Abstract
Butyrophilin 1a1 (Btn1a1), which is a member of the Ig superfamily, is highly expressed in the lactating mammary gland and is secreted into milk in association with lipid droplets. To determine the potential function of Btn1a1 in milk secretion, we ablated Btn1a1 in mice and analyzed the lactation phenotype of homozygous (Btn1a1-/-) animals. Two mutant mouse lines were generated in which expression of Btn1a1 was either disrupted or eliminated, respectively. The regulated secretion of milk–lipid droplets was severely compromised in both mutant mouse lines in comparison to wild-type animals. Large pools of triacylglycerol accumulated in the cytoplasm of secretory cells, and lipid droplets escaped from the apical surface with disrupted outer membranes. Luminal spaces became engorged with unstable lipid droplets, which coalesced to form large aggregates. The amount of lipid (wt/vol) was elevated, on average by 50%, during the first 10 days of lactation, and the diameter of the droplets was up to seven times larger than the normal diameter. In contrast, there was no significant difference between wild-type and null animals in the relative amounts of skim-milk proteins secreted from Golgi-derived secretory vesicles. Approximately half the pups suckling Btn1a1-/- animals died within the first 20 days, and weaning weights for the surviving pups were 60–80% of those suckling wild-type mice. Thus, expression of Btn1a1 is essential for the regulated secretion of milk–lipid droplets. We speculate that Btn1a1 functions either as a structural protein or as a signaling receptor by binding to xanthine dehydrogenase/oxidase.
Butyrophilin (Btn) genes constitute a subgroup of at least 10 genes in the Ig superfamily identified in human, mouse, and other species (1–5). Most family members structurally comprise two exoplasmic Ig folds (6), a membrane anchor, and a cytoplasmic tail containing short heptad repeats and a B30.2 domain (7). The exoplasmic domain is related to CD80/86 (8), suggesting a role for some family members in costimulation within the immune system. The B30.2 region is predicted to comprise two Ig-like folds (9), is present in a large number of proteins within and outside of the Btn family (7), and may function as a protein-binding domain (10–12).
The eponymous Btn gene (BTN1A1 in humans; Btn1a1 in mouse) is highly expressed in the secretory epithelium of the mammary gland during lactation (13, 14). Other homologues are predominantly expressed in skeletal muscle and the intestine (BTL-II, ref. 2) and erythroid cells (ERMAP, ref. 4). In contrast, BTN2A1 and 2 and BTN3A1, 2, and 3 are widely expressed in many tissues (3, 5), suggesting that the structural domains of Btn proteins may have both universal and tissue-specific functions. In addition, molecular mimicry between the IgI domain of BTN1A1, a milk protein and common dietary antigen, and a similar Ig fold in myelin oligodendrocyte glycoprotein may modulate autoimmune responses in a subset of patients with multiple sclerosis (15).
Despite their potential importance in health and disease, within and outside of the immune system, we do not understand the function of any Btn protein. Btn1a1 was originally named “butyrophilin” to reflect its specific association with milk fat (13), and circumstantial evidence supports the contention that it functions in the secretion of lipid droplets into milk (16). As a step toward determining the function of Btn1a1 in lactation, we produced mutant mouse lines with disrupted Btn1a1 alleles and describe herein the resultant phenotype of Btn1a1-/- animals.
Materials and Methods
Materials. Materials for electrophoresis were from Bio-Rad, and materials for Northern blotting were from Ambion (Austin, TX). Oxytocin was from Sigma, and restriction enzymes were from Boehringer Mannheim. All other reagents were of molecular biology grade or equivalent, from standard supply houses.
Generation of Mice with a Disrupted Btn1a1 Allele. Two strategies were used to ablate Btn1a1 in mice. In a first approach, the 3′ part of exon 2 and the contiguous 5′ region of intron B were replaced with a lacz/neo reporter/selection cassette (17) (Fig. 6 A and B, which is published as supporting information on the PNAS web site). One of four mouse lines with a disrupted Btn1a1 allele, called here BtnKO1, was backcrossed to C57BL/6 mice. Heterozygotes at the fifth and seventh back-crossed generations were intercrossed to provide Btn1a1+/+, Btn1a1+/-, and Btn1a1-/- mice for analysis.
In a second approach, Btn1a1 was ablated by Ingenious Targeting Laboratories (iTL) (Stony Brook, NY) on a fee for service basis. A total of 1.28 kb of 5′ flanking sequence and the gene through the 5′ part of exon 2 were replaced with the 1.8-kb iTLneo cassette (Fig. 6 A and C). One mouse line with a disrupted Btn1a1 allele, called here BtnKO2, was backcrossed once with C57BL/6 mice. Heterozygotes were intercrossed to provide Btn1a1+/+, Btn1a1+/-, and Btn1a1-/- mice for analysis.
Animal Care. Mice were maintained on Formulab Diet 5008 (PMI Nutrition International, Richmond, IN), and water was available ad libitum. The first full day after parturition was counted as day 1 of lactation, and litters were adjusted to eight pups each for comparison of lactation in Btn1a1+/+, Btn1a1+/-, and Btn1a1-/- mice.
Southern Blot Analysis. Homologous recombination was assessed by Southern hybridization of tail DNA digested with either EcoRV (BtnKO1) or KpnI (BtnKO2) (Fig. 6). cDNA probes were labeled with [α-32P]dCTP by random priming and hybridized overnight to the genomic DNA bound to either Zeta-Probe (Bio-Rad) (BtnKO1) or Hybond N+ (Amersham Pharmacia) (BtnKO2) membrane. For BtnKO1 DNA, the membranes were washed twice for 5 min each time with 2× SSC/0.1% (wt/vol) SDS at 65°C and once with 1× SSC/0.1% (wt/vol) SDS at room temperature. For BtnKO2 DNA, the membranes were washed once at room temperature for 10 min in 2× SSPE/0.5% (wt/vol) SDS and twice in 0.5× SSPE/0.5% (wt/vol) SDS at 60°C. The washed blots were exposed to x-ray film at -70 to -80°C.
Northern Blot Analysis. Total RNA was isolated from the fourth and fifth inguinal mammary glands on day 10 of the first or second lactations by using TRIzol reagent (Invitrogen). RNA was separated by electrophoresis in 1% (wt/vol) agarose containing 2.2 M formaldehyde and blotted to Zeta-Probe membrane in 10× SSC. cDNA, labeled by random priming with [α-32P]dCTP, was hybridized to the membrane-bound RNA overnight at 42°C in ULTRAhyb solution. The blots were then washed twice, for 5 min each time, with 2× SSC/0.1% (wt/vol) SDS and twice, for 15 min each time, with 0.1× SSC/0.1% (wt/vol) SDS at 42°C. Bound 32P-labeled cDNA was detected by using either a PhosphorImager (Molecular Dynamics) or x-ray film.
Preparation of Antipeptide Antibody to Btn1a1. A synthetic peptide (American Peptide, Sunnyvale, CA) comprising the C-terminal 21 aa of mouse Btn1a1 (14) was coupled to keyhole limpet hemocyanin and antibody produced in two New Zealand White rabbits as described (18). Antibody specificity was confirmed by Western blot (19).
Collection and Fractionation of Mouse Milk and Mammary Tissue. Mice were separated from their litters for 2 h and injected (i.p.) with 0.2 unit of oxytocin in physiological saline. Milk was expressed from all glands by using a hand-held vacuum device and immediately collected into capillary tubes (20). The milk was centrifuged, and the cream and skim milk fractions were collected by breaking the capillary tubes at the skim-milk/lipid interface. Milk–lipid globule membrane (MLGM) was prepared by sequentially freezing and thawing the cream samples in 62.5 mM Tris·HCl buffer, pH 6.8, containing proteinase inhibitors as described (19).
Lactating mice were asphyxiated in CO2, and the fourth and fifth inguinal mammary glands rapidly excised and homogenized in 10 mM Tris·HCl (pH 7.2)/140 mM NaCl containing proteinase inhibitors. Nuclei were removed by centrifugation at 500 × g for 5 min, and the postnuclear supernatant was separated into membrane and supernatant fractions by centrifugation at 100,000 × g for 1 h.
Microscopy. Lactating mammary tissue was fixed simultaneously with 2.5% (wt/vol) glutaraldehyde and 2% (wt/vol) OsO4 (21) and embedded in Epon. Sections (1–2 μm) for light microscopy were observed directly by differential interference contrast microscopy using a Leica DMIRE2 microscope. For electron microscopy, sections were stained with uranyl acetate and lead citrate and examined with a Zeiss EM10CA electron microscope.
Milk Lipid Assays. Fat percentage (vol/vol) was estimated by “creamatocrit” (22), and data were converted from volume to weight percentage values by using a factor of 0.61 ± 0.05 (20). Milk droplet size was determined from phase contrast micrographs of unfixed milk samples by using simple pci software, version 5.1 (Compix, Tualatin, OR). At least 200 droplets each were measured for wild-type and heterozygote samples, and a minimum of 60 or 120 droplets were measured for Btn1a1-/- samples from the BtnKO1 and BtnKO2 strains, respectively.
Protein Assays. Milk proteins were estimated by separating duplicate samples of skim milk by SDS/PAGE (19) and estimating the relative densities of the Coomassie blue-stained protein bands by using imagequant software (Version 5.2, Molecular Dynamics). One sample was separated, in duplicate, on each successive gel, to make adjustments for differences in staining intensities between gels. Total protein was assayed as described (23); BSA was used as a standard.
Statistical Analysis. Phenotypic differences between Btn1a1+/+, Btn1a1+/-, and Btn1a1-/- animals were tested for statistical significance by using Student's t test, as indicated in the figure legends.
Results
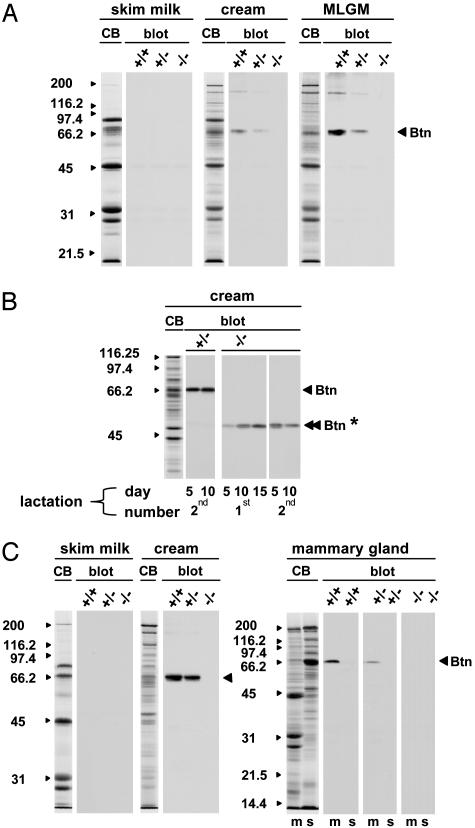
Analysis of total RNA from the lactating mammary tissue of BtnKO1 strain mice confirmed that Btn1a1 had been correctly targeted. Heterozygous mice expressed approximately half the amount of full-sized Btn1a1 mRNA as wild-type mice, and no full-length message was found in Btn1a1-/- animals (Fig. 6B Right). Lipid (cream) and MLGM fractions from the milk of Btn1a1+/- and Btn1a1-/- mice contained approximately half and undetectable amounts of Btn1a1 protein of the expected Mr (66,000), respectively (Fig. 1 A). No Btn1a1 was detected in the skim milk of wild type, heterozygous, or null animals (Fig. 1 A).
Fig. 1.
Analysis of Btn1a1 protein expression by immunoblot in Btn1a1+/+, Btn1a1+/-, and Btn1a1-/- animals. (A) BtnKO1 strain milk samples from day 10 of the first lactation. (B) BtnKO1 strain cream samples from one Btn1a1+/- and one Btn1a1-/- animal analyzed at the lactation stages indicated below the immunoblots. (C) BtnKO2 strain milk and tissue samples from day 10 of the first lactation. Samples were separated in 10% (wt/vol) polyacrylamide gels (30 μg per lane for skim milk and cream fractions, and 40 μg per lane for tissue fractions). CB, Coomassie blue-stained gels representative of each set of samples. Protein profiles were qualitatively similar in all three genotypes, with the exception of Btn1a1 protein in Btn1a1+/- and null mice. The sizes (kDa) of protein standards are indicated to the left of each panel. m, total tissue membrane; s, postmicrosomal supernatant; Btn, Btn1a1 protein; Btn*, fragment of Btn1a1 protein detected in BtnKO1 null mice.
Unexpectedly, however, we resolved mRNA that was ≈0.3 kb smaller than full-length message in some Northern blots of total RNA from Btn1a1+/- and Btn1a1-/- mice (for an example of a heterozygote, see arrowhead in Fig. 6B Right). In addition, immunoreactive protein with an Mr of ≈48,000 was detected in milk lipid fractions from some Btn1a1+/- and null animals (Fig. 1B, double arrowhead and asterisk). To explore the origin of this smaller mRNA and truncated protein, we analyzed total RNA of Btn1a1-/- mice by RT-PCR. DNA was amplified between exons 1B and 3 that was identical in sequence to that of Btn1a1 cDNA except that it lacked the region encoded by exon 2 (data not shown).
Thus, Btn1a1 transcripts of the null allele are adventitiously processed so that the 3′ end of exon 1B is spliced to the 5′ end of exon 3. Because exons 1B and 3 are in phase, a mutant form of Btn1a1 is synthesized lacking the IgI domain encoded by exon 2. Relative amounts of this mutant form of Btn1a1, estimated by quantitative densitometry of immunoblots, were highly variable in cream fractions between Btn1a1-/- animals and accounted for anything from trace amounts (Fig. 1 A) to ≈25% of wild-type values (Fig. 1B).
To assure complete ablation of Btn1a1, we produced a second mutant mouse line (BtnKO2), in which >2.7 kb of DNA encompassing the 5′-flanking region and gene sequence up to the 5′ region of exon 2 were replaced with a neomycin cassette (Fig. 6C). Northern and Western analysis confirmed that Btn1a1 mRNA and protein were effectively reduced to half the amounts of wild-type levels in heterozygotes and that they were eliminated in Btn1a1-/- animals (Figs. 1C and 6C Right).
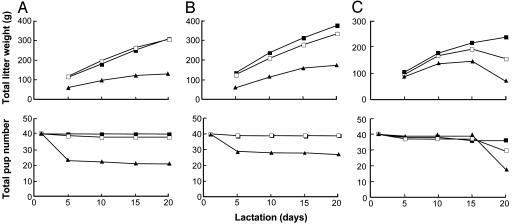
From the time that the first BtnKO1 strain null mice gave birth, it was obvious that lactation was compromised in Btn1a1-/- animals. Total litter weights of pups suckling Btn1a1-/- mothers were substantially lower than the litters maintained by either wild-type or Btn1a1+/- animals throughout lactation in both BtnKO1 and BtnKO2 mouse lines (Fig. 2A and B). Typically, only 50–60% survived until weaning on day 20 (48 of 80 pups in the examples shown in Fig. 2 A and B). Weaning weights for surviving pups suckling Btn1a1-/- mothers were 60–80% of pups suckling wild-type mothers (Table 1, which is published as supporting information on the PNAS web site).
Fig. 2.
Survival of BtnKO1 and BtnKO2 strain litters. Litters were adjusted to eight pups on day 1 of lactation, and the mothers were either not milked (A and B) or milked (C) with a hand-held vacuum device on days 5, 10, 15, and 20 of lactation (n = 5 animals per genotype). Btn1a1+/+ (filled squares), Btn1a1+/- (open squares), and Btn1a1-/- (filled triangles) are shown.
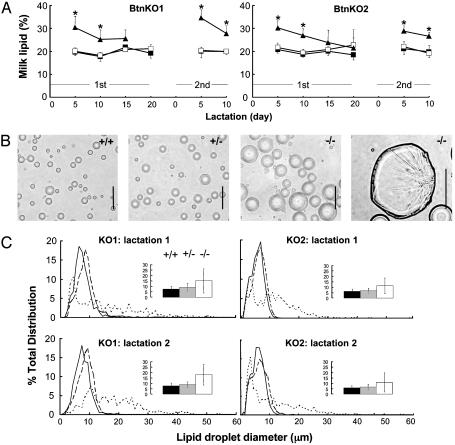
During the first 10 days of lactation, milk from both BtnKO1 and BtnKO2 null animals contained 25–35% (wt/vol) lipid, which on average was 50% higher than the levels of fat in Btn1a1+/+ and Btn1a1+/- mice (Fig. 3A, examples for the first lactation and half of the second). On average, lipid droplets from Btn1a1-/- mice were two to three times larger in diameter than those from wild type or heterozygous animals (Fig. 3C Insets) and they varied widely from <1 to >50 μm (Fig. 3 B and C). Some of the larger droplets appeared as misshapen globules of triacylglycerol that had lost the outer MLGM (Fig. 3B Right). The cream fractions from Btn1a1-/- mice were noticeably less stable than samples from wild-type and Btn1a1+/- animals when they were subjected to cycles of freezing and thawing to release MLGM from the lipid and significantly less membrane was recovered from the globules.
Fig. 3.
Analysis of lipid in the milk of Btn1a1+/+, Btn1a1+/-, and Btn1a1-/- animals. (A) Amount of milk lipid (%, wt/vol) in the first and first half of the second lactations of BtnKO1 and BtnKO2 strains, as indicated (values, mean ± SD, n = 5 animals per genotype). Btn1a1+/+ (filled squares), Btn1a1+/- (open squares), and Btn1a1-/- (filled triangles) are shown. (B) Phase contrast micrographs of lipid droplets from milk of the three genotypes of BtnKO2 strain mice, as indicated. (Bars: 25 μm.) (C) Profiles of lipid droplet size in milk from the three genotypes on day 10 of the first and second lactations of BtnKO1 and BtnKO2 lines as indicated (average of five lipid samples per genotype). Btn1a1+/+ (solid line), Btn1a1+/- (dashed line), and Btn1a1-/- (dotted line) are shown. (Insets) Mean diameter of lipid droplets from the same samples: Btn1a1+/+ (black bars), Btn1a1+/- (gray bars), and Btn1a1-/- (white bars) are shown (values are mean ± SD, n = 5 samples per genotype). *, Btn1a1-/- values significantly different from Btn1a1+/+ and Btn1a1+/- mice (P < 0.01).
Thus, neonatal pups born to Btn1a1-/- mothers appeared unable to get sufficient milk because large unstable lipid droplets interfered with the regulated expression of milk from the gland. To test this possibility, we milked mice every 5 days with a vacuum device to help clear the gland of coagulated lipid. Pup survival was enhanced for the first 15 days; thereafter, there was a precipitous drop in survival numbers (Fig. 2C). In the example shown in Fig. 2C, 22 of the 39 pups died between day 15 and 20, and a further 4 had died by day 25.
To assess possible effects on the secretion of milk proteins, we compared the relative amounts of the major skim milk proteins over a complete first lactation and half of a second by quantitative densitometry. The relative quantities of lactoferrin, α1-, β-, and γ-casein were essentially similar in all three genotypes in both BtnKO1 and BtnKO2 strains (data not shown and Fig. 7, which is published as supporting information on the PNAS web site).
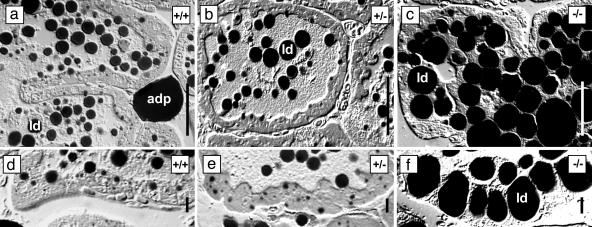
At the resolution of the light microscope the general architecture of the mammary tissue at peak lactation was broadly similar in wild-type and Btn1a1+/- mice from both mutant lines (an example of BtnKO2 is shown in Fig. 4 a, b, d, and e). However, in Btn1a1-/- mice, the secretory cells contained strikingly large lipid droplets, and many luminal spaces were completely filled with aggregated lipid (Fig. 4 c and f).
Fig. 4.
Light micrographs of lactating mammary tissue from Btn1a1+/+, Btn1a1+/-, and Btn1a1-/- BtnKO2 strain mice. Differential interference contrast micrographs are shown. Lipid droplets in milk and tissue (ld) and adipocytes (adp) stain black with OsO4. (Bars: 50 μm in a–c; 10 μm in d–f.)
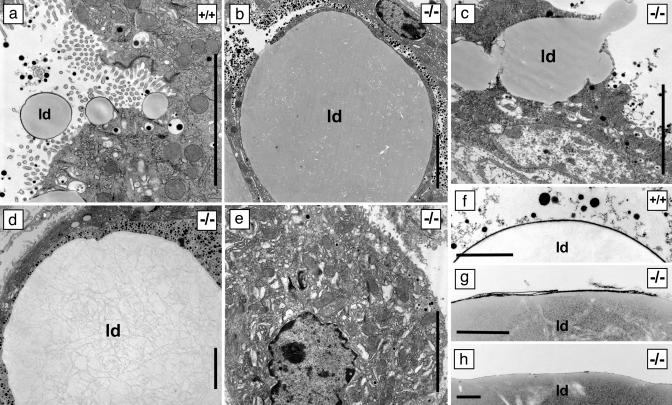
At the resolution of the electron microscope, the cytoplasm of Btn1a1-/- secretory cells was engorged with lipid that pushed organelles to the cellular periphery (compare Fig. 5 a and b). In some cells, the accumulated lipid appeared to escape directly by breaking through holes in the apical surface (Fig. 5c). Luminal spaces became filled with fat that coalesced into large aggregates with an internal structure presumably comprising detritus from the surfaces of merged lipid droplets (Fig. 5d). Many of the secreted droplets were coated with a fragmented membrane layer compared to wild type (Fig. 5 f and g), and areas of the larger lipid aggregates were solely bounded by an amorphous layer (Fig. 5h). In some cells that were devoid of lipid, organelles were structurally disorganized (Fig. 5e), presumably because the cells previously harbored large lipid droplets that had escaped from the apical surface.
Fig. 5.
Electron micrographs of lactating mammary tissue from Btn1a1+/+ and Btn1a1-/- BtnKO1 and BtnKO2 strain mice. (c, d, and f–h) BtnKO1. (a, b, and e) BtnKO2. (a–e) Tissue. (f–h) Milk–lipid droplet surface. ld, lipid droplet. (Bars: 5 μm in a–e; 1 μm in f–h.)
Discussion
The phenotype of the Btn1a1-/- mouse provides compelling evidence that expression of Btn1a1 in the lactating mammary gland is essential for regulated milk secretion. Furthermore, because lipid accumulates in the mammary cells of null mice and there are no obvious effects on the exocytosis of skim-milk proteins from secretory vesicles, the data support the possibility that Btn1a1 plays a direct role in the assembly, transport, or secretion of lipid droplets.
In the absence of Btn1a1, lipid droplets still form in the cytoplasm, suggesting that there is minimal inhibition of triacylglycerol synthesis and droplet assembly. Therefore, Btn1a1 most probably functions at some postsynthetic step, either in the transport of lipid droplets to the cell apex or in the formation of the MLGM and the expulsion of lipid from the cell. A possible role in lipid transport through the cytoplasm is unlikely because, at steady state, Btn1a1 is largely concentrated on the apical plasma membrane and around budding lipid droplets (13). In addition, in many species, the cytoplasmic domain of Btn1a1 is a major constituent of a protein complex, which forms between the surface of lipid droplets and the outer membrane bilayer as the droplets emerge from the cell (16). Taken together, these observations suggest that Btn1a1 plays a significant role in the terminal steps of lipid secretion itself.
An increasing body of evidence suggests that Btn1a1 functions by binding to the redox enzyme xanthine dehydrogenase/oxidase (Xdh). Xdh is incorporated into the MLGM and colocalizes with the cytoplasmic tail of Btn1a1 between the lipid droplet surface and outer membrane (16). Because of their close proximity, the two proteins can be cross-linked together with exogenous bifunctional cross-linking agents in either intact lipid droplets (A. U. Rao and I.H.M., unpublished data) or isolated membrane (24). Furthermore, the cytoplasmic domain of Btn1a1, expressed as a fusion protein with glutathione S-transferase, binds to Xdh in vitro (25). Most strikingly, Xdh+/- mice display many of the same deficiencies in lipid secretion as Btn1a1-/- mice, including the accumulation of lipid within the mammary epithelium and the secretion of large lipid droplets with fragmented outer membranes (26) (lactation in Xdh-/- mice could not be analyzed because they did not live beyond 6 weeks).
How might interaction between Btn1a1 and Xdh effect lipid secretion? Two possibilities deserve special consideration. First, Btn1a1 as an integral protein may function as a structural scaffold linking the outer membrane bilayer to Xdh and potentially other proteins such as adipophilin on the droplet surface (16, 27, 28). Progressive interactions between Btn1a1 and Xdh may proceed as a wave around the droplet as it emerges from the cell, forming a protein complex that stabilizes association of the droplet with the membrane, effectively precipitating formation of the MLGM (16). Second, Btn1a1 has the canonical structure of a receptor (6, 29) and there is increasing evidence that Xdh may serve as a signaling molecule through the generation of reactive oxygen species (30). Btn1a1 could therefore function as a signaling receptor that, through interactions with Xdh, stimulates the production of reactive oxygen intermediates, which either directly or indirectly regulate components of the secretory apparatus.
Ablation of Btn1a1 has several significant consequences besides obvious effects on lipid secretion. Most notably, triacylglycerol synthesis does not shut down as lipid accumulates in the cytoplasm despite the fact that the key lipogenic enzyme, acetyl–CoA carboxylase, is acutely and negatively regulated by the enzyme product, malonyl–CoA (31). Presumably, the synthesis of fatty acids and triacyglycerols in Btn1a1-/- mice continues at a rate sufficient to maintain the cytoplasmic pools of malonyl–CoA at subinhibitory concentrations. Alternatively, potential inhibition caused by high levels of malonyl–CoA may be ablated by an increase in levels of citrate, which competitively overrides inhibition caused by malonyl–CoA (31). Estimation of the extent to which lipid synthesis is depressed, if at all, in knockout animals will require appropriate comparisons of lipid synthetic rates in Btn1a1-/- and wild-type mammary tissue.
Another significant consequence of ablating Btn1a1 is the deleterious effect on litter survival. We presume that the pups die because they are unable to acquire normal amounts of milk because the alveolar luminae and ducts become variably clogged with unstable fat. A similar outcome was recently described for the transgenic akt mouse, which accumulates large quantities of milk lipid on expression of a constitutively active form of protein kinase B (Akt) (32).
The Btn1a1-/- mouse illustrates the importance of the MLGM for maintaining the stability of milk–lipid globules and the consequences to lactation and the survival of the neonate when this membrane is disrupted. Analysis of mutations within human BTN1A1 and XDH (33) may reveal some of the causes of lactation insufficiency in humans, a problem that, by some estimates, affects 5% of women worldwide (34). Further analysis of the functional domains of Btn1a1, including the Ig folds and the B30.2 domain, may also serve as a paradigm for understanding the functions of other members of the Btn family and related proteins.
Supplementary Material
Acknowledgments
We thank Ingenious Targeting Laboratories for generating the BtnKO2 mouse line, Charles River Laboratories for assistance with genotyping mice, and Liane Langbehn for maintenance of the mouse colonies. Iqbal Hamza, Anita Rao, and Abiy Zegeye all helped with various technical aspects of the project. Jan Ehrlich is thanked for help with the electron microscopy and Larry Douglas for assistance with the statistical analysis. This work was funded by grants from the National Science Foundation, the U.S. Department of Agriculture, the North Atlantic Treaty Organization, and the Maryland Agricultural Experiment Station (to I.H.M.). The U.K. Biotechnology and Biological Research Council provided support for the Gene Targeting Laboratory and A.J.H.S.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Btn1a1, butyrophilin 1a1; MLGM, milk–lipid globule membrane; Xdh, xanthine dehydrogenase/oxidase.
References
- 1.Shibui, A., Tsunoda, T., Seki, N., Suzuki, Y., Sugano, S. & Sugane, K. (1999) J. Hum. Genet. 44, 249-252. [DOI] [PubMed] [Google Scholar]
- 2.Stammers, M., Rowen, L., Rhodes, D., Trowsdale, J. & Beck, S. (2000) Immunogenetics 51, 373-382. [DOI] [PubMed] [Google Scholar]
- 3.Rhodes, D. A., Stammers, M., Malcherek, G., Beck, S. & Trowsdale, J. (2001) Genomics 71, 351-362. [DOI] [PubMed] [Google Scholar]
- 4.Ye, T.-Z., Gordon, C. T., Lai, Y.-H., Fujiwara, Y., Peters, L. L., Perkins, A. C. & Chui, D. H. K. (2000) Gene 242, 337-345. [DOI] [PubMed] [Google Scholar]
- 5.Ruddy, D. A., Kronmal, G. S., Lee, V. K., Mintier, G. A., Quintana, L., Domingo, R., Meyer, N. C., Irrinki, A., McClelland, E. E., Fullan, A., et al. (1997) Genome Res. 7, 441-456. [DOI] [PubMed] [Google Scholar]
- 6.Linsley, P. S., Peach, R., Gladstone, P. & Bajorath, J. (1994) Protein Sci. 3, 1341-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henry, J., Mather, I. H., McDermott, M. F. & Pontarotti, P. (1998) Mol. Biol. Evol. 15, 1696-1705. [DOI] [PubMed] [Google Scholar]
- 8.Bajorath, J., Peach, R. J. & Linsley, P. S. (1994) Protein Sci. 3, 2148-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seto, M. H., Liu, H.-L. C., Zajchowski, D. A. & Whitlow, M. (1999) Proteins 35, 235-249. [PubMed] [Google Scholar]
- 10.Schweiger, S., Foerster, J., Lehmann, T., Suckow, V., Muller, Y. A., Walter, G., Davies, T., Porter, H., van Bokhoven, H., Lunt, P. W., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 2794-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niikura, T., Hashimoto, Y., Tajima, H., Ishizaka, M., Yamagishi, Y., Kawasumi, M., Nawa, M., Terashita, K., Aiso, S. & Nishimoto, I. (2003) Eur. J. Neurosci. 17, 1150-1158. [DOI] [PubMed] [Google Scholar]
- 12.Zhai, L. M., Dietrich, A., Skurat, A. V. & Roach, P. J. (2004) Arch. Biochem. Biophys. 421, 236-242. [DOI] [PubMed] [Google Scholar]
- 13.Franke, W. W., Heid, H. W., Grund, C., Winter, S., Freudenstein, C., Schmid, E., Jarasch, E.-D. & Keenan, T. W. (1981) J. Cell Biol. 89, 485-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ogg, S. L., Komaragiri, M. V. S. & Mather, I. H. (1996) Mamm. Genome 7, 900-905. [DOI] [PubMed] [Google Scholar]
- 15.Guggenmos, J., Schubart, A. S., Ogg, S. L., Andersson, M., Olsson, T., Mather, I. H. & Linington, C. (2004) J. Immunol. 172, 661-668. [DOI] [PubMed] [Google Scholar]
- 16.Mather, I. H. & Keenan, T. W. (1998) J. Mammary Gland Biol. Neoplasia 3, 259-273. [DOI] [PubMed] [Google Scholar]
- 17.Nehls, M., Kyewski, B., Messerle, M., Waldschutz, R., Schuddekopf, K., Smith, A. J. H. & Boehm, T. (1996) Science 272, 886-889. [DOI] [PubMed] [Google Scholar]
- 18.Banghart, L. R., Chamberlain, C. W., Velarde, J., Korobko, I. V., Ogg, S. L., Jack, L. J. W., Vakharia, V. N. & Mather, I. H. (1998) J. Biol. Chem. 273, 4171-4179. [DOI] [PubMed] [Google Scholar]
- 19.Neira, L. M. & Mather, I. H. (1990) Protoplasma 159, 168-178. [Google Scholar]
- 20.Teter, B. B., Sampugna, J. & Keeney, M. (1990) J. Nutr. 120, 818-824. [DOI] [PubMed] [Google Scholar]
- 21.Franke, W. W., Krien, S. & Brown, R. M. (1969) Histochemie 19, 162-164. [DOI] [PubMed] [Google Scholar]
- 22.Lucas, A., Gibbs, J. A. H., Lyster, R. L. J. & Baum, J. D. (1978) Br. Med. J. 1, 1018-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith, P. K., Krohn, R. I., Hermanson, G. T., Mallia, A. K., Gartner, F. H., Provenzano, M. D., Fujimoto, E. K., Goeke, N. M., Olson, B. J. & Klenk, D. (1985) Anal. Biochem. 150, 76-85. [DOI] [PubMed] [Google Scholar]
- 24.Valivullah, H. M. & Keenan, T. W. (1989) Int. J. Biochem. 21, 103-107. [DOI] [PubMed] [Google Scholar]
- 25.Ishii, T.-N., Aoki, N., Noda, A., Adachi, T., Nakamura, R. & Matsuda, T. (1995) Biochim. Biophys. Acta 1245, 285-292. [DOI] [PubMed] [Google Scholar]
- 26.Vorbach, C., Scriven, A. & Capecchi, M. R. (2002) Genes Dev. 16, 3223-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heid, H. W., Schnölzer, M. & Keenan, T. W. (1996) Biochem. J. 320, 1025-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McManaman, J. L., Palmer, C. A., Wright, R. M. & Neville, M. C. (2002) J. Physiol. 545, 567-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jack, L. J. W. & Mather, I. H. (1990) J. Biol. Chem. 265, 14481-14486. [PubMed] [Google Scholar]
- 30.Meneshian, A. & Bulkley, G. B. (2002) Microcirculation 9, 161-175. [DOI] [PubMed] [Google Scholar]
- 31.Allred, J. B. & Reilly, K. E. (1997) Prog. Lipid Res. 35, 371-385. [DOI] [PubMed] [Google Scholar]
- 32.Schwertfeger, K. L., McManaman, J. L., Palmer, C. A., Neville, M. C. & Anderson, S. M. (2003) J. Lipid Res. 44, 1100-1112. [DOI] [PubMed] [Google Scholar]
- 33.Simmonds, H. A., Reiter, S. & Nishino, T. (1995) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S. & Valle, D. (McGraw–Hill, New York), Vol. 2, pp. 1781-1797. [Google Scholar]
- 34.Neifert, M. R. (2001) Pediatr. Clinics North Am. 48, 273-297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.