed using 0.05% 3,3′-diaminobenzidine tetrahydrochloride until adequate color development was seen. A negative control was obtained by omitting primaryantibody. Two pathologists (G. Alì and G. Fontanini) evaluated and graded immunohistochemical staining by scoring separately the percentage of positive tumor cells (< 10%; ≥ 10%) and their staining intensity (0/1+: 2+; 3+). The pathologists were blinded to clinico-pathologic characteristics of the patients. All experiments were approved by the “Comitato etico per la sperimentazione clinica del farma- co” of the Azienda Ospedaliera — Universitaria Pisana (Pisa, Italy).
Statistical analysis
Statistical analyses were performed using a paired, two-tailed Student's t test or Mann Whithney test (IHC) (GraphPad Prism 3.0, GraphPad Software, San Diego, CA, USA), and differences were considered to be statistically significant at a value of p <0.05. Data analysis was performed using GraphPad Prism 5.03 (GraphPad Software). All values were expressed as the means ± standard error (SE) or standard deviation (SD). Differences were considered significant at p value <0.05. Student's t-test was used to analyze the differences between control and treatment groups.
Supplementary Material
Acknowledgments
This study was in part supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) and by the Intramural Research Program of NIH, National Institute of Dental and Craniofacial Research, Z01DE00551.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Mulshine JL, Sullivan DC. Clinical practice. Lung cancer screening. N Engl J Med. 2005;352:2714–2720. doi: 10.1056/NEJMcp042630. [DOI] [PubMed] [Google Scholar]
- 2.Zakowski MF. Pathology of small cell carcinoma of the lung. Semin Oncol. 2003;30:3–8. doi: 10.1053/sonc.2003.50015. [DOI] [PubMed] [Google Scholar]
- 3.Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, et al. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- 4.Polizio AH, Chinchilla P, Chen X, Kim S, Manning DR, Riobo NA. Heterotrimeric Gi proteins link Hedgehog signaling to activation of Rho small GTPases to promote fibroblast migration. J Biol Chem. 2011;286:19589–19596. doi: 10.1074/jbc.M110.197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinchilla P, Xiao L, Kazanietz MG, Riobo NA. Hedgehog proteins activate pro angiogenic responses in endothelial cells through non-canonical signaling pathways. Cell Cycle. 2010;9:570–579. doi: 10.4161/cc.9.3.10591. [DOI] [PubMed] [Google Scholar]
- 6.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes & development. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes & development. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 8.Chuang PT, McMahon AP. Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature. 1999;397:617–621. doi: 10.1038/17611. [DOI] [PubMed] [Google Scholar]
- 9.Weaver M, Batts L, Hogan BL. Tissue interactions pattern the mesenchyme of the embryonic mouse lung. Dev Biol. 2003;258:169–184. doi: 10.1016/s0012-1606(03)00117-9. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz i Altaba A, Sanchez P, Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer. 2002;2:361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- 11.Hahn H, Christiansen J, Wicking C, Zaphiropoulos PG, Chidambaram A, Gerrard B, et al. A mammalian patched homolog is expressed in target tissues of sonic hedgehog and maps to a region associated with developmental abnormalities. J Biol Chem. 1996;271:12125–12128. doi: 10.1074/jbc.271.21.12125. [DOI] [PubMed] [Google Scholar]
- 12.Pepicelli CV, Lewis PM, McMahon AP. Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Curr Biol. 1998;8:1083–1086. doi: 10.1016/s0960-9822(98)70446-4. [DOI] [PubMed] [Google Scholar]
- 13.Watkins DN, Berman DM, Baylin SB. Hedgehog signaling: progenitor phenotype in small-cell lung cancer. Cell Cycle. 2003;2:196–198. [PubMed] [Google Scholar]
- 14.Park KS, Martelotto LG, Peifer M, Sos ML, Karnezis AN, Mahjoub MR, et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat Med. 2011;17:1504–1508. doi: 10.1038/nm.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carney DN, Cuttitta F, Moody TW, Minna JD. Selective stimulation of small cell lung cancer clonal growth by bombesin and gastrin-releasing peptide. Cancer Res. 1987;47:821–825. [PubMed] [Google Scholar]
- 16.Corjay MH, Dobrzanski DJ, Way JM, Viallet J, Shapira H, Worland P, et al. Two distinct bombesin receptor subtypes are expressed and functional in human lung carcinoma cells. J Biol Chem. 1991;266:18771–18779. [PubMed] [Google Scholar]
- 17.Alexander RW, Upp JR, Poston GJ, Gupta V, Townsend CM, Thompson JC. Effects of bombesin on growth of human small cell lung carcinoma in vivo. Cancer Res. 1988;48:1439–1441. [PubMed] [Google Scholar]
- 18.Toi-Scott M, Jones CL, Kane MA. Clinical correlates of bombesin-like peptide receptor subtype expression in human lung cancer cells. Lung Cancer. 1996;15:341–354. doi: 10.1016/0169-5002(95)00597-8. [DOI] [PubMed] [Google Scholar]
- 19.Thomas F, Arvelo F, Antoine E, Jacrot M, Poupon MF. Antitumoral activity of bombesin analogues on small cell lung cancer xenografts: relationship with bombesin receptor expression. Cancer Res. 1992;52:4872–4877. [PubMed] [Google Scholar]
- 20.Carney DN, Gazdar AF, Bepler G, Guccion JG, Marangos PJ, Moody TW, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985;45:2913–2923. [PubMed] [Google Scholar]
- 21.Douglas AE, Heim JA, Shen F, Almada LL, Riobo NA, Fernandez-Zapico ME, et al. The alpha subunit of the G protein G13 regulates activity of one or more Gli transcription factors independently of smoothened. J Biol Chem. 2011;286:30714–30722. doi: 10.1074/jbc.M111.219279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Offermanns S, Simon MI. Genetic analysis of mammalian G-protein signalling. Oncogene. 1998;17:1375–1381. doi: 10.1038/sj.onc.1202173. [DOI] [PubMed] [Google Scholar]
- 23.Chikumi H, Vazquez-Prado J, Servitja JM, Miyazaki H, Gutkind JS. Potent activation of RhoA by Galpha q and Gq-coupled receptors. J Biol Chem. 2002;277:27130–27134. doi: 10.1074/jbc.M204715200. [DOI] [PubMed] [Google Scholar]
- 24.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 25.Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 26.Ye RD. Regulation of nuclear factor kappaB activation by G-protein-coupled receptors. J Leukoc Biol. 2001;70:839–848. [PubMed] [Google Scholar]
- 27.Perona R, Montaner S, Saniger L, Sanchez-Perez I, Bravo R, Lacal JC. Activation of the nuclear factor-kappaB by Rho, CDC42, and Rac-1 proteins. Genes & development. 1997;11:463–475. doi: 10.1101/gad.11.4.463. [DOI] [PubMed] [Google Scholar]
- 28.Kasperczyk H, Baumann B, Debatin KM, Fulda S. Characterization of sonic hedgehog as a novel NF-kappaB target gene that promotes NF-kappaB-mediated apoptosis resistance and tumor growth in vivo. Faseb J. 2009;23:21–33. doi: 10.1096/fj.08-111096. [DOI] [PubMed] [Google Scholar]
- 29.Kim HJ, Hawke N, Baldwin AS. NF-kappaB and IKK as therapeutic targets in cancer. Cell Death Differ. 2006;13:738–747. doi: 10.1038/sj.cdd.4401877. [DOI] [PubMed] [Google Scholar]
- 30.Brennan J, O'Connor T, Makuch RW, Simmons AM, Russell E, Linnoila RI, et al. myc family DNA amplification in 107 tumors and tumor cell lines from patients with small cell lung cancer treated with different combination chemotherapy regimens. Cancer Res. 1991;51:1708–1712. [PubMed] [Google Scholar]
- 31.Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 32.Litingtung Y, Lei L, Westphal H, Chiang C. Sonic hedgehog is essential to foregut development. Nat Genet. 1998;20:58–61. doi: 10.1038/1717. [DOI] [PubMed] [Google Scholar]
- 33.Cuttitta F, Carney DN, Mulshine J, Moody TW, Fedorko J, Fischler A, et al. Bombesin-like peptides can function as autocrine growth factors in human small-cell lung cancer. Nature. 1985;316:823–826. doi: 10.1038/316823a0. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J, Chen J, Mokotoff M, Ball ED. Targeting gastrin-releasing peptide receptors for cancer treatment. Anticancer Drugs. 2004;15:921–927. doi: 10.1097/00001813-200411000-00001. [DOI] [PubMed] [Google Scholar]
- 35.Kiaris H, Schally AV, Sun B, Armatis P, Groot K. Inhibition of growth of human malignant glioblastoma in nude mice by antagonists of bombesin/gastrin-releasing peptide. Oncogene. 1999;18:7168–7173. doi: 10.1038/sj.onc.1203213. [DOI] [PubMed] [Google Scholar]
- 36.Saurin JC, Fallavier M, Sordat B, Gevrey JC, Chayvialle JA, Abello J. Bombesin stimulates invasion and migration of Isreco1 colon carcinoma cells in a Rho- dependent manner. Cancer Res. 2002;62:4829–4835. [PubMed] [Google Scholar]
- 37.Schally AV, Szepeshazi K, Nagy A, Comaru-Schally AM, Halmos G. New approaches to therapy of cancers of the stomach, colon and pancreas based on peptide analogs. Cell Mol Life Sci. 2004;61:1042–1068. doi: 10.1007/s00018-004-3434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lango MN, Dyer KF, Lui VW, Gooding WE, Gubish C, Siegfried JM, et al. Gastrin-releasing peptide receptor-mediated autocrine growth in squamous cell carcinoma of the head and neck. J Natl Cancer Inst. 2002;94:375–383. doi: 10.1093/jnci/94.5.375. [DOI] [PubMed] [Google Scholar]
- 39.Kim S, Hu W, Kelly DR, Hellmich MR, Evers BM, Chung DH. Gastrin-releasing peptide is a growth factor for human neuroblastomas. Ann Surg. 2002;235:621–629. doi: 10.1097/00000658-200205000-00003. discussion 629-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dierks C, Grbic J, Zirlik K, Beigi R, Englund NP, Guo GR, et al. Essential role of stromally induced hedgehog signaling in B-cell malignancies. Nat Med. 2007;13:944–951. doi: 10.1038/nm1614. [DOI] [PubMed] [Google Scholar]
- 41.Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455:406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 42.Yang L, Xie G, Fan Q, Xie J. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications. Oncogene. 2010;29:469–481. doi: 10.1038/onc.2009.392. [DOI] [PubMed] [Google Scholar]
- 43.Bruzzese F, Rocco M, Castelli S, Di Gennaro E, Desideri A, Budillon A. Synergistic antitumor effect between vorinostat and topotecan in small cell lung cancer cells is mediated by generation of reactive oxygen species and DNA damage-induced apoptosis. Mol Cancer Ther. 2009;8:3075–3087. doi: 10.1158/1535-7163.MCT-09-0254. [DOI] [PubMed] [Google Scholar]
- 44.Marinissen MJ, Chiariello M, Pallante M, Gutkind JS. A network of mitogen-activated protein kinases links G protein-coupled receptors to the c-jun promoter: a role for c-Jun NH2-terminal kinase, p38s, and extracellular signal-regulated kinase 5. Mol Cell Biol. 1999;19:4289–4301. doi: 10.1128/mcb.19.6.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pacifico F, Mauro C, Barone C, Crescenzi E, Mellone S, Monaco M, et al. Oncogenic and anti-apoptotic activity of NF-kappa B in human thyroid carcinomas. J Biol Chem. 2004;279:54610–54619. doi: 10.1074/jbc.M403492200. [DOI] [PubMed] [Google Scholar]
- 46.Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC. World Health Organization classification of tumours. Pathology and Genetics of tumours of the lung, pleura, thymys, and heart. IARC Press; Lyon, France: 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.