Abstract
Nanoparticles are promising platforms for the diagnosis and treatment of cancer. Diverse classes and shapes of materials have been investigated to establish design principles that achieve the effective partitioning of medical cargos between tumors and healthy tissues. Molecular targeting strategies combined with specific nanoparticle shapes confer tissue-specificity on the carriers, allowing the cell-specific delivery of cargos. We recently developed a filamentous platform technology in which the plant virus Potato virus X (PVX) was used as a scaffold. These particles are flexible 515 × 13 nm filaments that encourage passive tumor homing. Here we sought to advance the PVX platform by including a molecular targeting strategy based on cyclic RGD peptides, which specifically bind to integrins upregulated on tumor cells, neovasculature, and metastatic sites. Although the RGD-targeted filaments outperformed the PEGylated stealth filaments in vitro, enhanced tumor cell targeting did not translate into improved tumor homing in vivo in mouse tumor models. The RGD-PVX and PEG-PVX filaments showed contrasting biodistribution profiles. Both formulations were cleared by the liver and spleen, but only the stealth filaments accumulated in tumors, whereas the RGD-targeted filaments were sequestered in the lungs. These results provide insight into the design principles for virus-based nanoparticles that promote the delivery of medical cargos to the appropriate cell types.
Keywords: Nanoparticle shape, Cancer, Tumor targeting, Biodistribution, Integrins
Introduction
Cancer is the leading cause of death in the developed world and is also prevalent in the developing world. To improve survival and the quality of life, more efficacious therapies are required, especially to treat patients with tumors that resist conventional chemotherapy. Nanoparticles equipped with targeting ligands and medical cargo are promising approaches for cancer diagnosis and therapy because nanoparticle-based formulations enable the tissue-specific delivery of contrast agents for molecular imaging and/or toxic payloads for therapeutic intervention.17
There is mounting evidence that elongated, filamentous nanomaterials are advantageous for drug delivery. Non-spherical materials show increased margination toward the vessel wall, which improves the efficiency of tumor homing.6–8,10,14,15,18 Elongated materials present ligands more effectively to target cells and the flat vessel wall than their spherical counterparts.12,18,38 Furthermore, elongated materials are more likely to resist immune detection and macrophage uptake, thus contributing to synergistic target enhancement.3,39 Nevertheless, most platform technologies currently under development are spherical or elongated low-aspect-ratio materials. Filomicelles mimic the shape of filamentous viruses and are therefore exceptions, but these are micron-sized and therefore technically they are not nanomaterials.14 The synthesis of high-aspect-ratio materials remains challenging because it is impossible to avoid polydispersity and the combination of synthetic chemistry and nanotechnology cannot yet mimic what nature has achieved, i.e., self-assembly and programmability at the atomic level. Therefore, we have developed a bioinspired platform technology based on filamentous plant viruses, specifically Potato virus X (PVX), which is shown in Fig. 1.
Figure 1.
(a) Nicotiana benthamiana plants infected with Potato virus X (PVX) and (b) a negatively-stained transmission electron micrograph of PVX.
The PVX capsid is a flexible 515 × 13 nm protein-based filament that consists of 1270 identical copies of a coat protein unit suitable for genetic and chemical modification.19 The flexible nature of the material is advantageous because flexible nanomaterials can pass through restrictions and penetrate tissue more easily.20 We have shown that PVX-based filaments achieve more efficient partitioning between tumor and liver tissue compared to spherical nanoparticles.31 The shape-mediated enhanced tumor homing is reproducible in a variety of models, including human tumor xenografts of breast cancer, fibrosarcoma, squamous sarcoma and brain and colon cancers.31,32
A combination of prolonged circulation and the enhanced permeability and retention (EPR) effect are thought to contribute to passive tumor homing and accumulation.17 This phenomenon has been observed in many nanoparticle systems, including clinical formulations of Doxil® (a liposomal formulation of doxorubicin) and Abraxane® (an albumin nanoparticle formulation carrying paclitaxel). Both formulations increase the efficacy of their payloads, reflecting their beneficial physiochemical properties and the pathophysiological characteristics of the target tissue. Nevertheless, there is some ambiguity and controversy surrounding the EPR-mediated tumor homing of nanoparticles, especially in clinical applications.17 Therefore, nanoparticles have been modified to include targeting ligands that interact specifically with targets such as growth factor receptors,29,40 tumor matrix proteins,24,37 signatures of the tumor-associated endothelium4,36 or combinations thereof.
The overexpression of αvβ3, αvβ5 and α5β1 integrins on cancer cells, neovasculature and metastatic sites has been confirmed in many human malignancies, and changes in integrin expression and function are associated with disease progression.26,27,36 Integrins are validated targets for RGD peptide ligands and several nanoparticle systems carrying these peptides are under development. Here we describe the development and testing of integrin-targeted PVX nanofilaments modified with cyclic RGD peptides. The specificity of RGD-targeted PVX was compared to the PEG-PVX stealth formulation using a combination of in vitro and in vivo testing.
Materials and Methods
PVX Propagation and Synthesis
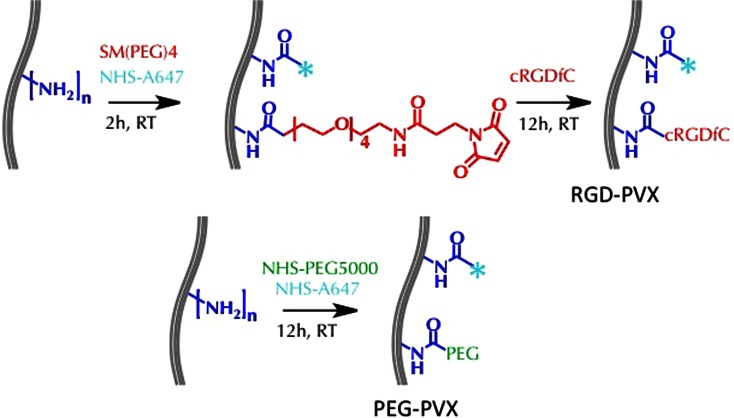
PVX was produced through farming in N. benthamiana plants using previously established protocols19 and extracted at yields of 20 mg of pure PVX from 100 g of infected leaf material. RGD-PVX particles were synthesized using a two-step protocol. First, PVX was covalently modified with Alexa Fluor 647 (A647) succinimdyl ester (NHS-A647, Life Technologies) and the bifunctional linker succinimidyl-[(N-maleimidopropionamido)-tetraethyleneglycol] ester [SM(PEG)4, Pierce] targeting surface-exposed lysine residues on the viral coat proteins (PVX consists of 1270 identical copies of a coat protein, each of which has an exposed lysine side chain). Subsequently, cyclic RGD peptide with cysteine functionality (cyclo(Arg-Gly-Asp-d-Phe-Cys (cRGDfC), Peptides International) was covalently attached to PVX through Michael addition targeting the terminal maleimide group of the bifunctional linker. Specifically, PVX particles at a concentration of 2 mg mL−1 in 0.1 M potassium phosphate buffer (pH 7.0) were incubated for 2 h at room temperature with 5000 molar excess of NHS-A647 and 10,000 molar excess of SM(PEG)4 in the presence of DMSO [at a final concentration of 10% (v/v)]. PVX formulations were then purified using 10-kDa cut-off centrifugal devices (Millipore) to separate unreacted dyes and linker molecules. Then, PVX rods were reacted overnight with 10,000 molar excess of cRGDfC in 0.1 M potassium phosphate buffer (pH 7.0) and purified as described above.
PEG-PVX stealth filaments were synthesized by reacting PVX (at 2 mg mL−1 in 0.1 M potassium phosphate buffer pH 7.0) with 5000 molar excesses of NHS-PEG5000 (NanoCS) and NHS-A647 (Life Technologies) overnight using a one-pot synthesis protocol. A647-labeled PEG-PVX formulations were then purified using centrifugal filters as described above. All formulations were resuspended in 0.1 M potassium phosphate buffer (pH 7.0) and stored at 4 °C until further processing.
UV/Visible Spectroscopy
UV/visible spectroscopy was performed using NanoDrop instrument to quantify labeling efficiency with fluorophores. The number of A647 labels per PVX was calculated based on Beer-Lambert law using the PVX and A647-specific extinction coefficients: εPVX = 2.97 mL mg−1 cm−1 at 260 nm, εA647 = 270,000 M−1 cm−1 at 650 nm.
Denaturing Gel Electrophoresis
SDS gel electrophoresis was carried out to analyze conjugation of RGD peptide and PEG chains to individual coat proteins. 10 μg denatured protein samples were analyzed on 4–12% NuPage gels (Life Technologies) in 1× MOPS SDS running buffer (Life Technologies). Protein bands were visualized under white light after staining the gels with SimplyBlue SafeStain (Life Technologies). Protein bands were analyzed using ImageJ software (http://imagej.nih.gov).
TEM
Diluted samples of PVX, RGD-PVX and PEG-PVX particles (20 μL, 0.1 mg mL−1) were negatively stained with of 2% (w/v) uranyl acetate for 2 min on a carbon-coated copper grid. Samples were analyzed using a Zeiss Libra 200FE transmission electron microscope operated at 200 kV.
Tumor Homing and Imaging Using Mouse Xenograft Models
All animal studies were carried out using IACUC-approved procedures. Human colon cancer xenografts were developed using HT-29 cells (ATCC) maintained in McCoy’s medium (Life Technologies) supplemented with 10% (v/v) fetal bovine serum (FBS), 1% (w/v) penicillin/streptomycin and 1% (w/v) glutamine (all from Life Technologies). Six-week-old male NCr-nu/nu nude mice maintained on an alfalfa-free diet (Teklad 2018S, Harlan Laboratories) were subcutaneously injected in the right flank with 2.5 × 106 cells in 50 μL medium mixed with an equal volume of matrigel (BD Matrigel, BD Biosciences, USA) using a microliter Hamilton 22 gauge syringe. One tumor was induced in each mouse. The animals were monitored daily and tumor homing studies commenced when tumors reached an average volume of 20–30 mm3 (typically within 12 days after the injection of HT-29 cells). Animals were assigned randomly into groups with n = 3 mice per formulation. PBS, PEG-PVX and RGD-PVX particles were administered intravenously via tail vein injection. At specific time points post-administration, mice were euthanized and major tissues (liver, lungs, kidney, heart, spleen, brain and tumors) were harvested and imaged in the Maestro™ fluorescence imager using combinations of yellow excitation/emission filters for A647 (excitation filter: 575–605 nm; and emission filter: 645 nm longpass). Region of interest (ROI) analysis was performed using the Maestro™ software to determine the fluorescence intensity from the respective tissues as compared to tissues from PBS injected mice. After imaging, tissues were frozen in OCT media for cryo-sectioning.
Immunofluorescence
Intratumoral localization of RGD-PVX and PEG-PVX particles with respect to integrins (αv), macrophages (CD68) and endothelium (CD31) was determined using immunofluorescence analysis of 10 μm thick tumors sections prepared from frozen tissues embedded in OCT medium (Fisher) using a Leica CM1850 cryostat. Tumor sections were fixed in 95% (v/v) ethanol for 20 min on ice and rinsed with cold PBS. Tumor sections were permeablized using phosphate buffered saline containing 0.02% (v/v) Tween-20 (PBST) twice for 10 min each and then blocked with 10% (v/v) goat serum (GS) in PBST for 60 min. Sections were then stained using a rabbit anti-integrin αv antibody (1:500, Millipore) or rat anti-mouse CD68 (1:250) antibody (Biolegend) with 1% (v/v) GS in PBST for 60 min at room temperature. After washing thoroughly with PBST, tumor sections were incubated with A488-conjugated goat anti-rabbit secondary antibody (1:2000, Life Technologies) and A555-conjugated goat anti-rat antibody (1:1000, Life Technologies) with 1% (v/v) GS in PBST for 60 min. Slides were then washed three-times with PBST and mounted with Fluoroshield with DAPI (Sigma Aldrich) and stored at −20 °C until imaged. Similarly, lung cryo-sections were stained to determine PVX localization within the lung tissues with respect to integrin and macrophages.
For CD31 staining, tumor sections were permeabilized using 0.2% (v/v) Triton-X-100 (EMD Chemicals) in PBS for 2 min. Slides were blocked using 10% GS (v/v) in PBS and then incubated with A488-conjugated rabbit anti-mouse CD31 antibody (Biolegend) for 60 min, washed with PBS and then mounted as above and stored at −20 °C until imaged. Analysis of sections was performed on an Olympus Fluoview FV1000 confocal microscope. Images were processed using ImageJ software (http://imagej.nih.gov).
Cell Uptake Using Flow Cytometry
HT-29 and RAW264.7 cells were grown to confluency in McCoy’s 5A media and DMEM media, respectively supplemented with fetal bovine serum (10% v/v) and penicillin/streptomycin (1% v/v) (all reagents from Life Technologies) at 37 °C and 5% CO2. Cells were washed three times with PBS and collected using enzyme-free Hank’s based Cell Dissociation Buffer (Fisher). Cells were then added to 96-well v-bottom plates (200,000 cells/200 μL/well) and incubated with 1 μg of RGD-PVX or PEG-PVX particles/well, in triplicate for 3 h at 37 °C and 5% CO2. Following incubation, cells were washed twice in FACS buffer [1 mM EDTA, 25 mM HEPES, 1% (v/v) FBS in PBS, pH 7.0] and fixed in 2% (v/v) paraformaldehyde (Electron Microscopy Sciences) in FACS buffer for 10 min at room temperature. Cells were washed twice after fixation, re-suspended in 400 μL FACS buffer and stored at 4 °C until analysis. Cells were analyzed using a BD LSRII Flow Cytometer and 10,000-gated events were recorded. Data were analyzed using FlowJo 8.6.3 software.
For competitive binding assays, HT-29 cells were first incubated with 1,000-fold excess of free RGD peptide for 30 min followed by incubation with RGD-PVX particles for 2 h. For competitive binding assays with RAW264.7 cells a series of conditions was tested: RAW264.7 cells were pre-incubated with 10, 100, 1000 and 10,000× excess of soluble RGD peptide for 30 min followed by addition of RGD-PVX filaments, which were allowed to interact with the macrophage cell line for 2 h. Cells were then collected, prepared and analyzed as described above.
Confocal Microscopy for Cellular Uptake
HT-29 cells and RAW264.7 cells were cultured as described above. For uptake studies, 25,000 HT-29 or RAW264.7 cells per well were cultured onto glass coverslips in a 24-well suspension culture plate for 24 h. After washing and replacing with fresh media, 1 μg of RGD-PVX or PEG-PVX particles/well were added into the culture media and incubated with cells for 3 h. Post-incubation, cells were washed three times with sterile saline and fixed for 5 min at room temperature with DPBS containing 4% (v/v) paraformaldehyde and 0.3% (v/v) glutaraldehyde. Cells were then washed three times with DPBS. RAW264.7 cell coverslips were then mounted with Fluoroshield with DAPI (Sigma Aldrich) for nuclear staining and sealed using nail polish.
Post fixation, HT-29 cells were permeablized with 0.2% (v/v) Triton-X 100 (Fisher Bioscience) for 2 min followed by blocking with 10% (v/v) GS for 60 min. Cells were then stained with mouse anti-human LAMP-1 antibody (1:250 dilutions) (Biolegend) with 1% (v/v) GS in DPBS for 60 min, washed three times and then counter stained with A488-tagged goat anti-mouse antibody (at 1:1000 dilution, Life Technologies) with 1% (v/v) GS in DPBS for 60 min at room temperature. Coverslips were washed three times with DPBS, and mounted with Fluoroshield with DAPI (Sigma Aldrich) resulting in nuclear staining and sealed using nail polish. Confocal images were captured on Olympus FluoViewTM FV1000 LSCM and data processed using ImageJ 1.44o software (http://imagej.nih.gov/ij).
Results and Discussion
Synthesis of Targeted RGD-PVX and Stealth PEG-PVX Filaments
PVX was produced by molecular farming in N. benthamiana plants with yields of 20 mg pure PVX per 100 g infected leaf material. The RGD-PVX derivative was generated by conjugating the cyclic RGDfC peptide (with a terminal cysteine residue) to solvent exposed lysine residues on PVX using a bifunctional PEG linker with a 24.6 Å spacer arm [SM(PEG)4]. The PEGylated stealth formulation was prepared using linear PEG with a molecular weight of 5 kDa because PVX modified with this PEG had previously achieved tumor homing in various animal models.31,32 Filament tracking in cells and tissues was achieved by conjugating the near-infrared dye Alexa Fluor 647 succinimidyl ester (NHS-A647) using the reaction scheme outlined in Fig. 2.
Figure 2.
Modification of PVX with PEG and RGD ligands. N-hydroxysuccinimide (NHS) chemistry was used to conjugate A647 and SM(PEG)4 bifunctional linkers to PVX lysine residues. The cRGDfC peptide (with terminal cysteine) was then conjugated using the maleimide handle to produce RGD-PVX. Similarly, A647 and PEG (5-kDa MW) were conjugated to PVX by NHS chemistry to yield PEG-PVX filaments.
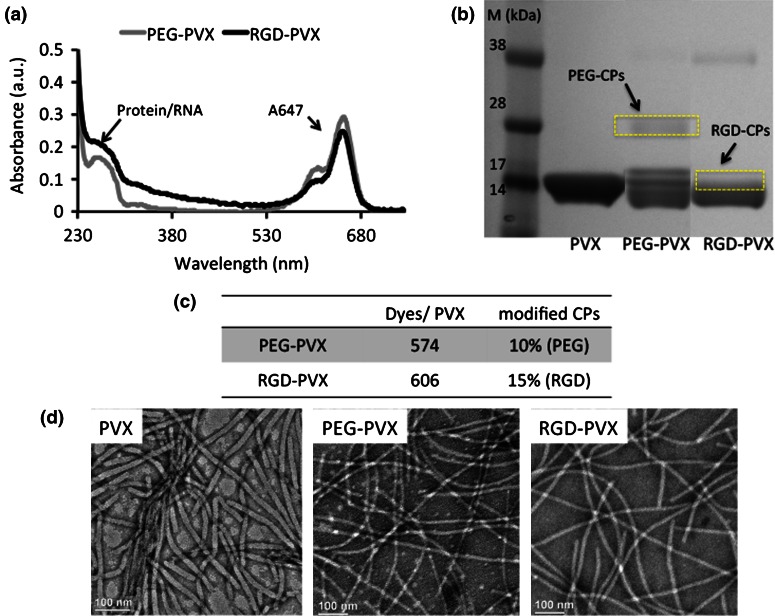
The PVX-based formulations were characterized by UV/Vis spectroscopy and denaturing gel electrophoresis (SDS-PAGE) to confirm the degree of modification. Based on the Beer-Lambert law and the extinction coefficients of the fluorophore and PVX, approximately 574 dye molecules were conjugated per PEG-PVX and approximately 606 per RGD-PVX (Figs. 3a, 3c). SDS-PAGE confirmed the covalent attachment of the RGD peptide and PEG chains (Fig. 3b). The PVX coat protein has a molecular weight of 19 kDa, so higher molecular weights indicate the addition of RGD and PEG. ImageJ software was used to determine the degree of labeling showing that ~10% of the PVX coat proteins in the PEG-PVX formulation were modified with PEG5000, and ~15% of the PVX coat proteins were modified with RGD in the RGD-PVX formulation. The RGD-PVX particles therefore displayed ~200 RGD peptides by way of a 400-Da intervening PEG spacer, whereas the PEG-PVX particles displayed ~130 PEG chains with a molecular weight of 5 kDa.
Figure 3.
Characterization of PEG-PVX and RGD-PVX particles: (a) UV/Vis spectroscopy was used to determine number of dye molecules attached and to determine the concentration of PEG-PVX and RGD-PVX particles. (b) SDS-PAGE was used to confirm the covalent conjugation of PEG and RGD peptides to the PVX coat proteins (CPs). ImageJ software was used to determine the percentage of modified coat proteins. (c) Quantification of dye molecules and PEG/RGD ligands per PVX filament based on UV/Vis and SDS-PAGE. (d) Transmission electron microscopy was used to confirm the structural stability of PVX particles after modification (left to right: PVX, PEG-PVX and RGD-PVX). Scale bar = 100 nm.
Biodistribution and Tumor Homing Properties
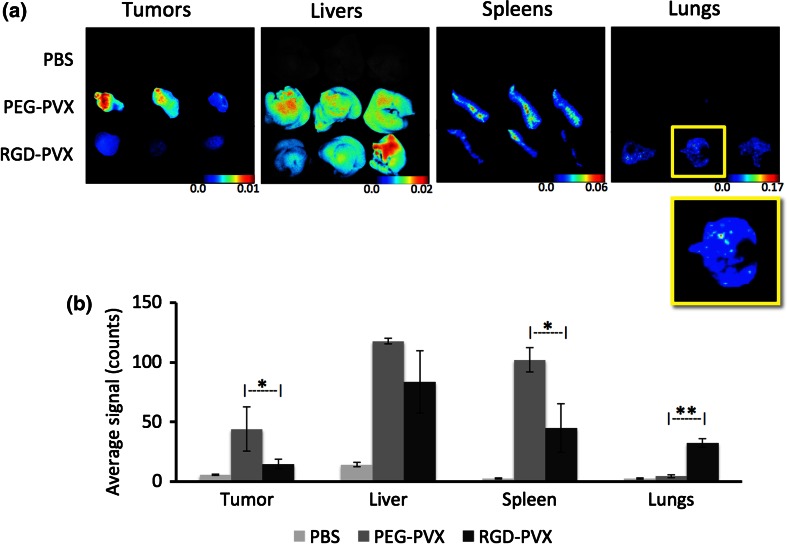
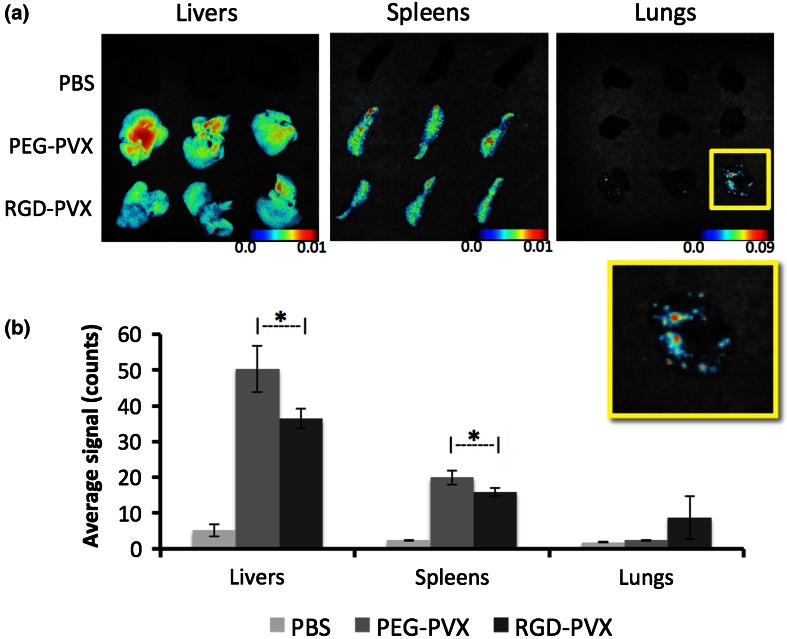
The biodistribution and tumor homing of the PVX formulations were studied using NCr-nu/nu nude mice with subcutaneous HT-29 xenografts. The nanoparticle formulations were administered intravenously as a single dose of 10 mg protein/kg body weight. At 24 h post-administration, the mice were sacrificed and the tumors, livers, spleens and lungs were removed for ex vivo quantitative tissue analysis using the Maestro™ fluorescence imaging system (Fig. 4).
Figure 4.
Comparative biodistribution of PEG-PVX and RGD-PVX particles injected intravenously into NCR-nu/nu mice with subcutaneous HT-29 tumor xenografts. Ex vivo Maestro analysis was carried out using dissected tissues 24 h post-administration. (a) Maestro images (yellow boxes show enlarged view of the lungs from animals treated with RGD-PVX, showing hot spots for PVX sequestration). (b) Quantitative analysis.
Both formulations were cleared by the liver and spleen as expected because these are the organs of the mononuclear phagocytic system (MPS), which has been shown to facilitate the clearance of several virus-based nanomaterials, including those based on Cowpea mosaic virus (CPMV), Tobacco mosaic virus (TMV) and bacteriophages Qβ and M13.5,13,21,28,34 The overall biodistribution of the PEG-PVX stealth filaments was consistent with our previous observations.31,32 Although the majority of the injected PEG-PVX particles were cleared by the liver and spleen, a substantial amount also accumulated in the tumor tissue. In stark contrast, the RGD-targeted PVX formulation showed negligible accumulation in the tumor but significantly enhanced uptake in the lungs, where no stealth filaments were found (Fig. 4).
Our central hypothesis was that the RGD-targeted formulation would show similar or better levels of tumor homing compared to the stealth particles and would be more efficiently localized in the tumor because targeting would promote uptake by integrin-positive tumor cells, as has been observed with other nanoparticle systems.30 However, our experiments showed the opposite outcome, with the stealth particles but not the targeted particles accumulating in the tumor. The ability of the RGD-targeted particles to bind selectively to integrin was confirmed in vitro (see below), therefore we were able to rule out the lack of target specificity and recognition as an explanation for these unexpected results.
We considered the possibility that the unanticipated biodistribution of the particles might be explained by differences in their pharmacokinetic profiles. As stated above, PEG-PVX was decorated with ~130 PEG molecules, each with a molecular weight of 5 kDa, whereas RGD-PEG was decorated with ~200 RGD molecules, each with a 400-Da intervening PEG spacer. Based on the available surface area of PVX (APVX = 21,033 nm2) and the Flory dimension of 5-kDa PEG (RF = 5.9 nm),9 we estimated that the PEG chains are presented in a mushroom conformation with nearly 70% surface area coverage. In contrast, the Flory dimension of the shorter PEG spacer (RF = 0.8 nm) achieves less than 10% surface coverage. The more effective stealth effect of PEG-PVX is likely to protect the particles from serum protein adsorption, thus evading the immune system more effectively and resulting in more efficient passive tumor accumulation. This is consistent with reports describing the prolonged circulation of PEGylated liposomal doxorubicin formulations, which promotes EPR-mediated tumor homing.22,23
We attempted to increase the molecular weight of the intervening PEG spacer in the RGD-PVX formulation to match the stealth formulation, but integrin recognition of RGD-modified PVX with an intervening PEG 5-kDa spacer was diminished (data not shown). The longer PEG chain in the mushroom conformation may block the interaction between the RGD ligand and integrin thus preventing molecular recognition.
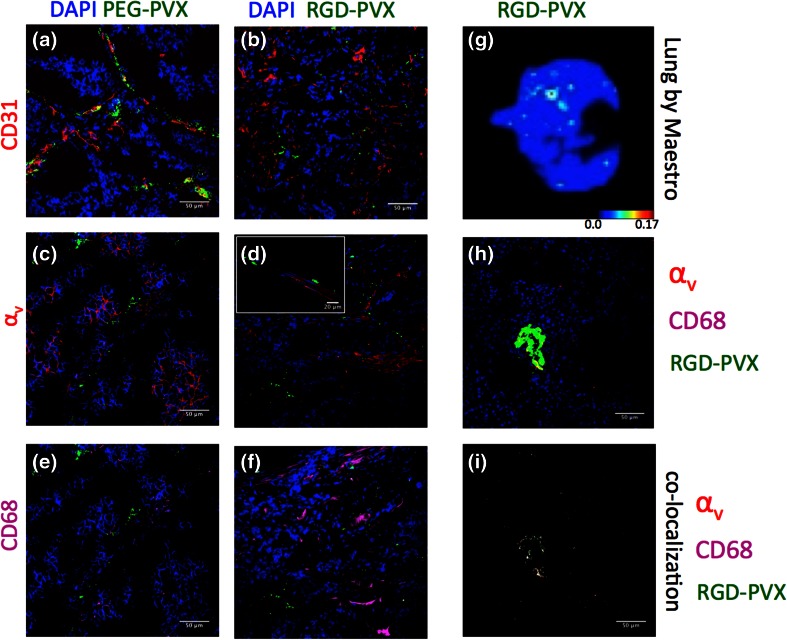
To gain further insight into the unexpected biodistribution profiles discussed above, we prepared tumor and lung sections from tumor xenograft mice and stained the sections using antibodies specific for the endothelium (CD31), αv integrin and macrophages (CD68) as shown in Fig. 5. Histological analysis showed that both formulations were generally located near the tumor endothelium, but there was no significant co-localization with the endothelial marker CD31 (Figs. 5a, 5b). The tumor sections stained positive for αv integrins as expected, since both HT-29 cells and the tumor endothelium overexpress αv integrins,11 and the integrin-specific antibody we used cross-reacts with human αv integrins (present on the cancer cells) and mouse αv integrins (present on the endothelium). Although fewer RGD-PVX particles were found in the tumor tissue, they were associated with αv integrin-positive sites within the tumor microenvironment (Fig. 5d). On the other hand, PEG-PVX stealth formulation did not colocalize with αv integrins, which is in line with our expectations (Fig. 5c). Neither formulation appeared to colocalize with CD68-positive macrophages (Figs. 5e, 5f). The immunofluorescence data therefore confirmed that the stealth particles accumulated more efficiently than the targeted particles in the tumor. While both formulations were localized near the endothelium, the RGD-targeted particles were colocalized with αv integrin-positive areas, indicating molecular specificity despite the diminished overall tumor accumulation.
Figure 5.
Immunofluorescence analysis of tumor and lung sections. The tumor sections were stained with antibodies specific for the endothelial marker CD31 (red, a + b), integrin α v [red, c + d (inset: higher magnification)], the tumor macrophage marker CD68 (pink, e + f) and DAPI (blue) to determine the localization of PEG-PVX and RGD-PVX within the tumor. (g–i) Whole lung (g) and lung sections (h, i) from mice treated with RGD-PVX were stained for α v integrin (red) and CD68+ macrophages (pink). Co-localization analysis was used to highlight the hotspots of RGD-PVX accumulation with integrins and CD68 macrophages (i). Scale bars = 50 μm.
Next, we investigated the histology of the lungs (Figs. 5g–5i). Only RGD-PVX was sequestered in the lungs, so tissue sections were prepared only from animals treated with the RGD-PVX formulation. Lung tissue sections were stained for αv integrins and macrophages (using the CD68 marker). RGD-PVX was accumulated in hotspots in the lungs, and these areas were positive for both αv integrins and CD68 (Figs. 5h, 5i).
RGD-tagged nanoparticles have been reported to target lung metastasis 16,25 but the mouse model used in our experiments does not present metastatic disease in the lungs. This was confirmed by staining for human cells within the lungs using an anti-human vimentin antibody, and no human cells were found (data not shown). To ensure that the biodistribution profile was not specific to our tumor mouse model, we repeated the experiments using healthy Balb/c mice (Fig. 6). The results were similar to those obtained in the tumor xenograft models, i.e., both types of particles were cleared by the liver and spleen whereas only the RGD-PVX particles were sequestered in the lungs.
Figure 6.
Biodistribution of PEG-PVX and RGD-PVX particles in healthy Balb/c mice. Tissues were collected for ex vivo Maestro™ fluorescence analysis 24 h after the administration of PEG-PVX and RGD-PVX. (a) Maestro images (yellow boxes show enlarged view of the lungs from animals treated with RGD-PVX, showing hot spots for PVX sequestration). (b) Quantitative analysis.
Our data indicate that targeted RGD-PVX filaments accumulate in the lungs in healthy animals as well as tumor xenograft models. The RGD-PVX filaments are sequestered in hotspots and are colocalized with αv integrins and the macrophage marker CD68. Integrins play a role in macrophage-mediated inflammatory responses.2 Alveolar macrophages and the bronchiolar epithelia of normal lungs express integrins, but integrin expression is more extensive during inflammation.33 These background levels of integrin within the lung microenvironment may explain the sequestration of RGD-targeted nanoparticles in the lungs. Carbon nanotubes can also accumulate in the lungs41,42 but little information is available about other nanoparticles accumulating in the lungs following intravenous injection. A recent study showed prolonged accumulation of RGD-nanoparticles in the lungs following systemic administration.1
Interactions with Cancer Cells and Macrophages
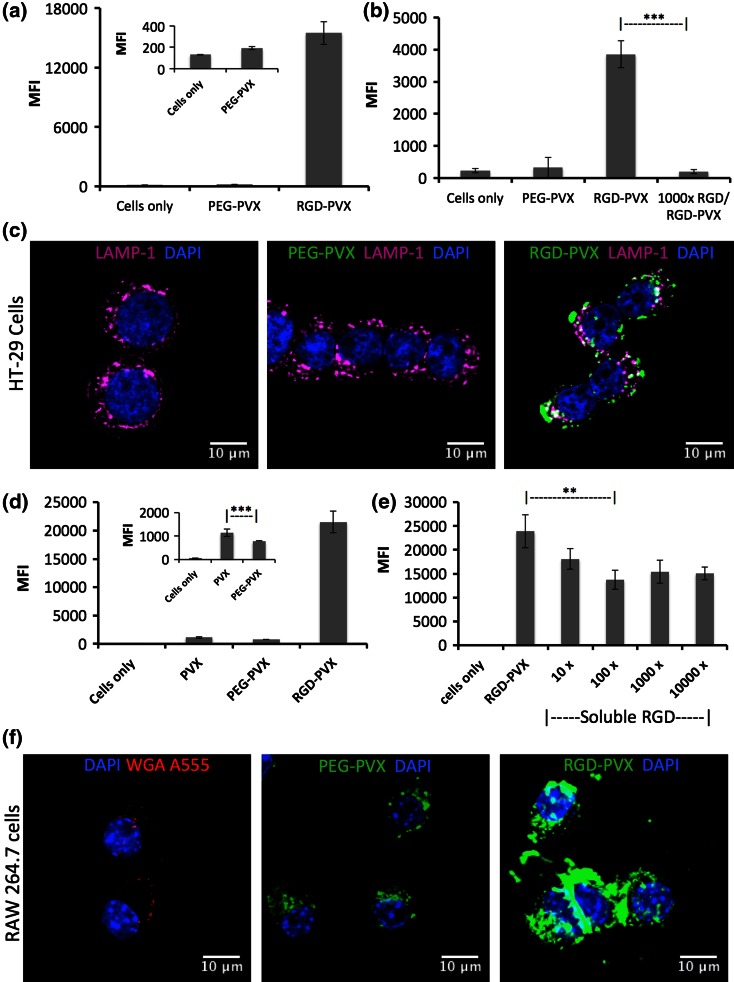
To gain further insight into the behavior of the PVX filaments in vivo, we conducted a series of in vitro experiments using HT-29 cells (a colon carcinoma cell line) and RAW264.7 cells (a mouse macrophage cell line). RGD-targeted and stealth PVX particles were incubated with each of the cell lines and then analyzed by flow cytometry and confocal microscopy. PEG shielding minimizes the non-specific interaction between PVX particles and cells, so few particles were taken up by either cell line (Figs. 7a, 7d). In contrast, the RGD-targeted particles showed significant interactions with both the HT-29 and RAW264.7 cell lines (Figs. 7a, 7d) and were readily taken up into both cell types (Figs. 7c, 7f). Competition binding assays were carried out to investigate the molecular specificity of the RGD-targeted formulation. We found that a molar excess of 1000 free RGD peptides was able to prevent the molecular interaction between RGD-PVX particles and HT-29 cells, confirming that the RGD-PVX particles target integrins expressed on the surface of these cells (Fig. 7b).
Figure 7.
The analysis of PEG-PVX and RGD-PVX interactions with and uptake into cells using flow cytometry and confocal microscopy. (a + d) Flow cytometry was used to determine the interactions between PVX particles and HT-29 cancer cells (a, b) or RAW264.7 macrophages (d, e). The mean fluorescence intensity (MFI) is plotted and error bars indicate the standard deviation (n = 3). (b + e) Competition binding assays using RGD-PVX and free RGD peptides. (c + f) Confocal microscopy shows that RGD-PVX is taken up more efficiently than PEG-PVX by HT-29 cells and RAW264.7 macrophages. Cells were stained with DAPI (blue) to show the nucleus and with the endosomal marker LAMP-1 (pink; in HT-29 cells) or WGA (red; RAW cells). Colocalization of the particles and the endosomal marker indicates cellular uptake. Scale bars = 10 μm.
Although stealth filaments were effectively shielded from phagocytosis, RGD-targeted filaments were also taken up by RAW264.7 cells (Figs. 7d–7f). PEG shielding reduces phagocytosis, but it is clear that the shorter PEG chain on the RGD-targeted formulation is unable to prevent non-specific clearance. To establish whether the RGD ligands also promote the uptake of RGD-PVX particles by the macrophage cell line, we repeated the competition binding assays, this time pre-incubating RAW264.7 cells with free RGD peptides at various molar excesses before adding the RGD-PVX particles. The presence of soluble RGD did not completely abolish the uptake of RGD-PVX particles but there was a significant reduction compared to the untreated control, suggesting that αv integrin plays a role in this process (Fig. 7e). This is consistent with other studies showing that RGD-modified particles were taken up by monocytes. In contrast to our study, nanoparticle–monocyte targeting was a successful way to target the nanoparticles to the tumor tissue by promoting monocyte delivery.35
Conclusions
We compared RGD-PVX and PEG-PVX filaments to determine their tumor homing properties based on biodistribution and cellular interaction in vitro and in tumor-bearing and healthy mice. The stealth PVX particles showed significant tumor homing capability, facilitated by their longevity in the circulation and therefore their prolonged bioavailability, as demonstrated by their slow phagocytic clearance. In contrast, RGD-targeted PVX filaments showed negligible tumor homing capability but were instead sequestered in the lungs. The lung-specific accumulation of the RGD-modified filaments probably reflects their ability to bind integrins expressed on alveolar macrophages and the bronchiolar epithelium.33 In future studies, one might consider utilizing the propensity of RGD-PVX filaments to target inflamed areas within the lung microenvironment to block integrin-mediated interactions with metastatic cells that facilitate tumor progression, but this would require further careful optimization.
Although molecular targeting may promote cellular uptake, the targeting ligand also affects the overall biodistribution of the carrier. We found that the addition of a targeting ligand may not necessarily achieve favorable tumor partitioning. Although RGD peptides are useful for preclinical and clinical investigation, their incorporation into PVX-based stealth filaments was only successful in vitro. Alternative design strategies, e.g., incorporation of the targeting ligands into more heavily PEGylated filaments, or the targeting of alternative molecular receptors such as growth factors that are up-regulated or selectively expressed within the tumor microenvironment, may be suitable strategies to achieve molecular targeting of the filamentous carrier. Overall, these studies provide an important insight into the design principles that are required for virus-based nanoparticles to achieve selective delivery of diagnostic or therapeutic cargos to tumor cells.
Acknowledgements
This work was supported by a grant from the National Science Foundation CMMI NM 1333651 (to N.F.S) and Mt. Sinai foundation (to N.F.S.) and an NIH T32 HL105338 Cardiovascular Research training grant (to A.M.W). We thank Dr. Richard M. Twyman for editing the manuscript.
Conflict of interest
Sourabh Shukla, Nicholas A. DiFranco, Amy M. Wen, Ulrich Commandeur, Nicole F. Steinmetz declare that they have no conflicts of interest.
Ethical Standards
No human studies were carried out by the authors for this article.
All animal studies were carried out in accordance with institutional guidelines and approved by Case Western Reserve University Institutional Animal Care and Use Committee.
Footnotes
This article is part of the 2015 Young Innovators Issue.
Nicole F. Steinmetz is an Assistant Professor of Biomedical Engineering at Case Western Reserve University, School of Medicine, where she is leading a research laboratory interfacing bio-inspired and molecular engineering approaches with medical research and technology development. Dr. Steinmetz trained at The Scripps Research Institute, La Jolla, CA, John Innes Centre, Norwich, UK (PhD in Bionanotechnology), and RWTH-Aachen University in Germany (Masters in Molecular Biotechnology). Dr. Steinmetz was named a 2014 Crain’s Cleveland Business 40 under 40 honoree; in 2011, Dr. Steinmetz was named a Mt. Sinai Scholar, she is a 2009 recipient of the NIH/NIBIB Pathway to Independence Grant (K99/R00), a previous American Heart Association Post-doctoral Fellow, and former Marie Curie Early Stage Training Fellow. Dr. Steinmetz serves on the Editorial Board of Wiley Interdisciplinary Reviews (WIREs) on Nanomedicine and Nanobiotechnology and on the Advisory Editorial Board for the ACS journal Molecular Pharmaceutics. Dr. Steinmetz has co-chaired the Gordon Conference of Physical Virology (2015), as well as symposia at ACS, MRS, and FNANO. Dr. Steinmetz has authored more than 70 peer-reviewed journal articles, reviews, and book chapters; she has authored and edited books on virus-based nanotechnology. 
References
- 1.An L, Hu H, Du J, Wei J, Wang L, Yang H, Wu D, Shi H, Li F, Yang S. Paramagnetic hollow silica nanospheres for in vivo targeted ultrasound and magnetic resonance imaging. Biomaterials. 2014;35:5381–5392. doi: 10.1016/j.biomaterials.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 2.Antonov AS, Antonova GN, Munn DH, Mivechi N, Lucas R, Catravas JD, Verin AD. alphaVbeta3 integrin regulates macrophage inflammatory responses via PI3 kinase/Akt-dependent NF-kappaB activation. J. Cell Physiol. 2011;226:469–476. doi: 10.1002/jcp.22356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnida, Janát-Amsbury MM, Ray A, Peterson CM, Ghandehari H. Geometry and surface characteristics of gold nanoparticles influence their biodistribution and uptake by macrophages. Eur. J. Pharm. Biopharm. 2011;77:417–423. doi: 10.1016/j.ejpb.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks PC, Montgomery AM, Rosenfeld M, Reisfeld RA, Hu T, Klier G, Cheresh DA. Integrin alpha v beta 3 antagonists promote tumor regression by inducing apoptosis of angiogenic blood vessels. Cell. 1994;79:1157–1164. doi: 10.1016/0092-8674(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 5.Bruckman MA, Randolph LN, VanMeter A, Hern S, Shoffstall AJ, Taurog RE, Steinmetz NF. Biodistribution, pharmacokinetics, and blood compatibility of native and PEGylated tobacco mosaic virus nano-rods and -spheres in mice. Virology. 2014;449:163–173. doi: 10.1016/j.virol.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai S, Vijayan K, Cheng D, Lima EM, Discher DE. Micelles of different morphologies—advantages of worm-like filomicelles of PEO-PCL in paclitaxel delivery. Pharm. Res. 2007;24:2099–2109. doi: 10.1007/s11095-007-9335-z. [DOI] [PubMed] [Google Scholar]
- 7.Chauhan VP, Popovic Z, Chen O, Cui J, Fukumura D, Bawendi MG, Jain RK. Fluorescent nanorods and nanospheres for real-time in vivo probing of nanoparticle shape-dependent tumor penetration. Angewandte Chemie. 2011;50:11417–11420. doi: 10.1002/anie.201104449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christian DA, Cai S, Garbuzenko OB, Harada T, Zajac AL, Minko T, Discher DE. Flexible filaments for in vivo imaging and delivery: persistent circulation of filomicelles opens the dosage window for sustained tumor shrinkage. Mol. Pharm. 2009;6:1343–1352. doi: 10.1021/mp900022m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Gennes PG. Polymers at an interface: a simplified view. Adv. Colloid Interface Sci. 1987;27:189–209. doi: 10.1016/0001-8686(87)85003-0. [DOI] [Google Scholar]
- 10.Decuzzi P, Godin B, Tanaka T, Lee S-Y, Chiappini C, Liu X, Ferrari M. Size and shape effects in the biodistribution of intravascularly injected particles. J. Controll. Release. 2010;141:320–327. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doshi N, Prabhakarpandian B, Rea-Ramsey A, Pant K, Sundaram S, Mitragotri S. Flow and adhesion of drug carriers in blood vessels depend on their shape: a study using model synthetic microvascular networks. J. Controll. Release. 2010;146:196–200. doi: 10.1016/j.jconrel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farkas ME, Aanei IL, Behrens CR, Tong GJ, Murphy ST, O’Neil JP, Francis MB. PET Imaging and biodistribution of chemically modified bacteriophage MS2. Mol. Pharm. 2013;10:69–76. doi: 10.1021/mp3003754. [DOI] [PubMed] [Google Scholar]
- 14.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat. Nanotechnol. 2007;2:249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentile F, Chiappini C, Fine D, Bhavane RC, Peluccio MS, Cheng MM-C, Liu X, Ferrari M, Decuzzi P. The effect of shape on the margination dynamics of non-neutrally buoyant particles in two-dimensional shear flows. J. Biomech. 2008;41:2312–2318. doi: 10.1016/j.jbiomech.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Nam HY, Choi JW, Yun CO, Kim SW. Efficient lung orthotopic tumor-growth suppression of oncolytic adenovirus complexed with RGD-targeted bioreducible polymer. Gene Ther. 2014;21:476–483. doi: 10.1038/gt.2014.18. [DOI] [PubMed] [Google Scholar]
- 17.Lammers T, Kiessling F, Hennink WE, Storm G. Drug targeting to tumors: principles, pitfalls and (pre-) clinical progress. J. Control Release. 2012;161:175–187. doi: 10.1016/j.jconrel.2011.09.063. [DOI] [PubMed] [Google Scholar]
- 18.Lee S-Y, Ferrari M, Decuzzi P. Shaping nano-/micro-particles for enhanced vascular interaction in laminar flows. Nanotechnology. 2009;20:495101. doi: 10.1088/0957-4484/20/49/495101. [DOI] [PubMed] [Google Scholar]
- 19.Lee KL, Uhde-Holzem K, Fischer R, Commandeur U, Steinmetz NF. Genetic engineering and chemical conjugation of potato virus X. Methods Mol. Biol. 2014;1108:3–21. doi: 10.1007/978-1-62703-751-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merkel TJ, Jones SW, Herlihy KP, Kersey FR, Shields AR, Napier M, Luft JC, Wu H, Zamboni WC, Wang AZ, Bear JE, DeSimone JM. Using mechanobiological mimicry of red blood cells to extend circulation times of hydrogel microparticles. Proc. Natl. Acad. Sci. USA. 2011;108:586–591. doi: 10.1073/pnas.1010013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molenaar TJ, Michon I, de Haas SA, van Berkel TJ, Kuiper J, Biessen EA. Uptake and processing of modified bacteriophage M13 in mice: implications for phage display. Virology. 2002;293:182–191. doi: 10.1006/viro.2001.1254. [DOI] [PubMed] [Google Scholar]
- 22.Nagayasu A, Uchiyama K, Kiwada H. The size of liposomes: a factor which affects their targeting efficiency to tumors and therapeutic activity of liposomal antitumor drugs. Adv. Drug Deliv. Rev. 1999;40:75–87. doi: 10.1016/S0169-409X(99)00041-1. [DOI] [PubMed] [Google Scholar]
- 23.Papahadjopoulos D, Allen TM, Gabizon A, Mayhew E, Matthay K, Huang SK, Lee KD, Woodle MC, Lasic DD, Redemann C, et al. Sterically stabilized liposomes: improvements in pharmacokinetics and antitumor therapeutic efficacy. Proc. Natl. Acad. Sci. USA. 1991;88:11460–11464. doi: 10.1073/pnas.88.24.11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park JH, von Maltzahn G, Zhang L, Schwartz MP, Ruoslahti E, Bhatia SN, Sailor MJ. Magnetic iron oxide nanoworms for tumor targeting and imaging. Adv. Mater. 2008;20:1630–1635. doi: 10.1002/adma.200800004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peiris PM, Toy R, Doolittle E, Pansky J, Abramowski A, Tam M, Vicente P, Tran E, Hayden E, Camann A, Mayer A, Erokwu BO, Berman Z, Wilson D, Baskaran H, Flask CA, Keri RA, Karathanasis E. Imaging metastasis using an integrin-targeting chain-shaped nanoparticle. ACS Nano. 2012;6:8783–8795. doi: 10.1021/nn303833p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pontes-Junior J, Reis ST, de Oliveira LC, Sant’anna AC, Dall’oglio MF, Antunes AA, Ribeiro-Filho LA, Carvalho PA, Cury J, Srougi M, Leite KR. Association between integrin expression and prognosis in localized prostate cancer. Prostate. 2010;70:1189–1195. doi: 10.1002/pros.21153. [DOI] [PubMed] [Google Scholar]
- 27.Pontes-Junior J, Reis ST, Bernardes FS, Oliveira LC, Barros EA, Dall’Oglio MF, Timosczuk LM, Ribeiro-Filho LA, Srougi M, Leite KR. Correlation between beta1 integrin expression and prognosis in clinically localized prostate cancer. Int. Braz. J. Urol. 2013;39:335–342. doi: 10.1590/S1677-5538.IBJU.2013.03.06. [DOI] [PubMed] [Google Scholar]
- 28.Prasuhn DE, Jr, Singh P, Strable E, Brown S, Manchester M, Finn MG. Plasma clearance of bacteriophage Qbeta particles as a function of surface charge. J. Am. Chem. Soc. 2008;130:1328–1334. doi: 10.1021/ja075937f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao J, Xue S, Pu F, White N, Jiang J, Liu ZR, Yang JJ. Molecular imaging of EGFR/HER2 cancer biomarkers by protein MRI contrast agents. J. Biol. Inorg. Chem. 2014;19:259–270. doi: 10.1007/s00775-013-1076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shuhendler AJ, Prasad P, Leung M, Rauth AM, Dacosta RS, Wu XY. A novel solid lipid nanoparticle formulation for active targeting to tumor alpha(v) beta(3) integrin receptors reveals cyclic RGD as a double-edged sword. Adv. Healthcare Mater. 2012;1:600–608. doi: 10.1002/adhm.201200006. [DOI] [PubMed] [Google Scholar]
- 31.Shukla S, Ablack AL, Wen AM, Lee KL, Lewis JD, Steinmetz NF. Increased tumor homing and tissue penetration of the filamentous plant viral nanoparticle Potato virus X. Mol. Pharm. 2013;10:33–42. doi: 10.1021/mp300240m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shukla S, Wen AM, Ayat NR, Commandeur U, Gopalkrishnan R, Broome AM, Lozada KW, Keri RA, Steinmetz NF. Biodistribution and clearance of a filamentous plant virus in healthy and tumor-bearing mice. Nanomedicine (Lond.) 2014;9:221–235. doi: 10.2217/nnm.13.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh B, Janardhan KS, Kanthan R. Expression of angiostatin, integrin alphavbeta3, and vitronectin in human lungs in sepsis. Exp. Lung Res. 2005;31:771–782. doi: 10.1080/01902140500324901. [DOI] [PubMed] [Google Scholar]
- 34.Singh P, Prasuhn D, Yeh RM, Destito G, Rae CS, Osborn K, Finn MG, Manchester M. Bio-distribution, toxicity and pathology of cowpea mosaic virus nanoparticles in vivo. J. Control Release. 2007;120:41–50. doi: 10.1016/j.jconrel.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith BR, Ghosn EE, Rallapalli H, Prescher JA, Larson T, Herzenberg LA, Gambhir SS. Selective uptake of single-walled carbon nanotubes by circulating monocytes for enhanced tumour delivery. Nat. Nanotechnol. 2014;9:481–487. doi: 10.1038/nnano.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutherland M, Gordon A, Shnyder SD, Patterson LH, Sheldrake HM. RGD-binding integrins in prostate cancer: expression patterns and therapeutic prospects against bone metastasis. Cancers. 2012;4:1106–1145. doi: 10.3390/cancers4041106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan M, Burden-Gulley SM, Li W, Wu X, Lindner D, Brady-Kalnay SM, Gulani V, Lu ZR. MR molecular imaging of prostate cancer with a peptide-targeted contrast agent in a mouse orthotopic prostate cancer model. Pharm. Res. 2012;29:953–960. doi: 10.1007/s11095-011-0635-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan J, Shah S, Thomas A, Ou-Yang HD, Liu Y. The influence of size, shape and vessel geometry on nanoparticle distribution. Microfluid Nanofluid. 2013;14:77–87. doi: 10.1007/s10404-012-1024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vacha R, Martinez-Veracoechea FJ, Frenkel D. Receptor-mediated endocytosis of nanoparticles of various shapes. Nano Lett. 2011;11:5391–5395. doi: 10.1021/nl2030213. [DOI] [PubMed] [Google Scholar]
- 40.Wei L, Li S, Yang J, Ye Y, Zou J, Wang L, Long R, Zurkiya O, Zhao T, Johnson J, Qiao J, Zhou W, Castiblanco A, Maor N, Chen Y, Mao H, Hu X, Yang JJ, Liu ZR. Protein-based MRI contrast agents for molecular imaging of prostate cancer. Mol. imaging Biol. 2011;13:416–423. doi: 10.1007/s11307-010-0342-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang S-T, Guo W, Lin Y, Deng X-Y, Wang H-F, Sun H-F, Liu Y-F, Wang X, Wang W, Chen M, Huang Y-P, Sun Y-P. Biodistribution of pristine single-walled carbon nanotubes in vivo. J. Phys. Chem. C. 2007;111:17761–17764. doi: 10.1021/jp070712c. [DOI] [Google Scholar]
- 42.Yang ST, Wang X, Jia G, Gu Y, Wang T, Nie H, Ge C, Wang H, Liu Y. Long-term accumulation and low toxicity of single-walled carbon nanotubes in intravenously exposed mice. Toxicol. Lett. 2008;181:182–189. doi: 10.1016/j.toxlet.2008.07.020. [DOI] [PubMed] [Google Scholar]