Abstract
An R120G missense mutation in the small heat shock protein α-B-crystallin (CryABR120G) causes desmin-related cardiomyopathy (DRM). DRM is characterized by the formation of aggregates containing CryAB and desmin, and it can be recapitulated in transgenic mice by cardiac-specific expression of the mutant protein. In this article, we show that expression of CryABR120G leads to the formation of electron-dense bodies characteristic of the DRMs and identify these bodies as aggresomes, which are characteristic of the neurodegenerative diseases. Cardiomyocytes transfected with adenovirus containing CryABR120G establish the necessity and sufficiency of CryABR120G expression for aggresome formation. The commonality of these aggresomes with oligomeric protein aggregates found in the amyloid-related degenerative diseases was corroborated by the presence of high levels of amyloid oligomers that may represent a primary toxic species in the amyloid diseases. These oligomeric amyloid intermediates are present also in cardiomyocytes derived from many human dilated and hypertrophic cardiomyopathies.
Human heart failure is the leading cause of death in the developed world, and it represents a final common endpoint for several disease entities, including hypertension, coronary artery disease, and the cardiomyopathies (1, 2). A lack of pathogenic commonality is underscored by the large number of mutations in different classes of cardiac proteins that have been linked to dilated and hypertrophic cardiomyopathy (HCM) (3). Mutations in the cytoskeletal and associated proteins can be causative because these proteins function in structural, sensor, and signaling roles in the normal and diseased cardiomyocyte (4). For example, up-regulation of the intermediate filament protein desmin occurs in cardiac disorders such as cardiac hypertrophy and congestive heart failure (CHF) (5), and desmin mutations are associated with desmin-related cardiomyopathy (DRM) and idiopathic dilated cardiomyopathy (6, 7). Mutations in other proteins also have been associated with the DRMs, and genetic evidence linking an R120G mutation in α-B-crystallin (CryAB, CryABR120G) to human DRM (8) prompted a series of experiments in which we showed that cardiac-restricted transgenic (TG) expression of CryABR120G was sufficient to cause heart failure in a mouse model (9). Although up-regulation of CryAB is associated with disease states including DRM, its synthesis probably represents a general cellular response to stress because CryAB has chaperone-like activity. Indeed, CryAB, which binds to both desmin and cytoplasmic actin, probably participates normally as a chaperone in intermediate filament formation and maintenance in the heart.
The α-crystallins (α-A and α-B) were of interest initially as major structural proteins present in the lens of the vertebrate eye. However, the discovery that they were related to the small heat shock proteins (hsps) in Drosophila (10) prompted reevaluation of their broader role(s), and it is now known that CryAB belongs to the small hsp family (11), which includes hsp20, hsp22, hspB2, B3, hsp25, hsp27, and the myotonic dystrophy kinase-binding protein. Although it was discovered and classified as a lens protein originally (12), CryAB is found also in nonlenticular cell types, with cardiac and skeletal muscles containing the highest CryAB levels among the nonlenticular tissues (13). CryAB binds both desmin and cytoplasmic actin, and it possesses molecular chaperone function in vitro (14–16). In vivo, CryAB exists as oligomers, with recent mass spectrometry data indicating that the predominant species is a 28-subunit oligomer (17). Like other chaperones, CryAB responds to stressful conditions by binding to unfolded proteins and preventing their denaturation and aggregation.
We hypothesized that the basis of the cardiomyopathic pathology caused by expression of CryABR120G is, at least partially, in the altered interactions of the protein with its partners and subsequent formation of the electron-dense bodies. In this article, we show that CryABR120G expression results in the formation of aggregates that are characteristic of DRM. These bodies are cardiomyocyte-based, and they are indistinguishable from aggresomes. As is the case for aggresome formation in the amyloidoses, development of the large electron-dense bodies found in later stages of CryABR120G DRM depends on microtubule-based transport. The data also show that the aggresomes present in diseased hearts contain an amyloid oligomer that may represent a primary toxic species in Alzheimer's disease and other amyloid-based degenerative diseases. Finally, we link the animal models to human disease by showing that these soluble amyloids are present in human dilated cardiomyopathy and HCM and that they are associated with the contractile apparatus. The data point to a pathogenic process for cardiovascular disease and provide a link to the degenerative amyloidoses.
Materials and Methods
TG Mice. Male FVB/N mice with cardiac-specific overexpression of normal CryAB (wild type) or CryAB containing the R120G missense mutation (CryABR120G), driven by the α-myosin heavychain promoter, have been described (9). TG mice were identified by PCR analysis of genomic DNA isolated from tail clips.
Cardiomyocyte Cultures and Adenovirus Infection. After isolation of the rat neonatal cardiomyocytes, cells were grown in two-wellchambered Permanox slides (Nalge) coated with either gelatin or Pronectin-F (BioSource International, Camarillo, CA). Replication-deficient recombinant adenoviruses were made by using the AdEasy system (18). The cells were normally transfected at an multiplicity of infection of 10. To distinguish the TG products from endogenous CryAB protein, a Flag epitope was introduced at the N termini of both the wild-type CryAB and CryABR120G cDNAs. The filter assay was performed essentially as described (19). Lysate from the transfected cells was centrifuged at 12,000 × g for10min, and the resultant pellet was diluted into 0.2 ml of 2% SDS and filtered through a 0.2-μm nitrocellulose membrane. The aggregate on the membrane was detected with anti-CryAB.
Immunohistochemistry. Immunohistochemical analyses were performed as described (9). All cell culture media were obtained from Life Technologies (Rockville, MD), unless indicated otherwise. Alexa 488-conjugated anti-rabbit and Alexa 568-conjugated anti-mouse antibodies, phalloidin–Alexa 594, and To-Pro-3 for nuclear staining were obtained from Molecular Probes. Anti-CryAB antibody (SPA-223) and anti-HSP25 (SPA-801) were obtained from Stressgen Biotechnologies (Victoria, Canada), anti-Flag was obtained from Stratagene, and SEC61α (G-20) and ubiquitin (P4D1) were obtained from Santa Cruz Biotechnology. Anti-cTnI antibody (MAB1691) was obtained from Chemicon, and antitubulin (ATN02) antibody was obtained from Cytoskeleton (Denver).
Results
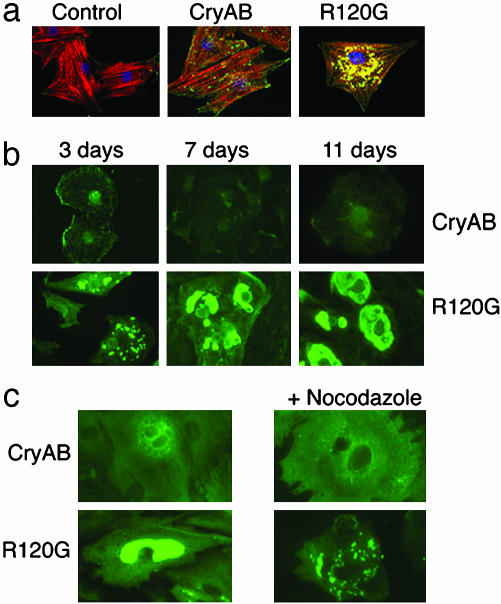
The Electron-Dense Bodies in CryABR120G-DRM Are Aggresomes. In the TG animals, the perinuclear location of the aggregates, as well as their appearance and association with disease, were reminiscent of a class of pathologies recognized as conformational diseases that are associated with protein misfolding and subsequent aggregation (20). Sequestration of the misfolded proteins is characteristic of this disease class, and it is a dynamic process with a well defined time course. The aggregates, which are transported retrogradely on the microtubules and accumulate in the perinuclear region, have been termed aggresomes (21, 22). Aggresomes have been detected in many diseases, including aging-related neurodegeneration and systemic amyloidosis (22, 23). To explore the hypothesis that the electron-dense bodies present in the DRMs are indistinguishable from and can be defined as aggresomes, we developed a cell culture-based assay system in which CryAB aggregate formation could be studied in detail. Adenovirus vectors containing either CryAB or CryABR120G cDNAs were used to express the respective proteins in neonatal rat cardiomyocytes (Fig. 1). In contrast to the CryAB transfected cardiomyocytes, cells that had been transfected with CryABR120G showed significant perinuclear aggregates (Fig. 1a).
Fig. 1.
Immunocytochemical analyses in CryAB- and CryABR120G-transfected rat neonatal cardiomyocytes. (a) CryAB and CryABR120G distribution in permeabilized rat neonatal cardiomyocytes. Control, empty adenoviral vector. (b) Expression of CryABR120G results first in the formation of small aggregates, which then coalesce around the nucleus and whose size increases in a time-dependent manner. (c) Formation of aggregates is microtubule-dependent. After 3 days in culture, 10 μM nocodazole was added.
Early aggresome formation is characterized by the presence of protoaggresomes that are distributed throughout the cytoplasm. These bodies are then transported to a perinuclear location by means of attachment as cargo to the dynein motor, which uses retrograde transport along the microtubule system (20). Aggregate formation in the CryABR120G-transfected myocytes was determined at 3, 7, and 11 days after transfection (Fig. 1b). In the CryAB-transfected myocytes, CryAB was relatively unorganized, and it appeared in a diffuse pattern throughout the cytoplasm and could be removed easily by mild treatment with Triton X-100. In the CryABR120G-transfected myocytes after 3 days, even after detergent treatment, CryABR120G was organized in small, intensely staining bodies, which at 3–11 days coalesced gradually into characteristic aggresome-like bodies, which were invariably perinuclear (Fig. 1b).
To determine whether formation of these bodies was microtubule-dependent, the microtubule-depolymerizing agent nocodazole was added to myocytes 3 days after transfection with either CryAB or CryABR120G (Fig. 1c). Microtubule depolymerization abrogated the formation of the perinuclear aggresomes. As expected, the drug did not prevent or reverse the formation of the protoaggresomes but, rather, prevented their subsequent perinuclear coalescence. Consistent with the requirements for aggresome formation, perinuclear aggregate formation in the cardiomyocytes depends on microtubule transport.
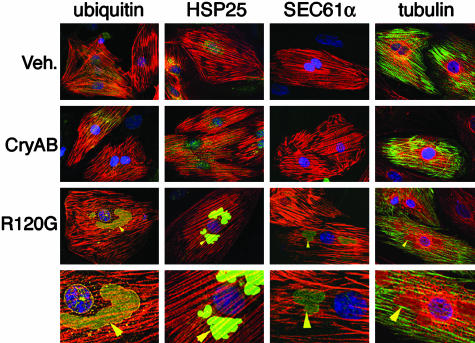
Aggresomes invariably recruit cytoplasmic components including chaperones and elements of the ubiquitin and proteasome pathways, presumably to aid in the clearance of the aggregated proteins (20). Consistent with the general composition of aggresomes in other cell types, the CryABR120G bodies in the transfected cardiomyocytes contained hsp25, ubiquitin, β-tubulin, and SEC61α, a membrane protein that is essential for protein translocation into endoplasmic reticulum cisternae (Fig. 2). The same immunostaining patterns were observed in sections derived from CryABR120G TG mice (data not shown), and they are consistent with the aggresome composition found in some degenerative diseases (21, 24, 25).
Fig. 2.
Aggresomal markers in the CryABR120G aggregates. The CryABR120G-induced aggregates are immunoreactive with antibodies for ubiquitin, HSP25, the endoplasmic reticulum complex marker protein SEC61α, and tubulin. For ubiquitin, HSP25, and SEC61α, actin is stained red (phalloidin). For tubulin, actin is stained green and tubulin is red. Nuclei are stained blue with To-Pro-3. The bottom row shows a ×2 magnification of the perinuclear regions of the row above. Arrowheads indicate immunopositive aggregates. Veh., empty adenovirus vector.
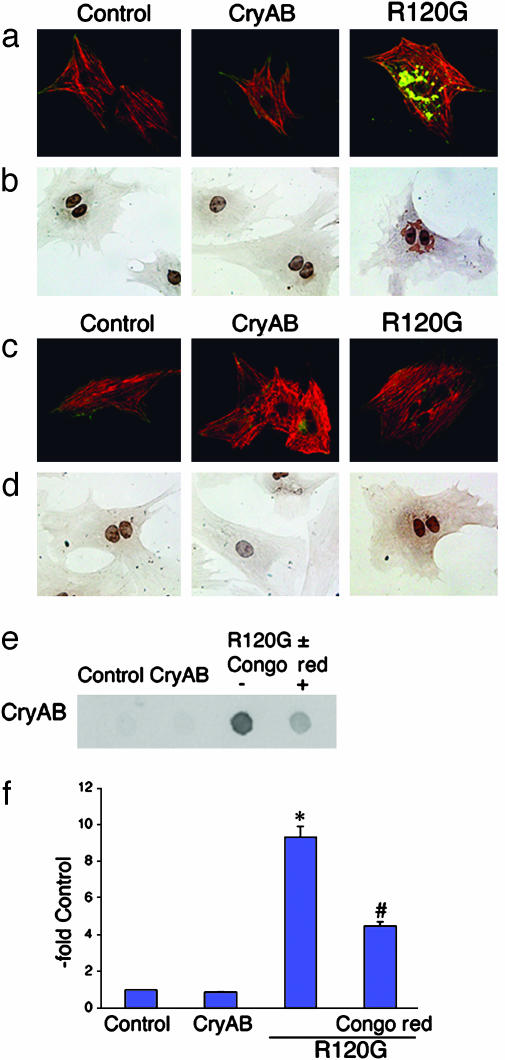
The Aggresomal Bodies in Cardiomyocytes Are Amyloidphilic. In an attempt to define further the relationship of these electron-dense bodies to the amyloid-based degenerative diseases, we tested their ability to react positively to the amyloidphilic dye Congo red (26, 27). Congo red-positive inclusions were abundant in CryABR120G-transfected cardiomyocytes and were undetectable in cells that were transfected with either empty adenovirus or with CryAB adenovirus (Fig. 3 a and b). Similar to the perinuclear location of the aggresomes in CryABR120G-transfected cardiomyocytes, Congo red staining was restricted largely to the perinuclear region. There are conflicting data with respect to the ability of Congo red to interfere with or enhance amyloid fibril formation, but a recent study (28) showed that although low concentrations led to enhanced fibril formation, high concentrations inhibited the process effectively. Continuous treatment of the cells with high concentrations of the dye prevented aggresome formation effectively, and no Congo red-positive bodies could be observed in the cells (Fig. 3 c and d). We established a biochemically based method for aggresome quantitation (see Materials and Methods) and quantitated the amount of aggregated protein present by filtration through nitrocellulose. The filter-based assay confirmed the effectiveness of Congo red inhibition of aggresome formation (Fig. 3 e and f).
Fig. 3.
CryABR120G aggregates show amyloid-like characteristics. (a) At 3 days after initial plating, rat neonatal cardiomyocytes expressing CryABR120G contain the characteristic aggregates. Control, empty adenoviral vector. (b) Perinuclear aggregates stain positive for Congo red. The nuclei are also stained. (c) Cultures were treated with Congo red starting at day 1. (d) Congo red staining of these cultures confirmed the lack of amyloid material and the effectiveness of the dye in preventing aggresome formation. CryAB (green) and cardiac TnI (red) are shown in a and c. (e and f) Quantitation of aggregates. The filter retardation assay confirms that Congo red treatment partially prevents aggregate formation. *, P < 0.001; #, P < 0.05.
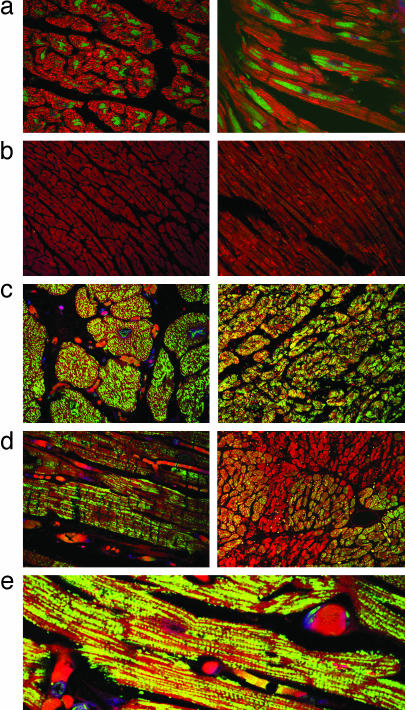
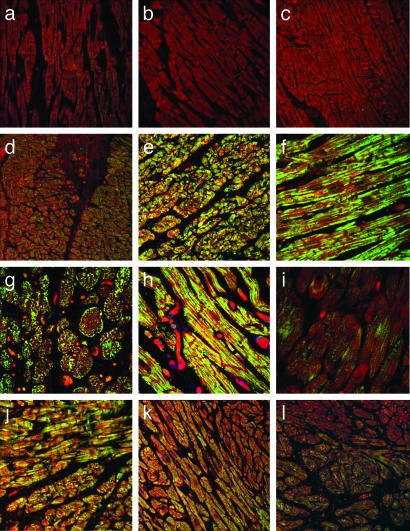
Amyloid Oligomers Are Present in CryABR120G Cardiomyocytes and in Human Heart Failure. The mechanisms underlying the toxicity of protein aggregates and amyloids are unclear. There is intense debate about what constitutes the pathogenic species and whether the mature fibrillar deposits or some other intermediate entity is responsible for the onset of pathogenesis (29). In at least some cases, it appears that toxicity lies not in the formation of the insoluble aggregates or fibrils but rather in soluble oligomeric intermediates (30, 31). By using an antibody that specifically recognizes a conformation-dependent epitope that is common to all types of soluble amyloid oligomers regardless of sequence (30), we probed for the presence of these structures in the CryABR120G mouse hearts. Immunoreactivity was abundant, distributed throughout the heart, and restricted to the diseased cardiomyocytes (Fig. 4a). Samples from two human hearts with no overt signs of cardiovascular disease showed little or no immunoreactivity with the antibody (Fig. 4b). However, samples from hearts with either HCM (Fig. 4c) or dilated cardiomyopathy (Fig. 4d) had abundant staining with decoration of the sarcomeres being sometimes apparent (Fig. 4e). Multiple samples from an additional three normal hearts and nine hearts from patients suffering from cardiovascular disease were tested subsequently to determine whether the cardiomyocyte-based soluble amyloid oligomers were widespread (Fig. 5). Although reactivity differed among the samples, the ventricular cardiomyocytes in all of the diseased hearts showed significant immunoreactivity to the amyloid oligomer antibody.
Fig. 4.
Amyloid oligomer is present in both mouse and human heart failure. (a) Transverse and longitudinal sections derived from the hearts of CryABR120G mice show the subcellular distribution of the amyloid oligomer, which correlates closely to the perinuclear location of the aggresomes. (b) Ventricular sections derived from normal human ventricles. (c) Sections derived from a 33-year-old woman suffering from nonobstructive HCM. (d) Ventricular sections derived from a 22-year-old male with idiopathic dilated cardiomyopathy. (e) Higher-magnification picture of ventricular section derived from a 37-year-old woman with nonischemic cardiomyopathy. Anti-oligomer antibody is shown in green, and phalloidin (actin staining) is shown in red.
Fig. 5.
Amyloid oligomer reactivity in human heart failure samples. (a–c) Samples derived from normal hearts. (d) A 50-year-old Caucasian male diagnosed with dilated cardiomyopathy (DCM) 1 year before death. (e) A 33-year-old Caucasian female with HCM. Complete heart block with pacemaker placement is shown. The patient died of an arrhythmia. (f) A 57-year-old African-American female with idiopathic DCM diagnosed 6 years before death. (g) A 37-year-old Caucasian male with idiopathic DCM. The patient died of CHF. (h) A 34-year-old African-American female with nonischemic cardiomyopathy diagnosed 3 years before death from CHF. (i) A 16-year-old Caucasian male diagnosed with idiopathic DCM 2 years before death. The patient died of an arrhythmia. (j) A 45-year-old Caucasian male with family history of HCM was diagnosed as being in the dilated stage of HCM 1.5 years before death. The patient died of CHF. (k) A 22-year-old African-American male with idiopathic DCM diagnosed 6 months before death. The patient died of CHF. (l) A 32-year-old Caucasian male with muscular dystrophy identified tentatively as Becker's. He died of pneumonia with complicating episodes of cardioverted ventricular tachycardia–fibrillation.
Discussion
The DRMs are characterized by the formation of electron-dense bodies containing the intermediate filament, desmin (8, 32). Although desmin-positive aggregates are a common feature in DRM (33), the CryABR120G-induced aggregates consist mainly of proteins that are found within aggresomes and show only minor immunoreactivity against an anti-desmin antibody (data not shown). Failure of CryABR120G to assist in intermediate filament organization is, therefore, unlikely to be the primary stimulus for aggregate formation in cardiomyocytes or other cell types (34). The R120G mutation is dominant negative (8, 9) and shows decreased β-sheet secondary structure of CryAB and an altered aromatic residue environment, resulting in defective chaperone activity (35). It is unlikely that a simple loss of function of chaperone activity is the major underlying pathogenic stimulus because the cardiac phenotype of the CryAB knockout is relatively benign (36). If the simple loss of CryAB chaperone function were the sole determinant for a developing cardiomyopathy, the absence of the protein would result in significant cardiovascular disease. We have examined the hearts of the CryAB null mice, and although there are some abnormalities that can be observed with light microscopy, there is no evidence of the electron-dense aggregates (R. Klevitsky and J.R., unpublished data).
CryABR120G protein shows impaired chaperone activity and abnormal supramolecular structure (34). Although these data are consistent with the hypothesis that CryABR120G is directly amyloidogenic, they do not prove it, and the causative mechanisms remain obscure because the protein can interact with many partners. For example, CryAB can interact directly with elements of the proteasome, causing pleiotropic effects on directed protein degradation. CryABR120G shows enhanced binding to the protein FBX4, an F box protein that is an integral part of the ubiquitin protein isopeptide ligase complex Skp-Cullin-F box (SCF), and the mutated protein recruits this component effectively to an insoluble cellular fraction, with a subsequent effect on ubiquitination patterns (37). CryAB can also bind to actin (16) as well as titin (38), and the protein has been detected at the M-line, which is consistent with its interaction with myomesin-2 (39). By using the hsp20 domain as bait in yeast two-hybrid screens, we have isolated titin, TnI, myomesin, and myosin-binding protein C, and analyses in which the R120G-hsp20 domain was compared with the wild-type sequence showed that the mutant fragment displayed higher affinity for a subset of the sarcomeric proteins, as evidenced by an increased rate of growth for the transformed yeast colonies under highly selective conditions (A.S. and J.R., unpublished data). Thus, the CryABR120G protein retains the ability to interact with the contractile proteins, and, similar to the situation for FBX4, this interaction is enhanced relative to the normal affinity of the protein. The stronger binding may provide a partial explanation for the pathogenicity of the mutant relative to the CryAB null mice, and a reasonable working hypothesis is that CryABR120G, which is defective for chaperone activity, binds tightly to the nascent contractile protein(s), preventing correct folding and insertion into a productive sarcomere. The presence of contractile protein fragments within the aggregates suggests that mutant CryAB binding may disturb contractile protein function directly, rendering the muscle particularly sensitive to the actions of CryABR120G. Thus, the pathogenesis of this mutation cannot be ascribed to a simple loss or gain of function, but it probably reflects a synergistic combination of both.
In general, pathological protein aggregates tend to be insoluble and metabolically stable under physiological conditions, and their accumulation is linked to cellular degeneration and organ failure (22). Cells have an active system that involves the sequestration of aggregated proteins into inclusion bodies, which are termed aggresomes (20). The protoaggresomes are first detectable at various sites in the cytoplasm. However, they rapidly accrete around the microtubule organizing center adjacent to the nucleus (20). This process is active, and it involves binding of the protoaggresomes to dynein motors and their subsequent retrograde transport along the microtubule network (23). Although composition can vary, the aggresomes are common cytopathological features in neurodegenerative, protein aggregation-related pathologies, and they usually contain components of the proteasome, molecular chaperones, ubiquitin conjugates, and intermediate filament proteins (23). Our data are consistent with the electron-dense bodies (known to be a hallmark of the DRMs) being classified as aggresomes, and a recent study (34) in which CryABR120G was found in aggresome-like aggregates by using transfections into a nonmuscle cell line is also consistent with the involvement of the mutant CryAB in aggresome formation. Thus, we propose that CryABR120G is responsible for aggresome formation in the cardiomyocyte and the CryAB-linked DRM is a subclass of the aggresomal diseases.
Amyloidoses are well characterized in many tissues, including the heart, and they are generally thought of as a heterogeneous syndrome characterized by extracellular proteinaceous fibrils that can form insoluble deposits (40, 41). These deposits, which now are known to include intracellular aggregates or inclusions as well, can be stained with the amyloidphilic dyes such as Congo red, and many of the misfolded proteins responsible for or present in the aggregates are known (42). Both hereditary and nonhereditary forms of systemic amyloidoses have been identified and can have diverse effects on cardiac function, resulting in dilative cardiomyopathy, restrictive cardiomyopathy, or diastolic dysfunction (43–46). Accumulation and aggregation of misfolded proteins is a hallmark of the amyloidoses, which include Alzheimer's disease (47), Huntington's disease (48), Parkinson's disease (25), and the spongiform encephalopathies (49). In many cases, the proteinaceous aggregates form as a consequence of mutation, modification, or other alteration in the primary structure of the causative protein(s), disturbing the tertiary structure or posttranslational assembly into oligomers (22, 29). Accumulating data suggest that the fibrillar deposits may not be the primary toxic agent but rather that the inherent pathogenicity resides in soluble oligomers that represent intermediates in the fiber formation pathway (30, 31). These soluble oligomers can be formed from many different sequences, but they exhibit a common conformation-dependent structure. The presence of these prefibrillar intermediates can be detected by the oligomer-specific antibody (30). Our data show that the antibody intensely stains the aggresomes in CryABR120G cardiomyocytes and reacts with cardiomyocytes derived from human heart-failure patients while showing little or no reactivity with samples derived from normal hearts, demonstrating that the cardiomyocytes from diverse cardiomyopathies contain structurally related amyloid oligomers.
The presence in heart-failure patients of protein that is related to the soluble oligomers formed by various amyloidogenic proteins is intriguing, and it implies that there may be common pathogenic mechanisms between at least a subset of the degenerative amyloidoses and cardiomyopathic diseases. In the narrow sense, our data support a rethinking of the DRMs as primarily desmin-based cardiomyopathies. More generally, the abundant cardiomyocyte-based staining in heart-failure patient samples, as well as the efficacy of Congo red in preventing the formation of the aggresomes, points to the potential commonalities that may exist. A rich literature that is broadly based on neurodegenerative processes exists with respect to protein misfolding, aggresomes, and the amyloidosis. We believe that these extensive data could be applied productively to many various cardiovascular pathologies.
Acknowledgments
This work was supported by National Institutes of Health Grants HL69799, HL60546, HL52318, HL56370, and HL66157 (to J.R.) and by a Beginning Grant-in-Aid from the American Heart Association (to A.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CHF, congestive heart failure; CryAB, α-B-crystallin; DRM, desmin-related cardiomyopathy; HCM, hypertrophic cardiomyopathy; hsp, heat shock protein; TG, transgenic.
References
- 1.Adams, K. F., Jr. (2001) Am. J. Med. 110, Suppl. 7A, 6S-13S. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Jones, D. M., Larson, M. G., Leip, E. P., Beiser, A., D'Agostino, R. B., Kannel, W. B., Murabito, J. M., Vasan, R. S., Benjamin, E. J. & Levy, D. (2002) Circulation 106, 3068-3072. [DOI] [PubMed] [Google Scholar]
- 3.Chien, K. R. (2003) J. Clin. Invest. 111, 175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knoll, R., Hoshijima, M., Hoffman, H. M., Person, V., Lorenzen-Schmidt, I., Bang, M. L., Hayashi, T., Shiga, N., Yasukawa, H., Schaper, W., et al. (2002) Cell 111, 943-955. [DOI] [PubMed] [Google Scholar]
- 5.Heling, A., Zimmermann, R., Kostin, S., Maeno, Y., Hein, S., Devaux, B., Bauer, E., Klovekorn, W. P., Schlepper, M., Schaper, W. & Schaper, J. (2000) Circ. Res. 86, 846-853. [DOI] [PubMed] [Google Scholar]
- 6.Li, D., Tapscoft, T., Gonzalez, O., Burch, P. E., Quinones, M. A., Zoghbi, W. A., Hill, R., Bachinski, L. L., Mann, D. L. & Roberts, R. (1999) Circulation 100, 461-464. [DOI] [PubMed] [Google Scholar]
- 7.Sjoberg, G., Saavedra-Matiz, C. A., Rosen, D. R., Wijsman, E. M., Borg, K., Horowitz, S. H. & Sejersen, T. (1999) Hum. Mol. Genet. 8, 2191-2198. [DOI] [PubMed] [Google Scholar]
- 8.Vicart, P., Caron, A., Guicheney, P., Li, Z., Prevost, M. C., Faure, A., Chateau, D., Chapon, F., Tome, F., Dupret, J. M., et al. (1998) Nat. Genet. 20, 92-95. [DOI] [PubMed] [Google Scholar]
- 9.Wang, X., Osinska, H., Klevitsky, R., Gerdes, A. M., Nieman, M., Lorenz, J., Hewett, T. & Robbins, J. (2001) Circ. Res. 89, 84-91. [DOI] [PubMed] [Google Scholar]
- 10.Ingolia, T. D. & Craig, E. A. (1982) Proc. Natl. Acad. Sci. USA 79, 2360-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klemenz, R., Frohli, E., Steiger, R. H., Schafer, R. & Aoyama, A. (1991) Proc. Natl. Acad. Sci. USA 88, 3652-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piatigorsky, J. (1984) Cell 38, 620-621. [DOI] [PubMed] [Google Scholar]
- 13.Iwaki, T., Kume-Iwaki, A. & Goldman, J. E. (1990) J. Histochem. Cytochem. 38, 31-39. [DOI] [PubMed] [Google Scholar]
- 14.Horwitz, J. (1992) Proc. Natl. Acad. Sci. USA 89, 10449-10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, K. & Spector, A. (1996) Eur. J. Biochem. 242, 56-66. [DOI] [PubMed] [Google Scholar]
- 16.Bennardini, F., Wrzosek, A. & Chiesi, M. (1992) Circ. Res. 71, 288-294. [DOI] [PubMed] [Google Scholar]
- 17.Aquilina, A. J., Benesch, J. L. P., Bateman, O. A., Slingsby, C. & Robinson, C. V. (2003) Proc. Natl. Acad. Sci. USA 100, 10611-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W. & Vogelstein, B. (1998) Proc. Natl. Acad. Sci. USA 95, 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wanker, E. (1999) Methods Enzymol. 309, 375-386. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Mata, R., Gao, Y. S. & Sztul, E. (2002) Traffic 3, 388-396. [DOI] [PubMed] [Google Scholar]
- 21.Johnston, J. A., Ward, C. L. & Kopito, R. R. (1998) J. Cell Biol. 143, 1883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopito, R. R. (2000) Trends Cell Biol. 10, 524-530. [DOI] [PubMed] [Google Scholar]
- 23.Kopito, R. R. & Sitia, R. (2000) EMBO Rep. 1, 225-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cummings, C. J., Mancini, M. A., Antalffy, B., DeFranco, D. B., Orr, H. T. & Zoghbi, H. Y. (1998) Nat. Genet. 19, 148-154. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka, K., Suzuki, T., Chiba, T., Shimura, H., Hattori, N. & Mizuno, Y. (2001) J. Mol. Med. 79, 482-494. [DOI] [PubMed] [Google Scholar]
- 26.Crystal, A. S., Giasson, B. I., Crowe, A., Kung, M. P., Zhuang, Z. P., Trojanowski, J. Q. & Lee, V. M. (2003) J. Neurochem. 86, 1359-1368. [DOI] [PubMed] [Google Scholar]
- 27.Koga, T., Taguchi, K., Kobuke, Y., Kinoshita, T. & Higuchi, M. (2003) Chemistry 9, 1146-1156. [DOI] [PubMed] [Google Scholar]
- 28.Kim, Y. S., Randolph, T. W., Manning, M. C., Stevens, F. J. & Carpenter, J. F. (2003) J. Biol. Chem. 278, 10842-10850. [DOI] [PubMed] [Google Scholar]
- 29.Bucciantini, M., Giannoni, E., Chiti, F., Baroni, F., Formigli, L., Zurdo, J., Taddei, N., Ramponi, G., Dobson, C. M. & Stefani, M. (2002) Nature 416, 507-511. [DOI] [PubMed] [Google Scholar]
- 30.Kayed, R., Head, E., Thompson, J. L., McIntire, T. M., Milton, S. C., Cotman, C. W. & Glabe, C. G. (2003) Science 300, 486-489. [DOI] [PubMed] [Google Scholar]
- 31.Hardy, J. & Selkoe, D. J. (2002) Science 297, 353-356. [DOI] [PubMed] [Google Scholar]
- 32.Goudeau, B., Dagvadorj, A., Rodrigues-Lima, F., Nedellec, P., Casteras-Simon, M., Perret, E., Langlois, S., Goldfarb, L. & Vicart, P. (2001) Hum. Mutat. 18, 388-396. [DOI] [PubMed] [Google Scholar]
- 33.Wang, X., Osinska, H., Gerdes, A. M. & Robbins, J. (2002) J. Card. Fail. 8, S287-S292. [DOI] [PubMed] [Google Scholar]
- 34.Chavez Zobel, A. T., Loranger, A., Marceau, N., Theriault, J. R., Lambert, H. & Landry, J. (2003) Hum. Mol. Genet. 12, 1609-1620. [DOI] [PubMed] [Google Scholar]
- 35.Bova, M. P., Yaron, O., Huang, Q., Ding, L., Haley, D. A., Stewart, P. L. & Horwitz, J. (1999) Proc. Natl. Acad. Sci. USA 96, 6137-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brady, J. P., Garland, D., Duglas-Tabor, Y., Robison, W. G., Jr., Groome, A. & Wawrousek, E. F. (1997) Proc. Natl. Acad. Sci. USA 94, 884-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.den Engelsman, J., Keijsers, V., de Jong, W. W. & Boelens, W. C. (2003) J. Biol. Chem. 278, 4699-4704. [DOI] [PubMed] [Google Scholar]
- 38.Golenhofen, N., Arbeiter, A., Koob, R. & Drenckhahn, D. (2002) J. Mol. Cell. Cardiol. 34, 309-319. [DOI] [PubMed] [Google Scholar]
- 39.Lutsch, G., Vetter, R., Offhauss, U., Wieske, M., Grone, H. J., Klemenz, R., Schimke, I., Stahl, J. & Benndorf, R. (1997) Circulation 96, 3466-3476. [DOI] [PubMed] [Google Scholar]
- 40.Glenner, G. G. (1980) New Engl. J. Med. 302, 1283-1292. [DOI] [PubMed] [Google Scholar]
- 41.Glenner, G. G. (1980) New Engl. J. Med. 302, 1333-1343. [DOI] [PubMed] [Google Scholar]
- 42.Buxbaum, J. N. (2004) Curr. Opin. Rheumatol. 16, 67-75. [DOI] [PubMed] [Google Scholar]
- 43.Lie, J. T. (1984) Appl. Pathol. 2, 341-356. [PubMed] [Google Scholar]
- 44.McCarthy, R. E., III, & Kasper, E. K. (1998) Clin. Cardiol. 21, 547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narang, R., Chopra, P. & Wasir, H. S. (1993) Cardiology 82, 294-300. [DOI] [PubMed] [Google Scholar]
- 46.Willens, H. J., Wolpowitz, A., Nitzberg, W. D. & Kessler, K. M. (1995) Am. Heart J. 130, 612-613. [DOI] [PubMed] [Google Scholar]
- 47.Fink, A. L. (1998) Folding Des. 3, R9-R23. [DOI] [PubMed] [Google Scholar]
- 48.DiFiglia, M., Sapp, E., Chase, K. O., Davies, S. W., Bates, G. P., Vonsattel, J. P. & Aronin, N. (1997) Science 277, 1990-1993. [DOI] [PubMed] [Google Scholar]
- 49.Hetz, C. & Soto, C. (2003) Cell Mol. Life Sci. 60, 133-143. [DOI] [PMC free article] [PubMed] [Google Scholar]