Abstract
Protein delivery to restricted plasma membrane domains is exquisitely regulated at different stages of the cell trafficking machinery. Traffic control involves the recognition of export/retention/retrieval signals in the endoplasmic reticulum (ER)/Golgi complex that will determine protein fate. A splice variant (SV), SV1, of the voltage- and Ca2+-activated K+ channel α-subunit accumulates the channel in the ER, preventing its surface expression. We show that SV1 insert contains a nonbasic, hydrophobic retention/retrieval motif, CVLF, that does not interfere with proper folding and tetramerization of SV1. Localization of proteins in the ER by CVLF is independent of its position; originally, on the first internal loop, SV1 insert or CVLF perform equally well if placed at the middle or end of the α-subunit intracellular carboxyl terminus. Also, CVLF is able to restrict the traffic of an independently expressed transmembrane protein, β1-subunit. CVLF is present in proteins across species and in lower organisms. Thus, CVLF may have evolved to serve as a regulator of cellular traffic.
Large-conductance, voltage- and Ca2+-activated K+ (MaxiK) channels are ubiquitously expressed, except in heart myocytes, and play important roles in regulating cell excitability including neurotransmission and smooth muscle tone. They are composed of the pore-forming α- and modulatory β-subunits (1, 2). The human MaxiK α-subunit (hSlo) is the product of a single gene with many possibilities for splice variation, which predicts the existence of multiple isoforms of the same channel with potentially diverse functions (3). Four modulatory β-subunit genes add to this diversity (2). Accordingly, MaxiK (or BK) channels have different electrical properties and levels of expression from tissue to tissue and within tissues (4, 5). In addition, their numbers in the plasma membrane can decrease under different physiological conditions such as pregnancy term (6) and coronary aging (7), resulting in higher muscle contractility.
Normal cell performance requires a delicate control of protein expression at the correct time and place. There are multiple mechanisms that regulate membrane protein localization and include those regulating transcription, protein synthesis, traffic, and targeting to specific regions or microdomains at the cell surface. Protein traffic is governed by the molecular recognition of signals, short stretches of amino acids embedded at various positions within proteins, at different stages of the secretory pathway beginning in the endoplasmic reticulum (ER). Traffic through the ER involves: (i) retention signals that inhibit forward movement of the protein, (ii) retrieval signals that return the escaped proteins to the initial resident compartment, and (iii) export signals that expedite protein exit from the ER organelle (8, 9). Retention/retrieval sequences have been identified for many luminal and monotopic (type I) transmembrane proteins, consisting mainly of carboxyl-terminal KDEL, RRXX, and KKXX motifs (8). However, just a handful of polytopic protein (type III transmembrane proteins) retention/retrieval sequences have been identified. These sequences consist of basic motifs (RXR, KR, RRKK, and RR) located in cytosolic regions of ion channels (KATP, KCNK, glutamate-gated/NMDA, and Kainate) (10–12) and of metabotropic glutamate receptor 1B (13), although a transmembrane motif (PLYFXXN) in acetylcholine receptor α1-subunit keeps unassembled subunits in the ER (14).
The amount of protein expressed on the cell membrane will depend, at the ER level, on an exquisite balance of protein export and retention mechanisms. The effectiveness of protein export is regulated not only by proper folding and assembly but also by forward signals (15, 16). In addition, export will occur if retention/retrieval signals are masked by association with subunits (10) or by SVs (12, 13, 17). We recently identified an SV, SV1, that localizes hSlo within the ER (18). SV1 contains a 33-aa insert after leucine-115 in the S1 transmembrane domain. The SV insert does not modify the hydrophobicity plot of S1 but instead replaces few hydrophobic amino acids and makes the S0–S1 intracellular linker longer. The hydrophobicity plot predicts that 27 aa are likely in the cytoplasmic linker of transmembrane segments S0 and S1 and that 6 aa are buried in transmembrane segment S1 (Fig. 1, shadowed amino acids) (18). However, homology analysis shows that none of the known ER retention motifs are present in the insert. We now report a hydrophobic ER retention/retrieval motif, CVLF, which inhibits the surface expression of the SV1 of MaxiK α-subunit. The same motif is present in other membrane proteins and inhibits the surface expression of an independently expressed β1-subunit, suggesting the easy access of this sequence to the cellular trafficking machinery and its potential role as a common ER retention/retrieval sequence.
Fig. 1.
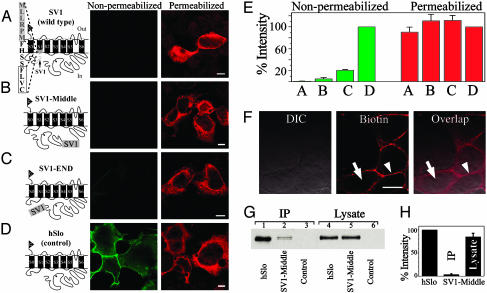
The retention capacity of the SV1 insert is prominently independent of its location. (A–D) hSlo constructs (c-Myc epitope, triangle) and corresponding expression patterns in nonpermeabilized and permeabilized conditions. Shown are the SV1 insert (gray box/dashed line) (A–C) and partial sequence at its carboxyl terminus (A). In SV1 (wild type) (A), CVLF (boxed) lies in the S0-S1 linker. SV1 (A), SV1-Middle (B), or SV1-END (C) showed no or insignificant surface labeling (nonpermeabilized) but were efficiently synthesized (permeabilized). (D) Insertless hSlo (control) with clear surface expression (nonpermeabilized). (E) Percent intensity with respect to hSlo. The number of cells are: 27 (bars A), 32 (bars B), 51 (bars C), and 77 (bars D). (F) DIC (Left) and biotin labeling (Center) (Texas Red avidin, Vector Laboratories) of surface proteins (arrowhead) in living cells; intracellular milieu free of biotin labeling (arrow). Right shows overlap (arrowhead, surface proteins; arrow, no intracellular labeling). (G) Immunoblots with anti-hSlo883–896 of IP surface proteins (lanes 1–3) and of cell lysates (lanes 4–6). hSlo molecular mass is ≈125 kDa. (H) Percent blot intensities normalized to corresponding hSlo values. n = 3. In this and following figures, pictures are single confocal sections taken at the middle of cells. (Bars = 10 μm.)
Methods
Constructs and Transfection. hSlo (U11058) and SV1 have an extracellular amino-terminal c-Myc epitope (18, 19). SV1-Middle and SV1-END contain the SV1 33-aa insert in the middle (between K588 and I589 with downstream L-K junctional amino acids) or the end of hSlo carboxyl terminus (Fig. 1). Human β1 (U25138) contains CVLF (β1+CVLF) or QKDG (β1+QKDG) followed by a c-Myc epitope at the intracellular carboxyl terminus. HEK293T cells were transfected with constructs in pcDNA3 and used after 3–4 days (18, 19). The day before immunocytochemistry, cells were transferred to chamber slides precoated with 0.1 mg/ml poly(d-lysine) and 0.1 mg/ml collagen.
Abs. Affinity-purified polyclonal Abs against hSlo (1–1.2 μg/ml anti-hSlo883–896, Alomone Labs, Jerusalem) (6, 20) and ERp72 (1 μg/ml, Calbiochem) (21, 22), polyclonal β1 Ab (1/1,000 anti-β185–102) (23), and anti-c-Myc monoclonal Ab (2 μg/ml clone 9E10) were used. In all experiments, a negative control was transfection with no DNA. Ab specificity was tested by preadsorbing each Ab with the corresponding antigenic peptide.
Nonpermeabilized and Permeabilized Labeling. Live cells were treated for 1 h (4°C) with Abs against extracellular epitopes of hSlo (anti-c-Myc) or β1 (anti-β185–102) constructs. Cells were fixed with 4% paraformaldehyde before permeabilization with 0.2% Triton X-100 and double labeling; incubation with primary Abs was overnight at 4°C. For hSlo constructs prelabeled with anti-c-Myc (nonpermeabilized), anti-hSlo883–896 was used. Cells were incubated for 1 h with biotinylated anti-mouse Ab, washed, and incubated for 1 h with 1 μg/ml fluorescein-avidin (Vector Laboratories) and 7.5 μg/ml Rhodamine Red-X-conjugated donkey anti-rabbit Ab (Jackson ImmunoResearch). For β1 constructs prelabeled with anti-β185–102 (nonpermeabilized), anti-c-Myc was used. Cells were washed and incubated with 6 μg/ml FITC-conjugated donkey anti-rabbit (Jackson ImmunoResearch) and 6 μg/ml Rhodamine Red-X-conjugated donkey anti-mouse Abs. Colabeling of Erp72 and hSlo was under permeabilized conditions with anti-c-Myc, anti-Erp72623–638, 6.5 μg/ml FITC-conjugated donkey anti-mouse, and Rhodamine Red-X-conjugated donkey anti-rabbit Abs. Cells were mounted by using ProLong (Molecular Probes).
Cell Surface Biotinylation and Immunoprecipitation (IP). Live cells were incubated for 1 h with 0.08 mg/ml EZ-Link Sulfo-NHS-LC-Biotin (Pierce) at 4°C. After washing, half of the cells were used for immunocytochemistry to check that only surface proteins were biotinylated, and half were used for IP. For IP, cells were lysed in 50 mM Tris, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and protease inhibitors (pH 8.0) and centrifuged for 15 min at 15,000 × g (24). Solubilized proteins (3 mg/ml) were immunoprecipitated with antibiotin beads. Beads were washed extensively and resuspended in SDS loading buffer before 7.5% SDS/PAGE and immunoblotting. Signals were analyzed as described in ref. 6
Soluble and Insoluble Fractions. Cells (2 × 105 per 100 μl) were solubilized in Tris-buffered saline (pH 8) containing 5 mM EDTA, 1 mM iodoacetamide, protease inhibitors, and 0%, 0.1%, or 1% Triton X-100. After 10 min on ice, lysates were centrifuged for 5 min (15,000 × g at 4°C). Solubilized fractions were saved, and insoluble fractions were washed and resuspended in equal volume as the soluble fractions (100 μl). Samples were boiled in SDS loading buffer with 1.4 M 2-mercaptoethanol, and 10 μl was fractionated on 7.5% SDS/PAGE and immunoblotted.
Nonreducing Versus Reducing Conditions. Cells were solubilized in 150 mM NaCl, 20 mM Hepes, 5 mM EDTA, and 1% CHAPS (pH 7.4) with protease inhibitors, and centrifuged at 15,000 × g for 10 min at 4°C. Solubilized fractions were precleared for 1 h with protein A-Sepharose beads and centrifuged at 100,000 × g for 30 min at 4°C. Soluble proteins (10 μg) were mixed with SDS loading buffer with or without 1.4 M 2-mercaptoethanol, run on 7.5% SDS/PAGE, and immunoblotted.
Sucrose Gradient. Cell lysates (150 μg of protein) or 150 μg of protein markers (Apoferritin, 440 kDa; β-amylase, 200 kDa; and alcohol dehydrogenase, 150 kDa) were layered on top of 5–50% continuous sucrose gradient prepared in 150 mM NaCl, 20 mM Hepes, 5 mM EDTA, and 1% CHAPS (pH 7.4) containing protease inhibitors. Samples were centrifuged for 18 h at 182,298 × g in a swinging rotor; 10 aliquots (400 μl each) were collected from the top of each gradient. Proteins of each fraction were concentrated by using PAGEprep protein clean-up resin (Pierce), reduced by 0.7 M 2-mercaptoethanol, boiled for 5 min, fractionated on 7.5% SDS/PAGE, and immunoblotted. Signals were detected by using infrared fluorescence (Li-Cor, Lincoln, NE).
Image Analysis and Statistics. Confocal sections were acquired every 0.25 or 0.5 μm and, unless otherwise stated, analyzed with image-pro plus (Media Cybernetics, Silver Spring, MD) or imagej (National Institutes of Health). Total protein expression was the mean intensity of cell outlines labeled under permeabilized conditions; surface expression was measured by using the same outlines superimposed on the same section labeled under nonpermeabilized conditions. All conditions including optical sectioning, number of sections, and exposures were identical for a given experiment. Values are means ± SE. Student's t test at P < 0.05 was considered significant.
Results
The Retention Capacity of the SV1 Insert Is Prominently Independent of Its Location. To determine whether SV1 insert has a potential retention/retrieval motif contained within its sequence, we first tested whether its retention ability was position-dependent. We created constructs with the 33-aa insert in three different locations within hSlo (Fig. 1 A–D). Cells that were transfected with the naturally occurring SV1 (18) showed no surface labeling (A, nonpermeabilized), whereas significant labeling was detected after permeabilization (Fig. 1 A, permeabilized). Similar results were observed when SV1 insert was relocated to the middle (B) or the end (C) of hSlo carboxyl terminus. As a control, cells transfected with insertless hSlo show prominent surface expression (Fig. 1D, nonpermeabilized).
Quantification of the labeling intensities (Fig. 1E) showed that SV1 (A), SV1-Middle (B), and SV1-END (C) have strikingly lower (≤21%) surface expression than hSlo (D) (nonpermeabilized), whereas no significant differences were observed in their total protein expression (permeabilized). SV1-Middle (Fig. 1B) and SV1-END (C) showed slightly higher surface expression when compared with SV1 (A) (nonpermeabilized). This slight increase in surface expression may be due to changes in accessibility of the SV1 retention signal to its receptor. Despite this modest escape observed when SV1 insert is outside of its native position, these results indicate that the retention capacity of the SV1 insert is largely independent of its location and, thus, could contain a retention/retrieval motif.
To verify our results with an alternative method, we used a biotinylation approach to measure surface protein expression. Fig. 1F displays an example of cells transfected with SV1-Middle that were biotinylated and fluorescently labeled. Single confocal images at the middle of the cells show that only surface proteins were biotinylated (labeling is restricted to the periphery of the cells; arrowheads) with no intracellular labeling (arrows) (see also overlap). Fig. 1G shows a Western blot of surface (IP, lanes 1–3) and total (lysate, lanes 4–6) protein obtained from cells expressing hSlo, SV1-Middle-transfected cells, and nontransfected cells (control, lanes 3 and 6). Consistent with optical experiments, SV1-Middle (lane 2) showed significantly lower surface expression than hSlo (lane 1), inasmuch as both had similar total protein (lanes 4 and 5). Quantification (Fig. 1H) shows that SV1-Middle surface expression (IP) is minimal (<5%) when compared with hSlo.
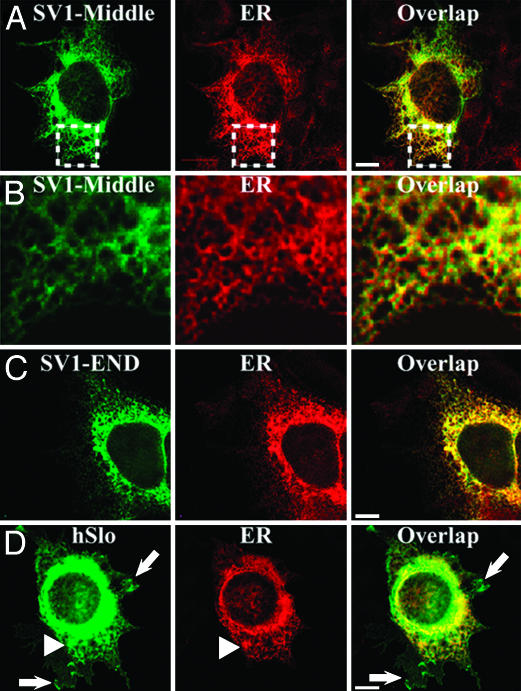
SV1 Insert Outside Its Natural Environment also Localizes hSlo Protein Within the ER. We reasoned that if SV1 contains an ER retention/retrieval signal, the constructs SV1-Middle and SV1-END should also localize the protein in the ER. To test this hypothesis, cells expressing SV1-Middle and SV1-END were double-labeled with anti-c-Myc and anti-ERp72 (ER marker) Abs under permeabilized conditions. Fig. 2A shows that SV1-Middle displays a network pattern (square) that strongly mimics ER expression. The overlap and higher magnification (Fig. 2B) display the striking similarity between the two expression patterns. Similarly, SV1-END (Fig. 2C) shows a strong colocalization with the ER marker. For comparison, cells were transfected with insertless hSlo and double-labeled under identical conditions (Fig. 2D). Consistent with normal protein trafficking, a significant amount of hSlo is localized within the ER (arrowheads), but, in this case, the synthesized hSlo can reach its destination on the cell surface (arrows). These experiments are a strong indication that the SV1 insert is sufficient to strongly promote the localization of the protein in the ER where the prototype ERp72 protein resides.
Fig. 2.
SV1 insert-containing constructs are predominantly retained within the ER. Cells expressing SV1-Middle, SV1-END, or hSlo were double-labeled with anti-c-Myc (green) and anti-ERp72 (red) under permeabilized conditions. (A) SV1-Middle had a network expression pattern (square) strongly mimicking the ER. Overlap illustrates a striking colocalization (yellow). (B) Higher magnification (squares in A). (C) SV1-END also shows strong colocalization with the ER marker. (D) hSlo shows ER colocalization (arrowheads) and significant expression at the periphery (arrows). Images were 3D subpixel deconvolved by using autoquant (representative of 15 cells; n = 3 experiments).
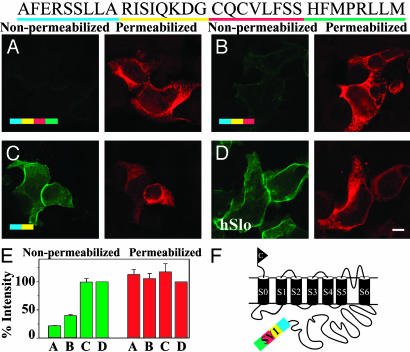
Identification of a Hydrophobic Retention/Retrieval Motif, CVLF, in SV1. To determine which amino acids of the SV1 insert (Fig. 3, sequence at top) are responsible for hSlo localization in the ER, we first made serial carboxyl-terminal deletions of the SV1-END construct (Fig. 3 A and F). Deletion of the last 8 aa (HFMPRLLM) caused a slight increase (≈20%) (Fig. 3E) in surface expression when compared with the whole insert (Fig. 3 B vs. A, nonpermeabilized). This result might indicate that the molecular environment provided by the spatial arrangement of these amino acids may facilitate, but is not critical for, full protein retention. In contrast, deletion of the next 8 aa (CQCVLFSS) induced a loss of the retention capability of the insert, allowing a drastic increase in surface expression (Fig. 3C, nonpermeabilized) similar to the WT channel (Fig. 3D).
Fig. 3.
Serial carboxyl-terminal deletions demonstrate that the majority of the SV1 retention capacity is confined to 8 aa of the SV1 insert. (A–D) Transfected cells labeled under nonpermeabilized (green) and permeabilized (red) conditions. (A) Full-length SV1-END served as reference of ER localization; amino acids were color-coded according to their location in the sequence shown at the top. Almost all protein is localized in the ER (permeabilized) with minimal surface expression (nonpermeabilized). (B) Removal of the last 8 aa (green bar) caused a slight increase in surface labeling (nonpermeabilized). (C) Deletion of the next 8 aa (red bar) significantly relieved retention (strong surface labeling, nonpermeabilized). (D) hSlo, control for unrestricted surface expression. (E) Percent intensities normalized to hSlo expression. Number of cells are as follows for four experiments: 51 (bars A), 45 (bars B), 43 (bars C), 54 (bars D). (F) SV1-end model.
Quantification of labeling intensities (Fig. 3E) shows that, although all constructs had similar total protein expression (red bars), their surface expression (green bars) clearly depended on the insert segment that they contained. Constructs containing the CQCVLFSS sequence showed <40% surface expression (Fig. 3E, green bars A and B), whereas the construct lacking this sequence lost its ability to retain the protein showing nearly 100% surface expression (green bar C) (similar to levels achieved by the insertless hSlo; green bar D). These results strongly indicated that the motif causing localization of hSlo in the ER was contained within the CQCVLFSS epitope.
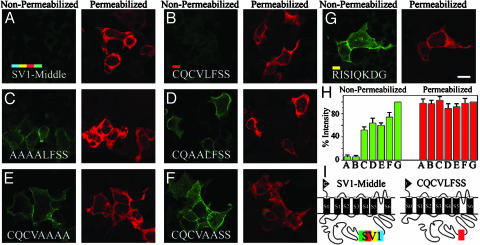
To test the hypothesis that CQCVLFSS contains a short string of amino acids responsible for the SV1 retention in the ER, we replaced the whole SV1-Middle insert (Fig. 4 A and I) with the CQCVLFSS sequence alone (Fig. 4 B and I). To further circumscribe the potential retention/retrieval motif, we simultaneously mutated two or four residues to alanines (Fig. 4 C–F) because retention/retrieval motifs are usually a string of two to four residues. Control experiments included (i) a construct containing the upstream sequence RISIQKDG (Fig. 4G), which should not retain protein in the ER as suggested by the serial deletion experiments (Fig. 3C), and (ii) parallel transfections with SV1-Middle (Fig. 4A). As predicted, the RISIQKDG epitope showed no ER retention/retrieval properties because the protein freely reached the surface (Fig. 4G). In contrast, when cells were transfected with CQCVLFSS (Fig. 4B), hSlo protein was practically absent from the surface, indicating that this fragment is sufficient to produce full retention.
Fig. 4.
Site-directed mutagenesis demonstrates that mutations within CQCVLFSS significantly relieve retention. (A) SV1-Middle as reference of SV1 retention/retrieval ability. Color code is the same as in Fig. 3. (B) CQCVLFSS insert (red box in model) localizes the protein in the intracellular compartment. (C–F) Alanine substitutions in the CQCVLFSS fragment relieved retention (nonpermeabilized). (G) Upstream RISIQKDG at same position as CQCVLFSS has unrestricted surface expression (control). (H) Percent intensity under nonpermeabilized and permeabilized conditions normalized to control values (G). Number of cells are as follows from three experiments: 45 (bars A), 49 (bars B), 45 (bars C), 47 (bars D), 57 (bars E), 49 (bars F) and 54 (bars G). (I) Models. (Bar = 20 μm.)
Results are quantified in Fig. 4H as a percentage of RISIQKDG expression. None of the constructs significantly affected total cell expression (permeabilized, red bars). Similar to SV1-Middle, CQCVLFSS showed <10% surface labeling (Fig. 4H, bars A and B, nonpermeabilized), whereas all alanine mutants tested (bars C–F) displayed >50% loss of the CQCVLFSS epitope's retention ability. Notice that alanine mutants had in common the substitution of either CV or LF by alanines, implicating the hydrophobic CVLF motif as the trafficking signal responsible for hSlo localization in the ER.
The CVLF Motif Is a Trafficking Signal That Inhibits Surface Expression of Polytopic Proteins. ER retention/retrieval sequences of monotopic and polytopic proteins are commonly formed by basic amino acids. Thus, it was remarkable to find that the motif of SV1 responsible for hSlo confinement in the ER could be a hydrophobic motif, CVLF. The CVLF hydrophobic motif is not predicted to form part of a transmembrane domain, but rather it would be located near the membrane facing the cytoplasm (Fig. 1A).
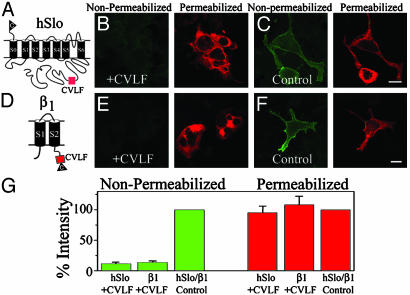
To assess the retention/retrieval nature of CVLF motif, we performed two tests: locate the CVLF motif at the middle of the carboxyl terminus of hSlo, away from its natural environment (Fig. 5A), and attach CVLF to the carboxyl terminus of an independently expressed transmembrane protein, the β1-subunit (Fig. 5D) (18). Immunolabeling experiments clearly demonstrate that CVLF can indeed act by itself as a retention/retrieval motif because no significant surface labeling was observed regardless of the background protein. Fig. 5B shows the tremendous reduction in hSlo surface expression (nonpermeabilized) by CVLF and its effective confinement in the intracellular compartment (permeabilized). As a control, we used hSlo+RISIQKDG, which does not have the ability to restrain the protein in the ER (Figs. 4G and 5C).
Fig. 5.
Hydrophobic CVLF motif is sufficient to inhibit surface expression of polytopic proteins, hSlo, and β1-subunit. (A) CVLF (box) position is the same as constructs in Fig. 4. hSlo+CVLF shows practically no surface labeling (nonpermeabilized) and retains protein intracellularly (permeabilized). (C) Substituting CVLF with RISIQKDG (control) at the same position shows strong surface labeling (nonpermeabilized). (D) β1-subunit with CVLF (box) and c-Myc epitope (triangle). (E) CVLF inhibited β1-subunit surface labeling (nonpermeabilized) and localized it intracellularly (permeabilized). (F) Substituting CVLF with the QKDG sequence (control) allowed protein surface expression (nonpermeabilized). (G) Percent intensity measurements were normalized to control constructs. Number of cells in three experiments are as follows: 38 (hSlo+CVLF), 52 (β1+CVLF), 42 (hSlo+RISIQKDG, hSlo control), and 69 (β1+QKDG, β1 control). (Bars = 20 μm.)
The same pattern was observed when the β1-subunit containing CVLF was used (Fig. 5D). Striking is the reduction in surface protein expression of β1+CVLF and its intracellular localization (Fig. 5E, nonpermeabilized vs. permeabilized). The control β1 protein contained QKDG (Fig. 5F), a string of amino acids in SV1 that does not cause retention (Figs. 3C and 4G), instead of CVLF. As illustrated in Fig. 5F, QKDG allowed the β1-subunit to readily reach the membrane (nonpermeabilized). Quantification shows that total expression was not modified in any of the constructs (Fig. 5G, permeabilized) and that the CVLF motif had the ability to restrain nearly 90% of the proteins in the ER. This level of effectiveness was similar to the one attained when the whole SV1 insert was located in the same position in hSlo (Fig. 1).
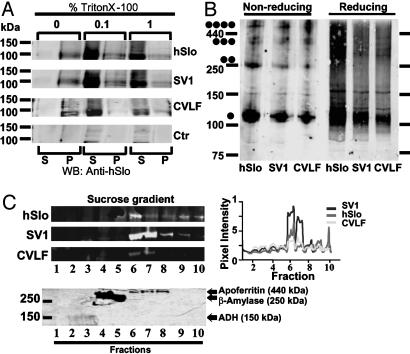
The CVLF Motif Does Not Interfere with Proper Folding and Tetramerization of the MaxiK Constructs. We used biochemical approaches to determine whether protein misfolding was responsible for the ER retention of the SV1 and hSlo+CVLF constructs. To assess folding integrity, hSlo-, SV1-, and hSlo+CVLF-expressing cells were analyzed for solubility in buffers containing different concentrations of the nonionic detergent Triton X-100. Insolubility in nonionic detergent is a useful indicator of gross misfolding of membrane proteins (25, 26). We found that, like hSlo, SV1 and hSlo+CVLF were soluble even at 0.1% Triton X-100 (Fig. 6A), consistent with properly folded SV1 and hSlo+CVLF membrane proteins.
Fig. 6.
The CVLF motif does not interfere with folding and tetramerization of channels. (A) Immunoblot of hSlo, SV1, hSlo+CVLF, and control (untransfected) cell lysates in 0%, 0.1%, and 1% Triton X-100. S, Solubilized fraction; P, nonsolubilized fraction (n = 2). (B) Immunoblots of hSlo, SV1, and hSlo+CVLF in nonreducing and reducing conditions. Dots mark expected sizes for tetramers, trimers, dimers, and monomers (n = 4). Parallel immunocytochemistry experiments of the samples in A and B showed the usual ER-retention properties of CVLF-containing constructs. (C) Immunoblot (Upper) and Coomasie blue gel (Lower) after nonreducing continuous sucrose gradient sedimentation. Plot shows hSlo, SV1, and hSlo+CVLF migration pattern similar to apoferritin (n = 2).
Alternatively, CVLF motif may prevent assembly and tetramerization of the MaxiK channels. Unassembled membrane proteins would then be retained in the ER and degraded by quality-control mechanisms. To determine whether retention of CVLF-containing constructs in the ER was due to defects in tetramerization, hSlo-, SV1-, and hSlo+CVLF-expressing cells were solubilized and immunoblotted under nonreducing and reducing conditions (Fig. 6B). Our results illustrate the existence of monomers, dimers, trimers, and tetramers for hSlo, SV1, and hSlo+CVLF under nonreducing conditions and monomers under reducing conditions. Compelling evidence for normal tetramerization was obtained by sucrose-density gradient (Fig. 6C), showing that the majority of hSlo, SV1, and hSlo+CVLF proteins appear in the fractions similar to apoferritin, which has a molecular mass (≈440 kDa) close to the estimated molecular mass of hSlo tetramers (≈500 kDa). From these findings, we can conclude that CVLF-containing constructs are folded properly and can form tetramers.
In summary, a retention/retrieval motif, CVLF, has been discovered that is unique in its lack of basic amino acids and in its hydrophobic nature.
Discussion
Previously, we isolated a Slo SV, SV1, that contains a 33-aa insert in S1 transmembrane domain and showed that SV1 is retained within the ER organelle acting as a dominant-negative modulator of hSlo surface expression (18). In this study, we found that the SV1 insert retention/retrieval property is not exclusive to its native position in hSlo or to hSlo protein but rather contains an ER retention/retrieval motif, CVLF. The CVLF hydrophobic motif is sufficient to prevent surface expression of independently expressed membrane proteins by trapping them in the intracellular compartments, likely the ER. In addition, CVLF-containing channels are tetramerized and have similar solubility to WT hSlo. These results suggest that retention by CVLF motif may be receptor-mediated.
Trafficking Signals Vary in Their Location and Composition. Trafficking signals of few polytopic proteins have been identified. The emerging picture is that the majority contain basic (RXR, KR, RRKK, and RR) retention/retrieval intracellular motifs (10–13). However, recent studies of the acetylcholine receptor indicate that an amphipatic ER retention/retrieval motif that lacks basic residues (PLYFxxN) may be located in the transmembrane region of the receptor (14). Also, a transmembrane segment of the ryanodine receptor Ca2+ channel seems to contain a sequence for ER localization; however, its molecular identity is not known yet (27). Further evidence suggesting the multiplicity of retention/retrieval sequences comes from studies using monotopic proteins (type I transmembrane proteins), such as the cytochrome P450 2C1 that contains a relatively sequence-independent retrieval transmembrane signal (28). The CVLF cytosolic motif reported here has two distinguishing characteristics: its lack of basic amino acids and its hydrophobic nature. Kyte–Doolittle hydrophobicity analysis (29) indicates that CVLF motif could be adjacent to the plasma membrane linked by four amino acids (SSHF) to the S1 transmembrane domain of MaxiK channels (Fig. 1A). Thus, it appears that retention/retrieval motifs of transmembrane proteins have evolved to a variety of retention/retrieval signals that are not always soluble linear motifs but may be more complex signals to assure the physiologically needed expression level in the plasma membrane.
The CVLF Motif Is Present in Proteins Across Species. The CVLF motif may be a common retrieval/retention signal as demonstrated by its ability to localize to the ER an independently expressed protein, the transmembrane β1-subunit of the MaxiK channel (Fig. 5). In fact, a database search revealed that the CVLF motif is found in proteins across species and in lower organisms such as bacteria and yeast. Moreover, CVLF motif is present in mammalian (human, pig, and mouse; P55072, P03974, and Q01853, respectively) amphibian (Xenopus laevis, P23787), and nematode (Caenorhabditis elegans, P54811) homologs of transitional ER ATPase, p97, a cytosolic protein bound to ER membranes that transports proteins from the ER into the cytosol (30) and is also involved in ER assembly (31). This evidence supports the view that CVLF may be another prototypical ER retention/retrieval motif distinct from the classical basic motifs, and suggests that CVLF may have ancient origins being selected through evolution.
In humans, 23 unrelated proteins were found that contain CVLF motif at different positions. Among them are GTP-binding proteins Rab8 (NP_005361) and Rab10 (NP_057215). Both Rab8 and Rab10 contain the CVLF motif and have 66% identity. Despite the high degree of homology between Rab8 and Rab10, the two proteins are localized to distinct cellular compartments. Whereas Rab8 is localized to the cell periphery, Rab10 is expressed in the perinuclear region (32). The cell periphery localization of Rab8 may be due to its ability to mask the CVLF motif. In this view, CVLF may behave similarly to the RKR motif of KATP channels (10) and the I–II loop of the Ca2+ channels (33). In KATP and Ca2+ channels, the masking of the ER retention signals is a prerequisite for surface expression. We attempted to mask CVLF by coexpression of the SV1 with hSlo (insertless) or β1 (18). These subunits were unable to mask the CVLF motif in the SV1. Further study could be directed to identify proteins that may release the SV1 from the ER.
Potential Mechanism of Retention and Quality Control by CVLF Motif. Different mechanisms could ensure the ER localization of CVLF-containing proteins like SV1: (i) CVLF may induce misfolding of the proteins that may lead to ER retention through aggregation or interaction with ER chaperones (8), (ii) CVLF may retain channels in the ER by inhibiting their tetramerization similar to KATP channels (10), or (iii) CVLF may act as retention and/or retrieval signal. Our results effectively exclude the first two mechanisms by illustrating that CVLF-containing channels are folded properly, can form tetramers (Fig. 6), and support retention and/or retrieval as mechanisms that can explain ER retention by CVLF. Further study is needed to investigate which mechanisms and receptors are responsible for the ER localization of CVLF-containing proteins. Because of the hydrophobic nature of CVLF, the mechanism involved in localizing proteins to the ER may include the recognition of a lipid environment. This mechanism could be rather important for proteins containing the CVLF motif in a transmembrane region. Supporting this view, lipid composition has been shown to be important in ER transport mediated by coatomer protein II that drives anterograde transport from the ER to the Golgi system (34).
Retention/retrieval signals of plasma membrane proteins can be a quality-control checkpoint for correct oligomeric assembly. This quality check mechanism is used by the KATP channel, whose basic retention/retrieval signals present in both α- and β-subunits are masked only when the channel is correctly coassembled into an octamer (10). Another example is the nicotinic acetylcholine receptor, whose ER retention/retrieval signal located in transmembrane domain 1 is buried in correctly assembled acetylcholine receptor pentamers. In this way, only correctly assembled proteins will be able to reach the plasma membrane, whereas unassembled subunits with exposed retention/retrieval signals would be confined to the ER. Retention/retrieval signals can also be a mechanism of modulating the expression levels of membrane proteins (18). Modulation of expression could also be achieved by changing the potency of trafficking signals. Interestingly, conserved substitutions to the CVLF motif to CILF can also be found in the database. A combinatorial analysis of arginine- and lysine-based hypothetical ER localization signals indicates that changes in local sequence context may give rise to a variety of signal strengths (35). In fact, when the SV1 insert or the CVLF motif is changed from its natural molecular environment, the strength of the retention signal is diminished by ≈5–20% (Figs. 1 and 4).
The MaxiK channel functions in different tissues can be influenced by their expression levels. How cells achieve a correct number of channels on the cell surface is poorly understood. Recently, we introduced down-regulation by SVs as a mechanism that may contribute to the diverse levels of MaxiK channel surface expression (18). It was proposed that an ER retention/retrieval signal embedded in the SV1 insert sequence significantly slows down the normal trafficking of the constitutive MaxiK channel protein by trapping it in the ER. The ER-restrained MaxiK channel would create a large intracellular reservoir of MaxiK channels that upon release can transiently increase the MaxiK channel surface expression. Further studies are necessary to determine whether CVLF can be masked by other membrane or cytoplasmic proteins that could lead to a transient release of MaxiK channels from the ER to the cell surface.
Acknowledgments
This work was supported by National Institutes of Health Grants HL54970 (to L.T.) and HD38983 (to E.S.) and the American Heart Association (M.M.Z.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, endoplasmic reticulum; hSlo, human MaxiK α-subunit; IP, immunoprecipitation; MaxiK, voltage- and Ca2+-activated K+ channel; SV, splice variant.
References
- 1.Toro, L., Meera, P., Wallner, M. & Tanaka, Y. (1998) News Physiol. Sci. 13, 112-117. [DOI] [PubMed] [Google Scholar]
- 2.Orio, P., Rojas, P., Ferreira, G. & Latorre, R. (2002) News Physiol. Sci. 17, 156-161. [DOI] [PubMed] [Google Scholar]
- 3.Graveley, B. R. (2001) Trends Genet. 17, 100-107. [DOI] [PubMed] [Google Scholar]
- 4.Latorre, R., Oberhauser, A., Labarca, P. & Alvarez, O. (1989) Annu. Rev. Physiol. 51, 385-399. [DOI] [PubMed] [Google Scholar]
- 5.McManus, O. B. (1991) J. Bioenerg. Biomembr. 23, 537-560. [DOI] [PubMed] [Google Scholar]
- 6.Song, M., Zhu, N., Olcese, R., Barila, B., Toro, L. & Stefani, E. (1999) FEBS Lett. 460, 427-432. [DOI] [PubMed] [Google Scholar]
- 7.Marijic, J., Li, Q.-X., Song, M., Nishimaru, K., Stefani, E. & Toro, L. (2001) Circ. Res. 88, 210-215. [DOI] [PubMed] [Google Scholar]
- 8.Ellgaard, L. & Helenius, A. (2003) Nat. Rev. Mol. Cell Biol. 4, 181-191. [DOI] [PubMed] [Google Scholar]
- 9.Mellman, I. & Warren, G. (2000) Cell 100, 99-112. [DOI] [PubMed] [Google Scholar]
- 10.Zerangue, N., Schwappach, B., Jan, Y. N. & Jan, L. Y. (1999) Neuron 22, 537-548. [DOI] [PubMed] [Google Scholar]
- 11.O'Kelly, I., Butler, M. H., Zilberberg, N. & Goldstein, S. A. (2002) Cell 111, 577-588. [DOI] [PubMed] [Google Scholar]
- 12.Scott, D. B., Blanpied, T. A., Swanson, G. T., Zhang, C. & Ehlers, M. D. (2001) J. Neurosci. 21, 3063-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan, W. Y., Soloviev, M. M., Ciruela, F. & McIlhinney, R. A. (2001) Mol. Cell. Neurosci. 17, 577-588. [DOI] [PubMed] [Google Scholar]
- 14.Wang, J. M., Zhang, L., Yao, Y., Viroonchatapan, N., Rothe, E. & Wang, Z. Z. (2002) Nat. Neurosci. 5, 963-970. [DOI] [PubMed] [Google Scholar]
- 15.Ma, D., Zerangue, N., Lin, Y. F., Collins, A., Yu, M., Jan, Y. N. & Jan, L. Y. (2001) Science 291, 316-319. [DOI] [PubMed] [Google Scholar]
- 16.Ma, D., Zerangue, N., Raab-Graham, K., Fried, S. R., Jan, Y. N. & Jan, L. Y. (2002) Neuron 33, 715-729. [DOI] [PubMed] [Google Scholar]
- 17.Jaskolski, F., Coussen, F., Nagarajan, N., Normand, E., Rosenmund, C. & Mulle, C. (2004) J. Neurosci. 24, 2506-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zarei, M. M., Zhu, N., Alioua, A., Eghbali, M., Stefani, E. & Toro, L. (2001) J. Biol. Chem. 276, 16232-16239. [DOI] [PubMed] [Google Scholar]
- 19.Meera, P., Wallner, M., Song, M. & Toro, L. (1997) Proc. Natl. Acad. Sci. USA 94, 14066-14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knaus, H. G., Eberhart, A., Koch, R. O. A., Munujos, P., Schmalhofer, W. A., Warmke, J. W., Kaczorowski, G. J., Garcia, M. L. & Koch, R. O. (1995) J. Biol. Chem. 270, 22434-22439. [DOI] [PubMed] [Google Scholar]
- 21.Mazzarella, R. A., Srinivasan, M., Haugejorden, S. M. & Green, M. (1990) J. Biol. Chem. 265, 1094-1101. [PubMed] [Google Scholar]
- 22.Kuznetsov, G., Bush, K. T., Zhang, P. L. & Nigam, S. K. (1996) Proc. Natl. Acad. Sci. USA 93, 8584-8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wanner, S. G., Koch, R. O., Koschak, A., Trieb, M., Garcia, M. L., Kaczorowski, G. J. & Knaus, H. G. (1999) Biochemistry 38, 5392-5400. [DOI] [PubMed] [Google Scholar]
- 24.Meier, T. & Leying, H. (1996) in A Laboratory Guide to Biotin-Labeling in Biomolecule Analysis, eds. Meier, T. & Fahrenholz, F. (Birkhauser, Basel), pp. 83-96.
- 25.Manganas, L. N., Wang, Q., Scannevin, R. H., Antonucci, D. E., Rhodes, K. J. & Trimmer, J. S. (2001) Proc. Natl. Acad. Sci. USA 98, 14055-14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquardt, T. & Helenius, A. (1992) J. Cell Biol. 117, 505-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhat, M. B. & Ma, J. (2002) J. Biol. Chem. 277, 8597-8601. [DOI] [PubMed] [Google Scholar]
- 28.Szczesna-Skorupa, E. & Kemper, B. (2000) J. Biol. Chem. 275, 19409-19415. [DOI] [PubMed] [Google Scholar]
- 29.Kyte, J. & Doolittle, R. F. (1982) J. Mol. Biol. 157, 105-132. [DOI] [PubMed] [Google Scholar]
- 30.Ye, Y., Meyer, H. H. & Rapoport, T. A. (2001) Nature 414, 652-656. [DOI] [PubMed] [Google Scholar]
- 31.Roy, L., Bergeron, J. J. M., Lavoie, C., Hendriks, R., Gushue, J., Fazel, A., Pelletier, A., Morre, D. J., Subramaniam, V. N., Hong, W., et al. (2000) Mol. Biol. Cell 11, 2529-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen, Y., Holcomb, C. & Moore, H. H. (1993) Proc. Natl. Acad. Sci. USA 90, 6508-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bichet, D., Cornet, V., Geib, S., Carlier, E., Volsen, S., Hoshi, T., Mori, Y. & De Waard, M. (2000) Neuron 25, 177-190. [DOI] [PubMed] [Google Scholar]
- 34.Matsuoka, K., Orci, L., Amherdt, M., Bednarek, S. Y., Hamamoto, S., Schekman, R. & Yeung, T. (1998) Cell 93, 263-275. [DOI] [PubMed] [Google Scholar]
- 35.Zerangue, N., Malan, M. J., Fried, S. R., Dazin, P. F., Jan, Y. N., Jan, L. Y. & Schwappach, B. (2001) Proc. Natl. Acad. Sci. USA 98, 2431-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]