Abstract
Plant metabolism is a complex set of processes that produce a wide diversity of foods, woods, and medicines. With the genome sequences of Arabidopsis and rice in hands, postgenomics studies integrating all “omics” sciences can depict precise pictures of a whole-cellular process. Here, we present, to our knowledge, the first report of investigation for gene-to-metabolite networks regulating sulfur and nitrogen nutrition and secondary metabolism in Arabidopsis, with integration of metabolomics and transcriptomics. Transcriptome and metabolome analyses were carried out, respectively, with DNA macroarray and several chemical analytical methods, including ultra high-resolution Fourier transform-ion cyclotron MS. Mathematical analyses, including principal component analysis and batch-learning self-organizing map analysis of transcriptome and metabolome data suggested the presence of general responses to sulfur and nitrogen deficiencies. In addition, specific responses to either sulfur or nitrogen deficiency were observed in several metabolic pathways: in particular, the genes and metabolites involved in glucosinolate metabolism were shown to be coordinately modulated. Understanding such gene-to-metabolite networks in primary and secondary metabolism through integration of transcriptomics and metabolomics can lead to identification of gene function and subsequent improvement of production of useful compounds in plants.
Plants produce a huge array of compounds used for foods, medicines, flavors, and industrial materials. These plant metabolites are synthesized and accumulated by the networks of proteins encoded in the genome of each plant. However, even after the completion of the genome sequencing of Arabidopsis (1) and rice (2, 3), function of those genes and networks of gene-to-metabolite are largely unknown. To reveal the function of genes involved in metabolic processes and gene-to-metabolite networks, the metabolomics-based approach is regarded as a direct way (4–7). In particular, integration of comprehensive gene expression profile with targeted metabolite analysis is shown to be an innovative way for identification of gene function for specific product accumulation in plant (8) and microorganisms (9). However, to depict a whole-cellular process of metabolism, integration of comprehensive gene expression analysis (transcriptomics), and nontargeted metabolite profiling (metabolomics) is needed. Bioinformatics designed suitably for data mining helps the integration efficiently.
The gene expression profiling can be achieved by DNA array analysis. For metabolomics, a nontargeted, high-throughput analytical system is required. Traditionally, GC-MS has been used to detect >300 metabolites in plant tissues (5, 6). Fourier transform-ion cyclotron MS (FT-MS) is a system for metabolome analysis in which crude plant extract is introduced by means of direct injection without prior separation of metabolites by chromatography (10). The mass resolution (>100,000) and accuracy (<1 ppm) of FT-MS is extremely high; hence, complex mixtures of metabolites can be separated based on differences in their isotopic masses, and the elemental composition of these metabolites are determined based on their accurate mass determination.
In the present study, we explored whole-cellular processes at the levels of transcriptome and metabolome under sulfur (S) deficiency and related stresses in Arabidopsis by applying a DNA array and a combination of FT-MS and targeted analysis. S is one of the essential macronutrients for plants. Understanding of plants' mechanisms to adapt to S deficiency is important for improvement of crop yield (11–13). Moreover, S-containing metabolites such as glucosinolates (GLSs) and alliins, defense compounds of plants to herbivores and pathogens, determine quality of human foods (14). Hence, regulation of S metabolism is important for improvement of quality and quantity of plants. In the present paper, we depicted a whole picture of regulation of plant metabolism, especially in relation to S nutrition, by combining a huge amount of multidimensional omics data to find specific key pathways indispensable for the regulation of metabolism.
Materials and Methods
Plant Materials. Arabidopsis thaliana ecotype Col-0 was grown hydroponically under control (1.5 mM sulfate/7 mM nitrate), S-deficient (-S) (30 μM sulfate/7 mM nitrate), nitrogen (N)-deficient (-N) (1.5 mM sulfate/350 μM nitrate), or S- and N-deficient (-SN) (30 μM sulfate/350 μM nitrate) conditions at 22°C under fluorescent light (16 h light/8 h dark) according to ref. 15. We confirmed by the experiment in which concentrations of sulfate and nitrate in culture media were variously changed that plants grew apparently normal at 30 μM sulfate and 350 μM nitrate although expression of a -S-responsive gene changed (15). Rosette leaves were harvested at day 17 after imbibition before bolting. Roots were harvested at day 24 after imbibition because biomass of roots was too small to be subjected to analysis at day 17 after imbibition. Samples were immediately frozen with liquid nitrogen and stored at -80°C until use. Previously, we reported the transcript profiles in leaves and roots of Arabidopsis, which was grown on agarose-solidified control medium (1.5 mM sulfate) for 3 weeks, transferred to control, -S (0 mM sulfate) or O-acetyl-l-serine (OAS)-supplemented (1.5 mM sulfate/1 mM OAS) media, and grown for 48 h (16). In this study, the identical plant materials were used for both transcriptome and metabolome analyses.
Transcriptome Analysis. A cDNA macroarray contains ≈13,000 ESTs corresponding to ≈9,000 genes of Arabidopsis. ESTs were obtained from aboveground organs, flower buds, roots, green siliques, and liquid-cultured seedlings (17). Hybridization, signal detection, and data processing were conducted according to ref. 16. The normalized signal intensities of each clone obtained from repetitive hybridizations were averaged, and the fold change under stress condition was calculated as the ratio of the average intensity in treated samples to that in the appropriate control sample. The statistical program sam (18) was used to identify the ESTs whose expression was significantly changed by the treatment (16).
Metabolome Analysis. High-, middle- and non-polar extracts of the plant materials were subjected to FT-MS (APEX III FT-ICMS, Bruker Daltonics, Billerica, MA) according to ref. 10. Approximately 3,000 peaks were detected with a single sample. Data processing was carried out according to ref. 10. Fold change of each mass peak was calculated as the ratio of signal intensity in treated samples to that in the appropriate control sample. Metabolites were identified based on elemental composition calculated from an accurate m/z value by using discovarray software (Phenomenome Discoveries, Saskatoon, SK, Canada). Inorganic anions, amino acids, and thiols were also analyzed by using capillary electrophoresis and HPLC according to ref.16. Organic acids and sugars were analyzed by using capillary electrophoresis under the same conditions for inorganic anion analysis. Citrate and phosphoenolpyruvate had the same migration time and could not be measured as a single compound. Then, citrate/phosphoenolpyruvate content was calculated as citrate content. It was also the case for 2-oxoglutarate, succinate, and malate. The content of these compounds was calculated as 2-oxoglutarate content.
Mathematical Analyses of Multidimensional Transcriptome and Metabolome Data. Principal component analysis (PCA) was conducted with genelinker gold (Predictive Patterns Software) by using log2 value of fold change. For batch-learning self-organizing map (BL-SOM) analysis, average normalized signal intensities obtained from array experiment and signal intensities obtained from FT-MS were used. The sum of squares of (average) signal intensities in 14 samples for each EST or metabolite was set equal to 1, which gave relative intensities in each sample. BL-SOM analysis was conducted according to ref. 19.
Results and Discussion
Acquisition of Transcriptome and Metabolome Data. Arabidopsis was grown for 3 weeks under control, -S, -N, or -SN conditions. In S- and/or N-starved plants, the root/shoot ratio in biomass increased (data not shown), as expected in plants under nutrient deficiencies (20). These plants looked quite healthy; they did not show chlorosis or apparent stunted growth, suggesting that these plants were well adapted to these conditions. The transcriptome and metabolome in leaves and roots of these plants and also of plants subjected to 2-day S deficiency and OAS supplementation (16) were analyzed (Fig. 1A).
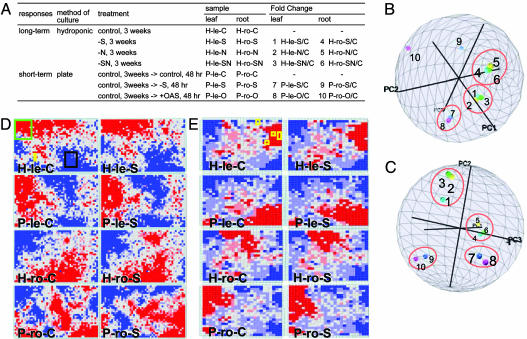
Fig. 1.
Experimental design and global profiling of transcripts and metabolites. (A) Plant materials. H, hydroponic culture; P, plate culture; le, leaf; ro, root; C, control medium; S, -S medium; N, -N medium; SN, -SN medium; O, OAS-supplemented medium;/C, fold change calculated as the ratio of the gene expression level/metabolite content in a treated sample to that in the appropriate control sample. The numbers given to fold change (1 to 10) correspond to those in other figures and tables. (B and C) PCA analyses on transcriptome (B) and metabolome (C). Each small globe represents treatment (fold change). Proportions of the first, second, and third components are 35.8%, 14.6%, and 13.3% (B) and 27.6%, 18.8%, and 15.8% (C), respectively. (D and E) BL-SOM analyses on transcriptome (D) and metabolome (E). Each cell in the lattice represents a cluster into which several ESTs or metabolites are classified. Red, pink, pale blue, and blue indicated that relative level of EST expression or metabolite accumulation in the cell was greater than the average relative levels of all ESTs or metabolites in the sample + SD, greater than the average, smaller than the average, and smaller than the average - SD, respectively. In D, green, black, and yellow rectangles indicate the regions containing the genes for photosynthesis, the pentose-phosphate pathway, and GLS metabolism, respectively. In E, yellow rectangles indicate the regions containing GLS molecular species. Among 14 samples, the representatives are shown.
Expression profiles of ≈13,000 ESTs and nontargeted metabolic profiles of ≈3,000 mass peaks were analyzed by cDNA macroarray and by FT-MS, respectively. Targeted metabolite analyses for amino acids, thiols, organic acids, sugars, and inorganic anions were also performed by using HPLC and capillary electrophoresis. Changes in gene expression or metabolite accumulation were shown as fold change, the ratio of expression level or metabolite content in a treated sample to that in the appropriate control sample, respectively.
Data Mining by Bioinformatics. To find out networks between transcriptome and metabolome, data were simplified and visualized by PCA and BL-SOM analyses.
PCA. PCA was conducted by using ESTs or mass peaks as variable (Fig. 1 B and C). In both transcriptome and metabolome, treatments (fold change) were clustered according to the plant organ sampled, method of plant culture, and period of stress, indicating several features of global regulation: (i) Long-term -S, -N, and -SN had similar effects on transcript/metabolite profiles, (ii) transcript/metabolite profiles were quite different between long-term -S and short-term -S, (iii) OAS, a key intermediate of S assimilation (16), had similar effects to short-term -S, suggesting that OAS is a regulator of global transcript/metabolite profiles under short-term -S. Similar clustering patterns between transcriptome and metabolome indicated that the global transcript profile and metabolite profile were strongly related to each other. It is notable that in the transcriptome, short-term -S and OAS supplementation resulted in similar changes in leaves (fold changes 7 and 8) but not in roots (fold changes 9 and 10), presumably because unusually high OAS content in roots (16) had stronger influence on root gene expression. However, in the metabolome, these two treatments resulted in similar profiles in leaves and roots, suggesting that environmental stress can be coped with by changing global gene expression to minimize the changes in the metabolome. S deficiency is supposed to be caused by shortage of S nutrition relative to N nutrition, because several responses under limited S supply; changes in gene expression and protein accumulation, are diminished when the N supply is also limited. From this point of view, it was not surprising if changes in transcriptome and metabolome under -N might have been in opposite direction to those under -S. However, similar changes occurred under -S and -N, suggesting kinds of general responses to nutritional deficiency (21) in regulation of global transcriptome and metabolome (see below).
BL-SOM analysis. BL-SOM analyses were conducted to classify the ESTs and metabolites according to their patterns of expression and accumulation, respectively. BL-SOM is an improved method of the original SOM (22, 23) with regard to the fact that the initial weight vectors are set by PCA and the learning process is designed to be independent of the order of input of vectors, and hence the result is reproducible (19, 24). Approximately 13,000 ESTs were classified into a 31 × 50 lattice according to their relative expression level in 14 samples (Fig. 1D). Interestingly, functionally related ESTs were clustered in the neighboring cells of the lattice (Fig. 1D and Table 1, which is published as supporting information on the PNAS web site). For example, among the ESTs involved in photosynthesis and in the pentose-phosphate pathway, 64% and 48% of ESTs were classified into the region surrounded by green and black rectangles, respectively. Genes involved in GLS biosynthesis were clustered in the region surrounded by the yellow rectangle, suggesting that GLS metabolism may be coordinately regulated at mRNA accumulation level encoding GLS biosynthetic enzymes. On the other hand, the genes involved in a metabolic pathway in which posttranscriptional control is operated, such as S assimilation (11–13), were scattered in the map (data not shown). Approximately 3,000 metabolites were also classified into a 23 × 25 lattice by BL-SOM analysis (Fig. 1E and Table 2, which is published as supporting information on the PNAS web site). GLSs were classified into the region surrounded by the yellow rectangles, supporting the idea of coordinated regulation of GLS metabolism.
Intercalation into Metabolic Map. Expression data of a total of ≈1,300 ESTs exhibiting statistically significant change by the treatments were put on metabolic maps. Besides sulfate and nitrate assimilation pathways, genes involved in GLS metabolism and carbon metabolism were modulated. Changes in gene expression and metabolite accumulation of our particular interests are shown in Figs. 2 (GLSs) and 3 (central primary metabolism). Secondary metabolism is well studied in the context of regulation by hormonal treatment, biotic and abiotic stress, light, and so on. In this study, we found a regulatory link between secondary metabolism; GLS metabolism, primary metabolism, and S and N nutrition.
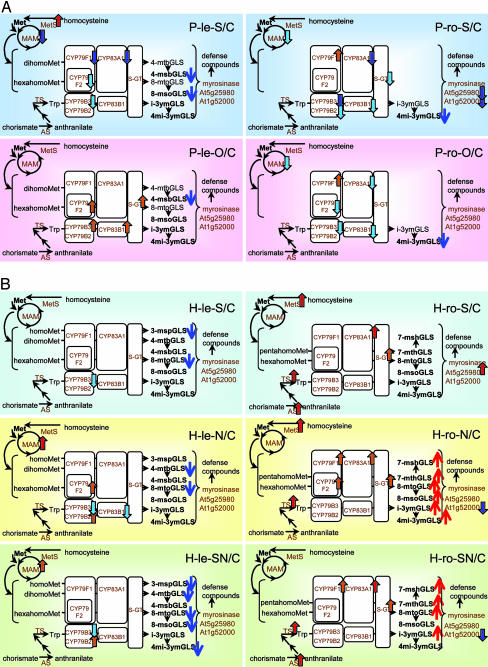
Fig. 2.
Changes in GLS metabolism. (A) Short-term response. (B) Long-term response. Changes in expression of the genes involved in GLS biosynthesis and degradation (14) are shown: MAM, methylthioalkylmalate synthase; CYP, cytochrome P450; S-GT, S-glucosyltransferase; MetS, Met synthase; AS, anthranilate synthase; TS, tryptophan synthase; red bordered arrow, statistically significant up-regulation; orange bordered arrow, up-regulation by >1.3-fold; pale blue bordered arrow, down-regulation by <0.77-fold; blue bordered arrow, statistically significant down-regulation. Accumulation of GLSs are also shown: 3-msp, 3-methylsulfinylpropyl; 4-mtb, 4-methylthiobutyl; 4-msb, 4-methylsulfinylbutyl; 7-mth, 7-methylthioheptyl; 7-msh, 7-methylsulfinylheptyl; 8-mto, 8-methylthio-octyl; 8-mso, 8-methylsulfinyloctyl; i-3ym, 3-indolylmethyl; 4mi-3ym, 4-methoxyindol-3-ylmethyl; red arrow, increase by >2-fold; blue arrow, decrease by <0.5-fold. Metabolites detected in the samples are indicated in bold.
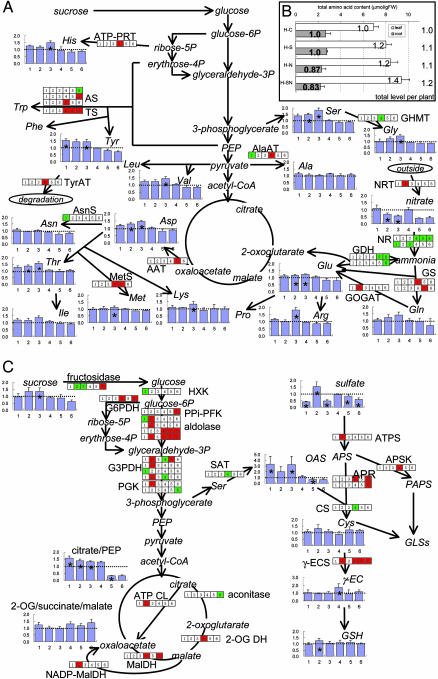
Fig. 3.
Changes in primary metabolism under long-term nutritional stresses. Statistically significant changes in gene expression (SAM, the false discovery rate <10%) are shown in the boxes (1, H-le-S/C; 2, H-le-N/C; 3, H-le-SN/C, 4, H-ro-S/C; 5, H-ro-N/C; 6,H-ro-SN/C) as red (induced) or green (repressed). Fold change values in metabolite accumulation are shown in bar graphs. (A) Nitrate assimilation and amino acid metabolism. (B) Total free amino acid content in leaves and roots. H-C, hydroponic, control; H-S, hydroponic, -S; H-N, hydroponic, -N; H-SN, hydroponic, -SN. The numbers in the bars indicate fold change relative to the appropriate control sample. Total amino acid contents per plant were calculated as the sum of the contents in leaves and roots and fold changes relative to control are shown at the right of the graph. (C) Carbon metabolism and sulfate assimilation. *, Statistically significant change relative to the appropriate control sample (two-sided t test, P < 0.05). Detailed information of the genes shown is summarized in Table 3. PEP, phosphoenolpyruvate; APS, adenosine 5′-phosphosulfate; APR, APS reductase; PAPS, phosphoadenosine 5′-phosphosulfate; γ-EC, γ-glutamylcysteine; γ-ECS, γ-glutamylcysteine synthetase; GDH, glutamate dehydrogenase; GSH, glutathione; 2-OG, 2-oxoglutarate; GS/GOGAT, glutamine synthetase/glutamate synthase.
Glucosinolate metabolism. GLSs are synthesized from several amino acids such as chain-elongated Met and Trp thorough a number of reactions (Fig. 2): chain elongation of Met by methylthioalkylmalate synthase-1 (MAM-1), aldoxime formation catalyzed by cytochromes P450 from CYP79 family, aldoxime oxidation by CYP83s, thiohydroximic acid formation by conjugation to an S donor and after C-S bond cleavage, desulfoGLS formation by S-glucosyltransferase (S-GT), and GLS formation by sulfotransferase. GLSs are degraded by a thioglucosidase, myrosinase. GLSs content and gene expression for GLSs biosynthesis and degradation changed in a treatment-specific manner (Fig. 2). One GLS molecule contains two or three S atoms and one N atom, hence GLSs can play a role as an S storage source (11, 25). Under short-term -S, GLS biosynthesis was down-regulated in gene expression and GLSs accumulation both in leaves and roots (Fig. 2 A). In roots, OAS mimicked S deficiency in GLS biosynthesis. On the other hand, in leaves treated by OAS, change in gene expression was apparently in opposite direction to that in -S leaves. In regulation of GLS biosynthetic genes in leaves, unknown factor(s) specific to OAS treatment might be dominant to factor(s) common to -S and +OAS treatments. As mentioned above, however, global changes in transcriptome were similar in -S leaves and +OAS leaves. Actually, for instance, gene expression in jasmonic acid biosynthesis, carbon metabolism, and photosynthesis showed similar trend in both treatments (16). Under long-term nutrient deficiencies, both roles of GLSs as defense compounds and as an S storage source determined metabolic balance of GLSs. Because nutrient-starved roots may possibly be susceptive of pathogen attack, they try to accumulate more GLSs against attack. Indeed, this is the case from gene expression profile and metabolite accumulation shown in Fig. 2B. GLS accumulation in roots under nutrient starvation was achieved by gene induction of GLS biosynthetic enzymes and possibly also by translocation of GLSs from leaves to roots. Unknown transporters might be responsible for the change in distribution of GLSs between organs. In case of -S, GLSs once synthesized or transported were probably degraded in roots by myrosinase, a degrading enzyme of GLSs, to meet S demand for S-containing primary metabolites. Under -N and -SN conditions, myrosinase gene was down-regulated to store extra S relative to N as GLSs. In H-ro-N six GLS molecular species detected accumulated by 2.6- to 13.7-fold compared with H-ro-C, whereas in H-ro-SN they accumulated by 1.7- to 6.2-fold, probably due to less S supply than in H-ro-N. The changes in gene expression and metabolite accumulation observed in -SN were of additives of those in -S and -N, although the effects by -N were more dominant than -S. Under long-term nutrient deficiencies, general response (induction of GLS biosynthetic genes) and specific response (induction of myrosinase genes in -S and repression in -N and -SN) determine GLS contents in roots. Although GLSs are indispensable defense compounds for plants, they are not always desirable for human foods (14); for example, some GLS molecular species result in odors that diminish the value of canola oil produced from oil-seed rape. Metabolic engineering for custom designed GLS profiles has a strong interest (14). Our findings on GLSs metabolism may lead to improvement of crop quality by genetic manipulation or by physiological treatments without genetic manipulation. High GLSs content in vegetative organs and low GLSs content in the edible part for humans may be a desirable trait of Brassica crops.
Primary metabolism. Under long-term deficiencies, although environmental sulfate and nitrate concentrations were reduced to one fiftieth and one twentieth, respectively, the changes of intracellular levels of these ions were at most 2-fold (Fig. 3 A and C). The content of total free amino acids per plant changed only slightly (Fig. 3B), suggesting the mechanism to keep primary assimilation product levels. Metabolite distribution among organs changed (Fig. 3B); however, proportion of each amino acid content did not change much (data not shown). In the N assimilation pathway (Fig. 3A and Table 3, which is published as supporting information on the PNAS web site), nitrate reductase (NR) genes were down-regulated not only by -N but also by -S. It is known that NR transcription is induced by nitrate (26–28), which is one of the specific responses to N nutrition. On the other hand, NR transcription is also known to be induced by photosynthetic product (21). Photosynthetic activity was reduced both under -S and -N in green algae Chlamydomonas, and this response is considered to be general under nutrient deficiencies (21). In our study, reduction of photosynthetic activity might result in down-regulation of NR genes as one of general responses. Gene encoding Glu dehydrogenase, which plays primarily a catabolic role of Glu (29), was down-regulated under -N and -SN in roots. Genes for the Gln synthetase/Glu synthase cycle responsible for ammonium assimilation, and several amino acids biosynthetic genes seemed to be induced under nutrient deficiencies to maintain amino acid levels (Fig. 3A). In the S assimilation pathway (Fig. 3C), OAS accumulation and adenosine 5′-phosphosulfate reductase gene induction under -S and -SN were observed as expected as specific responses to -S (Fig. 3C, see literatures cited in ref. 16). The gene encoding γ-glutamylcysteine synthetase (γ-ECS) was upregulated in roots under -S, -N, and -SN (Fig. 3C). Interestingly, this gene is responsible for cell division in root meristem (30). Activity of cell division in root meristem might be enhanced under nutritional deficiencies to elongate roots to capture more sulfate or nitrate ions. In fact, root biomass increased in S- and/or N-starved plants (see above). Changes in carbon metabolism were complicated (Fig. 3C). Metabolite levels are regulated by changes in gene expression, protein accumulation, and enzyme activity. In turn, these processes are regulated by metabolite levels. Such reciprocal regulations compose complicated networks for maintenance of metabolic balance, especially in primary metabolism. In the present study using plants adapted to nutrient deficiency, we observed the steady state of transcriptome and metabolome after equilibrium of dynamic changes in these processes. Time-course experiments after transfer to deficient conditions will help us to understand the processes.
In conclusion, in the present study, we showed global and individual regulations under S deficiency and related nutritional stresses by integration of transcriptome and metabolome analyses. The present study opens an avenue to investigate the gene-to-metabolite network more precisely, aiming for functional genomics and better biotechnological application.
Supplementary Material
Acknowledgments
We thank the Japanese Consortium for Arabidopsis thaliana DNA Array, with cooperation of Kazusa DNA Research Institute, for providing macroarray filters used in this study. A part of the experiments was conducted in the Radioisotope Research Center of Chiba University. This work was supported in part by Core Research for Evolutional Science and Technology of the Japan Science and Technology Agency and by Grants-in-Aid from the Ministry of Education, Science, Culture, Sports, and Technology of Japan.
Abbreviations: FT-MS, Fourier transform-ion cyclotron MS; GLS, glucosinolate; OAS, O-acetyl-l-serine; PCA, principal component analysis; BL-SOM, batch-learning self-organizing map; C, control; -S, S-deficient; -N, N-deficient; -SN, S- and N-deficient; MAM-1, methylthioalkylmalate synthase-1; S-GT, S-glucosyltransferase; NR, nitrate reductase.
See Commentary on page 9949.
References
- 1.Arabidopsis Genome Initiative (2000) Nature 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 2.Yu, J., Hu, S., Wang, J., Wong, G. K., Li, S., Liu, B., Deng, Y., Dai, L., Zhou, Y., Zhang, X., et al. (2002) Science 296, 79-92. [DOI] [PubMed] [Google Scholar]
- 3.Goff, S. A., Ricke, D., Lan, T. H., Presting, G., Wang, R., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H., et al. (2002) Science 296, 92-100. [DOI] [PubMed] [Google Scholar]
- 4.Tretheway, R. N., Krotzky, A. J. & Willmitzer, L. (1999) Curr. Opin. Plant Biol. 2, 83-85. [DOI] [PubMed] [Google Scholar]
- 5.Fiehn, O., Kopka, J., Dörmann, P., Altmann, T., Trethewey, R. N. & Willmitzer, L. (2000) Nat. Biotechnol. 18, 1157-1161. [DOI] [PubMed] [Google Scholar]
- 6.Roessner, U., Luedemann, A., Brust, D., Fiehn, O., Linke, T., Willmitzer, L. & Fernie, A. R. (2001) Plant Cell 13, 11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sumner, L. W., Mendes, P. & Dixon, R. A. (2003) Phytochemistry, 62, 817-836. [DOI] [PubMed] [Google Scholar]
- 8.Goossens, A., Häkkinen, S. T., Laakso, I., Seppänen-Laakso T., Biondi, S., De Sutter, V., Lammertyn F., Nuutila A. M., Söderlund, H., Zabeau, M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8595-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Askenazi, M., Driggers, E. M., Holtzman, D. A., Norman, T. C., Iverson, S., Zimmer, D. P., Boers, M. E., Blomquist, P. R., Martinez, E. J., Monreal, A. W., et al. (2003) Nat. Biotechnol. 21, 150-156. [DOI] [PubMed] [Google Scholar]
- 10.Aharoni, A., Ric De Vos, C. H., Verhoeven, H. A., Maliepaard, C. A., Kruppa, G., Bino, R. & Goodenowe, D. B. (2002) OMICS 6, 217-234. [DOI] [PubMed] [Google Scholar]
- 11.Hawkesford, M. (2000) J. Exp. Bot. 51, 131-138. [PubMed] [Google Scholar]
- 12.Leustek, T., Martin, M. N., Bick, J.-A. & Davies, J. P. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 141-165. [DOI] [PubMed] [Google Scholar]
- 13.Saito, K. (2000) Curr. Opin. Plant Biol. 3, 188-195. [PubMed] [Google Scholar]
- 14.Mikkelsen, M. D., Petersen, B. L., Olsen, C. E. & Halkier, B. A. (2002) Amino Acids (Vienna) 22, 279-295. [DOI] [PubMed] [Google Scholar]
- 15.Hirai, M. Y., Fujiwara, T., Chino, M. & Naito, S. (1995) Plant Cell Physiol. 36, 1331-1339. [PubMed] [Google Scholar]
- 16.Hirai, M. Y., Fujiwara, T., Awazuhara, M., Kimura, T., Noji, M. & Saito, K. (2003) Plant J. 33, 651-663. [DOI] [PubMed] [Google Scholar]
- 17.Asamizu, E., Nakamura, Y., Sato, S. & Tabata, S. (2000) DNA Res. 7, 175-180. [DOI] [PubMed] [Google Scholar]
- 18.Tusher, V. G., Tibshiriani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanaya, S., Kinouchi, M., Abe, T., Kudo, Y., Yamada, Y., Nishi, T., Mori, H. & Ikemura, T. (2001) Gene 276, 89-99. [DOI] [PubMed] [Google Scholar]
- 20.Eaton, S. V. (1935) Bot. Gaz. 97, 68-100. [Google Scholar]
- 21.Grossman, A. & Takahashi, H. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 163-210. [DOI] [PubMed] [Google Scholar]
- 22.Kohonen, T. (1990) Proc. IEEE 78, 1464-1480. [Google Scholar]
- 23.Kohonen, T., Oja, E., Simula, O., Visa, A. & Kangas, J. (1996) Proc. IEEE 84, 1358-1384. [Google Scholar]
- 24.Abe, T., Kanaya, S., Kinouchi, M., Ichiba, Y., Kozuki, T. & Ikemura, T. (2003) Genome Res. 13, 693-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blake-Kalff, M. M. A., Harrison, K. R., Hawkesford, M. J., Zhao, F. J. & McGrath, S. P. (1998) Plant Physiol. 118, 1337-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheible, W.-R., González-Fontes, A., Lauerer, M., Müller-Röber, B., Caboche, M. & Stitt, M. (1997) Plant Cell 9, 783-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stitt, M. (1999) Curr. Opin. Plant Biol. 2, 178-186. [DOI] [PubMed] [Google Scholar]
- 28.Wang, R., Guegler, K., LaBrie, S. T. & Crawford, N. M. (2000) Plant Cell 12, 1491-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coruzzi, G. & Last, R. (2000) in Biochemistry and Molecular Biology of Plants, eds. Buchanan, B. B., Gruissem, W. & Jones, R. L. (Am. Soc. Plant Physiol., Rockville, MD), pp. 358-410.
- 30.Vernoux, T., Wilson, R. C., Seeley, K. A., Reichheld, J. P., Muroy, S., Brown, S., Maughan, S. C., Cobbett, C. S., Van Montagu, M., Inze, D., et al. (2000) Plant Cell 12, 97-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.