Abstract
Human Rad51 (hRad51) protein plays a key role in homologous recombination and DNA repair. hRad51 protein forms a helical filament on single-stranded DNA (ssDNA), which performs the basic steps of homologous recombination: a search for homologous double-stranded DNA (dsDNA) and DNA strand exchange. hRad51 protein possesses DNA-dependent ATPase activity; however, the role of this activity has not been understood. Our current results show that Ca2+ greatly stimulates DNA strand exchange activity of hRad51 protein. We found that Ca2+ exerts its stimulatory effect by modulating the ATPase activity of hRad51 protein. Our data demonstrate that, in the presence of Mg2+, the hRad51-ATP-ssDNA filament is quickly converted to an inactive hRad51-ADP-ssDNA form, due to relatively rapid ATP hydrolysis and slow dissociation of ADP. Ca2+ maintains the active hRad51-ATP-ssDNA filament by reducing the ATP hydrolysis rate. These findings demonstrate a crucial role of the ATPase activity in regulation of DNA strand exchange activity of hRad51 protein. This mechanism of Rad51 protein regulation by modulating its ATPase activity is evolutionarily recent; we found no such mechanism for yeast Rad51 (yRad51) protein.
Homologous recombination (HR) is critical for maintaining genome stability. Its major function in the cell is to repair DNA double-stranded breaks (DSB), the most lethal type of DNA damage (1–3). HR is also important for chromosome disjunction during meiosis (4, 5). Malfunction of HR leads to genome instability, causing genetic diseases (6). A key protein of human HR, human Rad51 (hRad51), forms a helical nucleoprotein filament on single-stranded DNA (ssDNA) (7), which perform several of the most basic steps of HR: a search for homologous double-stranded DNA (dsDNA) and DNA strand exchange (8). hRad51 protein is an evolutionarily conserved protein; its orthologs are ubiquitous among pro- and eukaryotes. Biochemical studies demonstrated that hRad51 protein, like all its orthologs, possesses ATPase activity, which is activated by ssDNA and to a lesser extent by dsDNA (8). However, it is thought that binding of ATP, but not necessarily ATP hydrolysis, is important for the filament assembly and several critical steps of DNA strand exchange promoted by the proteins of Rad51 family (1). Still, for bacterial RecA protein, ATP hydrolysis plays several important functions promoting the disassembly of the filament, unidirectional branch migration, bypass of heterologous DNA regions, and four-strand DNA strand exchange (9, 10). In contrast, no specific role of ATP hydrolysis has yet been discovered for eukaryotic orthologs, including hRad51 protein (2). Therefore, the role of the ATPase activity of hRad51 protein remains obscure.
In vitro, hRad51 protein can promote DNA strand exchange in the presence of ATP and Mg2+ (11, 12). However, DNA strand exchange activity of hRad51 seemed to be weak, in comparison with that of bacterial and yeast orthologs, and limited mostly to AT-rich sequences. Recently, it was demonstrated that hRad51 protein can be stimulated by ammonium sulfate, indicating that intrinsic DNA strand exchange activity of hRad51 protein is much higher than it was thought (13). Here, we examined the effect of Ca2+, along with several other divalent ions, on DNA strand exchange activity of hRad51 protein. It is known that Ca2+ is a ubiquitous regulator of cell proliferation, differentiation, and apoptosis in vertebrates (14). Importantly, elevation in Ca2+ concentration in response to DNA damage in human cells has been reported (15–19), and dependence of meiosis on Ca2+ has been observed, particularly of meiosis I, when it is thought HR takes place (20, 21). Our current results show that Ca2+ greatly stimulates DNA strand exchange activity of hRad51 protein. We analyzed the mechanism of hRad51 protein activation by Ca2+ and found that Ca2+ exerts its stimulatory effect by modulating the ATPase activity of hRad51 protein. In the presence of Mg2+, hRad51 behaves as a self-inactivating ATPase. Due to relatively rapid ATP hydrolysis and slow dissociation of ADP, the hRad51-ATP-ssDNA filament becomes quickly converted to an inactive hRad51-ADP-ssDNA form. Ca2+, by slowing down the ATP hydrolysis rate, restores the active hRad51-ATP-ssDNA filament. These findings demonstrate a previously unknown mechanism of regulation of DNA strand exchange activity of hRad51 protein and reveal a crucial role of the ATPase activity in this regulation.
Materials and Methods
Proteins and DNA. Yeast and human Rad51, RecA protein, and human replication protein A (hRPA) were purified as described (13, 22, 23). Single-stranded binding protein was purchased from United States Biochemical. The oligonucleotides were: no. 71, 94-mer, 5′-CTTTAGCTGCATATTTACAACATGTTGACCTACAGCACCAGATTCAGCAATTAAGCTCTAAGCCATCCGCAAAAATGACCTCTTATCAAAAGGA; no. 5, 32-mer, 5′-CCATCCGCAAAAATGACCTCTTATCAAAAGGA; no. 6, 32-mer, complementary to no. 5; no. 90, 90-mer, 5′-CGGGTGTCGGGGCTGGCT TA ACTATGCGGCATCAGAGCAGAT TGTACTGAGAGTGCACCATATGCGGTGTGAAATACCGCACAGATGCGT. Oligonucleotides, pUC19 DNA, and εDNA were prepared as described (24, 25). φX174 ssDNA and dsDNA were purchased from New England Biolabs and Invitrogen, respectively. In this study, the DNA concentrations are expressed as moles of nucleotide.
Strand Exchange with DNA Oligonucleotide Substrates. To form nucleoprotein filaments, hRad51 or yeast Rad51 (yRad51) protein (2 μM) was incubated with ssDNA (94-mer, no. 71) (6 μM) in buffer containing 33 mM Hepes (pH 7.0), 2 mM ATP, 100 μg/ml BSA, and indicated divalent ions for 10 min at 37°C. Reactions were initiated by addition of 32P-labeled dsDNA (32-mer, nos. 5 and 6) (4 μM) and carried out for 5 min. Products were analyzed by electrophoresis in 10% polyacrylamide gels and quantified as described (24).
D-Loop Formation. To form filaments, hRad51 or yRad51 protein (1 μM) was incubated with 32P-labeled ssDNA (90-mer, no. 90) (3 μM) in buffer containing 25 mM Tris·acetate (pH 7.5), 1 mM ATP (unless indicated otherwise), 100 μg/ml BSA, 1 mM DTT, 20 mM KCl (added with the protein stock), and indicated divalent ions for 10 min at 37°C. D-loop formation was initiated by addition of pUC19 dsDNA (50 μM). Aliquots were deproteinized by addition of SDS to 1%, and proteinase K to 880 μg/ml; incubated for 15 min at 37°C; mixed with a 1/10 vol of loading buffer (70% glycerol, 0.1% bromophenol blue); analyzed by electrophoresis in 1% agarose-TAE (40 mM Tris·acetate, pH 8.0 and 1 mM EDTA) gels; and quantified as described (24). The yield was expressed as a percentage of the total plasmid DNA.
Three-Strand Exchange Reaction. Nucleoprotein filaments were formed by incubating hRad51 protein (7.5 μM) with φX174 ssDNA (30 μM) in buffer containing 25 mM Tris·acetate (pH 7.5), 250 mM NaCl, 2 mM ATP, 1 mM DTT, 1 mM MgCl2, and, when indicated, 2 mM CaCl2 for 5 min at 37°C. Then hRPA (2 μM) was added, and incubation was continued for 5 min. Reactions were initiated by addition of linear φX174 dsDNA (30 μM) (DNA form I cleaved by ApaLI endonuclease). Aliquots (7 μl) were deproteinized by adding stop solution (3 μl) containing 4.8% SDS and 7.3 mg/ml proteinase K, incubated for 15 min at 37°C, and analyzed by electrophoresis in 1% agarose-TAE gels. A 1-kb DNA ladder (Invitrogen) was used as DNA migration markers. DNA bands were stained with ethidium bromide and quantified by using an Image Station 440CF (Kodak). The reaction with RecA protein was performed as described (26).
Fluorimetric Assay. Fluorescence of M13 εDNA was excited at 300 nm and monitored at 408 nm by using a FluoroMax-3 spectrofluorometer (Jobin Yvon Inc., Edison, NJ). hRad51 or yRad51 protein (0.33 μM) was added to εDNA (3 μM) in buffer containing 33 mM Hepes (pH 7.0), 2 mM ATP or 2 mM ADP, 1 mM DTT, and 5 mM of the indicated divalent ions at 23°C. An ATP-regenerating system (3 mM phosphoenolpyruvate, and pyruvate kinase, 20 units/ml) was added, when indicated. In salt-titration experiments, before addition of NaCl, the filaments were incubated for 10 min. For each set of conditions, the fluorescent intensity of hRad51–εDNA complexes in the absence of NaCl is expressed as 1; the fluorescent intensity of εDNA is expressed as 0. The changes in fluorescence due to the presence of NaCl were subtracted.
Isolation and Analysis of hRad51-ATP/ADP-ssDNA Complexes. ATP hydrolysis was initiated by adding hRad51 protein (10 μM) to ssDNA (30 μM) (no. 71, 94-mer) in buffer containing 25 mM Tris·acetate (pH 7.5), 100 μM ATP, 6 μCi [α-32P]ATP (1 Ci = 37 GBq), 1 mM DTT, 5 mM MgCl2, and/or 5 mM CaCl2 at 37°C. Aliquots (10 μl) were loaded onto nylon filters (Nytran, Schleicher & Schuell) and quickly washed twice (100 μl each) with buffer containing 25 mM Tris·acetate (pH 7.5) and either 5 mM MgCl2 or 5 mM CaCl2 in a Minifold apparatus (Schleicher & Schuell) under vacuum. The hRad51-ATP/ADP-ssDNA complexes were immediately eluted from the filters into buffer (200 μl) containing 25 mM Tris·acetate (pH 7.5), 50 mM EDTA, and 1% SDS; aliquots (3 μl) were analyzed by using TLC on PEI-Cellulose plates in 0.3 M KH2PO4 (pH 7.5), and the products were quantified by using a Storm 840 PhosphorImager (Molecular Dynamics). We evaluated a contribution of ATP hydrolysis occurring on filters during the isolation of complexes. By measuring the kinetics of such hydrolysis, we estimated that this hydrolysis could contribute to no more than 3% of protein-bound ADP. In another control, we determined the curve of ADP accumulation in the presence of Mg2+, as described above, except that we washed the filters with buffer containing Ca2+, which inhibits the hRad51 ATPase activity ≈4-fold. The obtained curve of ADP accumulation was virtually indistinguishable from the original one, indicating that the contribution due to ATP hydrolysis on filters was insignificant.
hRad51 ATPase Assay. Reactions were carried out in buffer containing 33 mM Hepes (pH 7.0), 2 mM ATP, 3 μCi [γ-32P]ATP, 6 μM ssDNA (no. 71, 94-mer), 1 mM DTT, 10 mM MgCl2 or/and 5 mM CaCl2, and 2 μM hRad51 protein at 37°C. The extent of ATP hydrolysis was determined by using TLC as described above.
Results
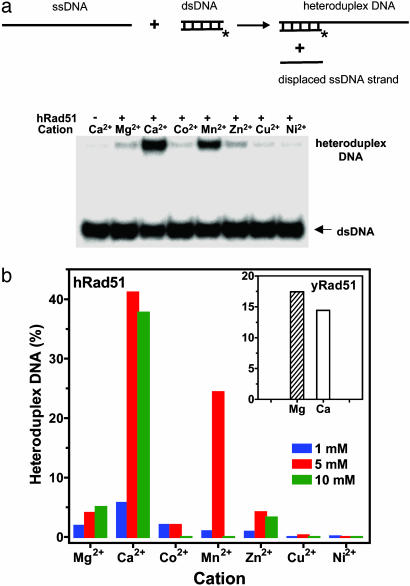
Ca2+ Stimulates DNA Strand Exchange Activity of hRad51 Protein. Using oligonucleotide substrates, we tested the effect of several divalent cations on DNA strand exchange activity of hRad51 protein. We found that the extent of DNA strand exchange was much greater in the presence of Ca2+ than with other cations including Mg2+, a traditional hRad51 protein cofactor (Fig. 1). Mn2+ was the second best cofactor. In contrast, Mg2+ was a better cofactor than Ca2+ for yRad51 protein (Fig. 1b Inset).
Fig. 1.
Effect of divalent cations on DNA strand exchange activity of hRad51 protein. (a) The hRad51 nucleoprotein filaments were formed on ssDNA (94-mer, no. 71); DNA strand exchange was initiated by addition of dsDNA (32-mer, nos. 5 and 6). Asterisks denote the 32P label. The products of DNA strand exchange resolved by gel-electrophoresis are shown at the Bottom. The cation concentrations were 5 mM. (b) The effect of cation concentrations on the extent of DNA strand exchange. (Inset) The effect of Ca2+ (10 mM) and Mg2+ (20 mM) on DNA strand exchange promoted by yRad51 protein is shown as a graph.
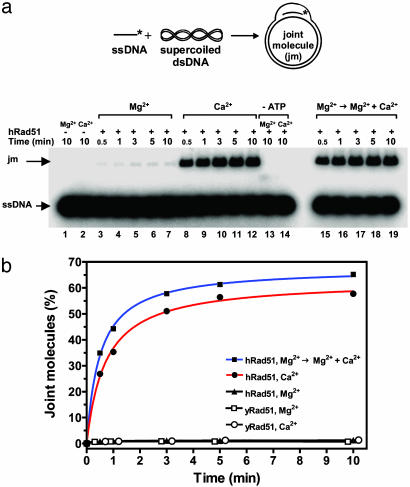
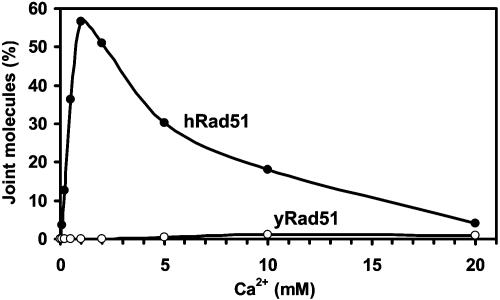
We further investigated the effect of Ca2+ on hRad51 by using D-loop assay, in which hRad51 protein promoted formation of joint molecules (D-loops) between ssDNA and pUC19 super-coiled plasmid dsDNA. In this assay, Ca2+ greatly stimulated joint molecule formation (Fig. 2a, denoted “Ca2+”; Fig. 2b, filled circles). The productive reaction required ATP (Fig. 2a, denoted “-ATP”). Mn2+ supported the reaction too, but to smaller extent than Ca2+ (data not shown). In contrast, Mg2+ supported this reaction poorly (Fig. 2a, denoted “Mg2+”; Fig. 2b, triangles). However, under physiological concentrations [0.5–1 mM of free ion (27)] Mg2+ did not prevent the stimulation of hRad51 protein by Ca2+, even if it was added to the Rad51-ssDNA filament before Ca2+ (Fig. 2a, denoted “Mg2+ → Mg2+ + Ca2+”; Fig. 2b, filled squares). We investigated the effect of Ca2+ concentration on the yield of joint molecules (Fig. 3). The yield of joint molecules was highest at 1 mM Ca2+ concentration, approximately stoichiometric to ATP. We tested the effect of Ca2+ on D-loop formation promoted by yRad51 protein, which normally shows low efficiency in this assay under standard conditions (Fig. 2b, open squares). No stimulation of D-loops was observed in the range of Ca2+ concentrations from 0.1–20 mM (Fig. 3, open circles; Fig. 2b, open circles). Thus, the stimulatory effect of Ca2+ is apparently specific for hRad51 protein.
Fig. 2.
Ca2+ stimulates D-loop formation promoted by hRad51 protein. (a) The scheme and the time course of D-loop formation analyzed in a 1% agarose gel. An asterisk denotes the 32P label. hRad51 protein was preincubated with ssDNA (90-mer, no. 90) either for 10 min at 1 mM MgCl2 (denoted “Mg2+”); for 10 min at 1 mM CaCl2 (denoted “Ca2+”); or for 10 min at 1 mM MgCl2, and then for 10 min at 1 mM MgCl2 and 1 mM CaCl2 (denoted “Mg2+ → Mg2+ + Ca2+”). The reactions in which ATP was omitted are denoted as “-ATP”. The reactions were carried out for the time period indicated by the numbers above the gel. (b) The results of a and the time coarse of D-loop formation promoted by yRad51 protein in the presence of 10 mM Ca2+ (open circles) and 20 mM Mg2+ (open squares) are shown as a graph.
Fig. 3.
Effect of Ca2+ concentrations on the efficiency of D-loop formation. The human and yeast Rad51-ssDNA filaments (filled and open circles, respectively) were formed by incubation of the protein with 32P-labeled ssDNA (90-mer, no. 90) in the presence of indicated Ca2+ concentrations. JM formation was initiated by addition of pUC19 dsDNA and carried out for 10 min.
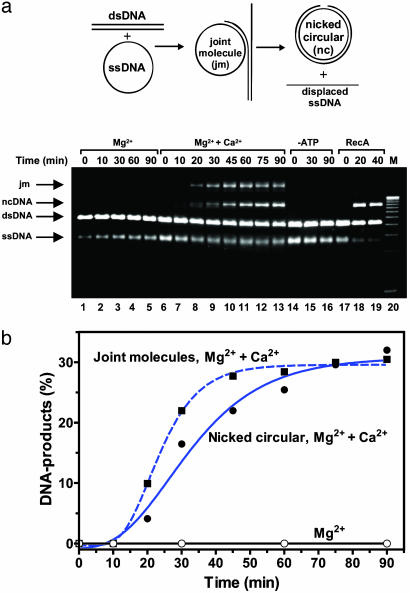
The stimulatory effect of Ca2+ was also strong in the threestrand assay, in which hRad51 protein promotes DNA strand exchange between circular ssDNA and linearized dsDNA of bacteriophage φX174 (Fig. 4). As in the case of yeast and bacterial hRad51 orthologs, the presence of RPA, an ssDNA-binding protein, was essential to stimulate joint molecule (JM) formation and, especially, to produce nicked-circular (NC) DNA product. In the presence of RPA, Ca2+ and ATP, hRad51 protein produced DNA JM, early intermediates of the reaction (Fig. 7, which is published as supporting information on the PNAS web site). In addition to Ca2+, Mg2+ was also required for NC product formation, but not for JM formation. Mg2+ might act by either supporting optimal DNA binding of RPA, or stimulating DNA branch migrating activity of hRad51 protein itself. We found that efficient NC formation, but not JM formation, required also NaCl, which decreases an excessively high coaggregation observed in this reaction (data not shown). Although Mg2+ and NaCl play a stimulatory role in this reaction, only Ca2+ was an essential ion for both JM and NC formation. As in two other assays, ATP was also essential for DNA strand exchange activity of hRad51 protein (Fig. 4a, lanes 14–16).
Fig. 4.
Ca2+ stimulates DNA three-strand exchange promoted by hRad51 protein. (a) The scheme and the time course of DNA stand exchange between φX174 circular ssDNA and linear dsDNA promoted by hRad51 protein analyzed in a 1% agarose gel. The reactions were carried out at 1 mM Mg2+ (denoted “Mg2+”); 1 mM Mg2+ and 2 mM Ca2+ (denoted “Mg2+ + Ca2+”); 1 mM Mg2+ and 2 mM Ca2+, but without ATP (denoted “-ATP”); DNA strand exchange promoted by RecA protein is shown as a control (denoted “RecA”). “M” denotes DNA migration markers. (b) The results are shown as a graph. JM and NC formation in the presence of both Ca2+ and Mg2+ are indicated by squares and circles, respectively. JM formation in the presence of Mg2+ is indicated by open circles.
Thus, using three different assays, we have demonstrated that Ca2+ is an important cofactor for DNA strand exchange activity of hRad51 protein. In contrast, under tested conditions, Mg2+ was a better cofactor than Ca2+ for bacterial RecA (28) and yRad51 protein (Fig. 1b Inset).
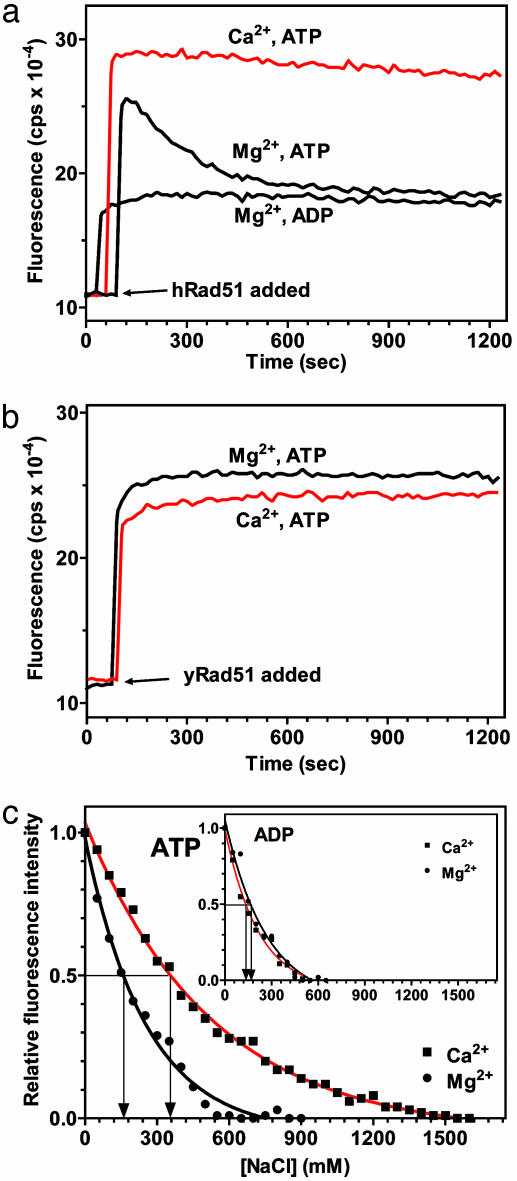
Ca2+ Promotes Formation of the High-Affinity State of the hRad51-ssDNA Filament. We next investigated the mechanism of the hRad51 protein stimulation by Ca2+. In the D-loop assay, we observed that the stimulation was strongest when Ca2+ was present during hRad51-ssDNA filament assembly, but was diminished when Ca2+ was added at later steps, i.e., after dsDNA substrate addition (Fig. 8, which is published as supporting information on the PNAS web site). This observation implied that Ca2+ acts during filament assembly. It was previously shown that DNA within the hRad51-ssDNA filament formed in the presence of an ATP nonhydrolyzable analogue is significantly stretched (29). This DNA stretching is an attribute of the “active” high-affinity ssDNA binding state of the filament, distinguishing it from the more compact “inactive” filament, e.g., one formed in the presence of ADP (30). Formation of the active filament can be detected by an increase in fluorescence of chemically modified etheno-ssDNA (εDNA), when it is used as a binding substrate (22, 31, 32). By using this approach, we found that, in the presence of Mg2+, the initial increase in fluorescence produced by hRad51 protein binding was followed by its gradual decline to the level observed for an inactive hRad51-ADP-εDNA filament (Fig. 5a). Similar results have been reported (33). The decline in fluorescence was unique to hRad51 protein; we did not observe it for yRad51 protein (Fig. 5b). In contrast, in the presence of Ca2+, hRad51 protein binding produces a stable increase in the fluorescence of εDNA, an attribute of an active filament (Fig. 5a). Because hRad51 protein is a DNA-dependent ATPase, we suggested that, in the presence of Mg2+, but not Ca2+, the hRad51-ATP-ssDNA filament is converted into an inactive hRad51-ADP-ssDNA form because of the slow dissociation of ADP, the product of ATP hydrolysis. In contrast, the hRad51-ADP-ssDNA filament could not have been produced due to rebinding to the filament of ADP accumulated in solution during the experiment because (i) hRad51 could have hydrolyzed only 0.05% of ATP, and (ii) the ATP-regeneration system did not prevent the fluorescence decline.
Fig. 5.
Ca2+ increases the stability of the hRad51-ssDNA filament. (a) The fluorescence of hRad51-ATP-εDNA complexes declines in the presence of Mg2+, but not Ca2+. (b) The fluorescence of yRad51-ATP-εDNA complexes does not show such a decline in the presence of Mg2+. (c) hRad51-ATP-εDNA complexes show higher resistance to NaCl in the presence of Ca2+ than in the presence of Mg2+ whereas (Inset) hRad51-ADP-εDNA complexes show the same resistance in the presence of Ca2+ and Mg2+.
Ca2+ Increases Salt Resistance of the hRad51–ssDNA Complex. Another attribute of the active filament, as previously demonstrated for RecA protein, is its higher resistance against disruption by NaCl compared with an inactive filament (25). If the stimulatory role of Ca2+ is to preserve an active hRad51-ATP-ssDNA filament, one can expect that the filament in the presence of Ca2+ is more salt resistant than in the presence of Mg2+. Indeed, by using the fluorimetric assay described above, we found that the salt titration midpoint (STMP) for hRad51-ATP-εDNA complex, which is defined as the NaCl concentration at which one-half of the nucleoprotein complexes is dissociated, was significantly greater in the presence of Ca2+ (360 mM NaCl) than in the presence of Mg2+ (160 mM NaCl) (Fig. 5c). Moreover, the STMP of the hRad51-ATP-εDNA filament in the presence of Mg2+ was close to that of the hRad51-ADP-εDNA filament (165 mM NaCl) (Fig. 5c Inset). The stabilizing effect of Ca2+ was specific for the hRad51-ATP-εDNA; Ca2+ did not increase the STMP of the hRad51-ADP-εDNA filament (Fig. 5c Inset) or the RecA-ATP-εDNA filament (34), as compared with Mg2+. These results are consistent with the role of Ca2+ in preventing the conversion of the hRad51-ATP-ssDNA filament into an inactive hRad51-ADP-ssDNA form.
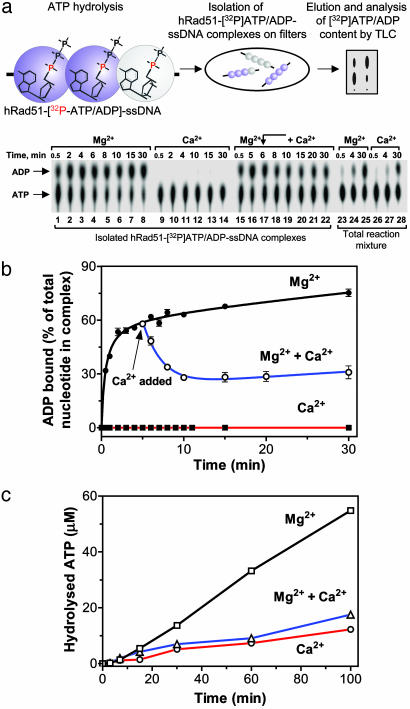
The hRad51-ATP-ssDNA Filament Self-Converts into an Inactive hRad51-ADP-ssDNA Form in the Presence of Mg2+, but Not Ca2+. To directly detect the effect of Ca2+ on the conversion of the hRad51-ATP-ssDNA filament into a hRad51-ADP-ssDNA form, we developed a filter-binding assay (Fig. 6a). This assay allowed us to isolate on nylon filters hRad51-ATP/ADP-ssDNA complexes formed in the presence of [α-32P]ATP and to analyze by TLC the composition of the filament-bound nucleotides. We validated the assay by establishing that the amounts of labeled nucleotides retained on filters in the presence of either Ca2+ or Mg2+ were nearly stoichiometric with respect to hRad51 protein, indicating essentially quantitative recovery of the complexes (data not shown). By using the assay, we detected rapid accumulation of the hRad51-ADP-ssDNA complexes in the presence of Mg2+ (55% of total complexes in 4 min); the accumulation of ADP in total reaction mixture (not subjected to filtration) was much slower (16% of the cofactors in 4 min) (Fig. 6a, lanes 3 and 24; Fig. 6b, closed circles). This accumulation of an inactive filament cannot be explained by rebinding of ADP from the reaction mixture because addition of the ATP regeneration system to the reaction mixture, which completely eliminated accumulation of ADP in solution, did not prevent it (Fig. 9, which is published as supporting information on the PNAS web site). Therefore, we concluded that accumulation of the inactive hRad51-ADP-ssDNA filament was caused by slow dissociation of ADP, the product of ATP hydrolysis. This conclusion is in accord with the report of Fishel and coworkers (35) that ADP dissociation is the rate-limiting step of ATP hydrolysis by hRad51 protein. In contrast, in the presence of Ca2+, the filament remains in an active hRad51-ATP-ssDNA form; the amount of protein-bound ADP did not exceed 3–4% after 30 min of incubation (Fig. 6a, lanes 9–14; Fig. 6b, filled squares). Moreover, Ca2+ partially restored the hRad51-ATP-ssDNA filament, which was initially converted to a hRad51-ADP-ssDNA form by incubation in the presence of Mg2+ (Fig. 6a, lanes 15–22; Fig. 6b, open circles).
Fig. 6.
Ca2+ averts conversion of the hRad51-ATP-ssDNA filament into a hRad51-ADP-ssDNA form by inhibiting ATP hydrolysis. (a) The scheme and results of analysis by filter-binding and TLC of accumulation of the hRad51-ADP-ssDNA complexes in the presence of Mg2+ (lanes 1–8); Ca2+ (lanes 9–14); or Mg2+ followed by addition of Ca2+ (lanes 15–22). The content of ATP and ADP in the reaction mixtures (without filter binding) containing either Mg2+ or Ca2+ is shown in lanes 23–25 and 26–28, respectively. (b) Graphical representation of three independent experiments, as that in a.(c) Inhibition by Ca2+ of ATP hydrolysis by the hRad51-ssDNA filament.
If the formation of an inactive ADP-containing filament is the main cause for the low DNA strand exchange activity of hRad51 protein in the presence of Mg2+, one might expect that hRad51 protein should be more active in the presence of nonhydrolyzable ATP analogues. Indeed, we found that hRad51 protein efficiently promotes joint molecule formation between ssDNA and supercoiled plasmid dsDNA in the presence of Mg2+ and either AMP-PNP or AMP-PCP (Fig. 10, which is published as supporting information on the PNAS web site).
Ca2+ Inhibits Conversion of the hRad51 Filament into an Inactive ADP-Bound Form by Slowing Down Its ATP Hydrolysis Rate. To determine whether Ca2+ prevents accumulation of inactive hRad51-ADP-ssDNA filaments by accelerating ADP dissociation or by slowing down its generation i.e., by inhibiting ATP hydrolysis, we measured the ATP hydrolysis rate. Fig. 6c shows that Ca2+ alone or in combination with Mg2+ lowered the rate of the reaction ≈4- and 3.3-fold, respectively, as compared with Mg2+. This result demonstrates that Ca2+ acts primarily by slowing down ATP hydrolysis, thereby increasing the fraction of ATP within the hRad51-ssDNA filament. Ca2+ may also stimulate ADP dissociation; this possibility, suggested by the very low amounts of protein-bound ADP in the presence of Ca2+ (Fig. 6a, lanes 9–14), warrants further investigation.
Discussion
Previously, it was thought that only ATP binding, but not necessarily hydrolysis, plays a functional role for the eukaryotic Rad51 protein orthologs. Here, we have discovered that ATP hydrolysis in the presence of Mg2+ leads to self-inactivation of hRad51, turning it into an inactive ADP-bound form. This interpretation agrees with the previously published electron microscopy data: extended active hRad51-ssDNA filaments have been observed in the presence of a nonhydrolyzable nucleotide analogue, 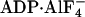 (29) whereas, under standard conditions in the presence of ATP and Mg2+, only inactive filaments have been observed (7, 36). The current results demonstrate that Ca2+ by modulating the ATPase activity of hRad51 protein helps to preserve the active hRad51-ATP-ssDNA filament and thereby strongly stimulates DNA strand exchange activity of this protein. Thus, this finding relates two important characteristics of hRad51 protein: the ATPase activity and the ability to form an active nucleoprotein filament on ssDNA.
(29) whereas, under standard conditions in the presence of ATP and Mg2+, only inactive filaments have been observed (7, 36). The current results demonstrate that Ca2+ by modulating the ATPase activity of hRad51 protein helps to preserve the active hRad51-ATP-ssDNA filament and thereby strongly stimulates DNA strand exchange activity of this protein. Thus, this finding relates two important characteristics of hRad51 protein: the ATPase activity and the ability to form an active nucleoprotein filament on ssDNA.
The well known precedents of the NTPase-dependent self-inactivation include G proteins (37). RecA protein also shows some features of self-inactivating ATPase (38). Like hRad51 protein, it hydrolyzes ATP producing ADP during hydrolysis. In a complex with ADP, RecA protein has affinity for ssDNA lower than in a complex with ATP, forming structurally distinct filaments that are inactive in DNA strand exchange. However, there is an important difference in the biochemical behavior between RecA and hRad51 protein. In the case of RecA protein, ADP produced during ATP hydrolysis is readily replaced from the protein-complexes by free ATP; thus, self-inactivation of RecA protein is not occurring during ATP hydrolysis, as long as the pool of free ATP is not sufficiently depleted (34). In contrast, our results demonstrate that ADP product remains stably associated with hRad51 protein even in the presence of high ATP concentrations and the ATP-regeneration system. Thus, hRad51 protein evolved into a truly self-inactivating ATPase. The described differences in the biochemical properties of hRad51 and RecA proteins are fully consistent with the effect of Ca2+ on DNA strand exchange promoted by these two proteins: although Ca2+ was shown to inhibit the ATPase activity of RecA protein, it does not stimulate its DNA strand exchange activity (28). Our results show that Ca2+ does not stimulate DNA strand exchange activity of yRad51 protein either, indicating that the mechanism of hRad51 protein regulation by modulating its ATPase activity is evolutionarily recent.
The results presented here demonstrate an important role of the ATPase activity in maintaining of the active hRad51 nucleoprotein filament. The significance and universality of this structure for HR has been recognized in early studies (39, 40). Recently, a report appeared in which the effect of ammonium sulfate and also KCl was investigated on DNA strand exchange promoted by hRad51 protein (36). By using electron microscopy, it was demonstrated that high salt, 100 mM ammonium sulfate, induces formation of an extended form of hRad51-ssDNA filament in the presence of ATP and Mg2+ (36). Consistent with the biochemical data on the stimulation of DNA strand exchange by high salt (13), these results reemphasize the importance of the extended high-affinity ssDNA-nucleoprotein filament for DNA strand exchange promoted by the proteins of the Rad51/RecA family.
Our results show that, in vitro, Ca2+ is both necessary and sufficient to support DNA strand exchange activity of hRad51 in the presence of ATP. The only step of DNA strand exchange that required also Mg2+ and RPA seems to be branch migration. RPA or its bacterial single-stranded binding protein homologue, SSB, is a ubiquitous stimulator of all eukaryotic and prokaryotic proteins of the Rad51/RecA family. RPA plays two important functions during DNA strand exchange: it disrupts secondary structures in ssDNA, facilitating the formation of contiguous Rad51 filament, and binds to the displaced ssDNA strand during strand exchange, stabilizing the joint molecules and preventing reinitiation (41, 42). Although both Mg2+ and Ca2+ support RPA binding to ssDNA (data not shown), it is possible that Mg2+ is a better cofactor for optimal ssDNA binding by RPA. However, it is also possible that Mg2+ stimulates hRad51 protein specifically during DNA branch migration. The efficiency of three-strand DNA exchange reaction under described conditions in the presence of Ca2+ was similar to that in the presence of ammonium sulfate (13), if compared back to back (data not shown).
In vertebrates, Ca2+ is a ubiquitous modulator of cell proliferation, differentiation, and apoptosis (14). This in vitro study, which demonstrates that Ca2+ is a cofactor of hRad51 protein, calls for investigation of a possible in vivo role of Ca2+ in HR.
The current paper demonstrates a previously unidentified role of hRad51 protein ATPase activity. Modulation of the ATPase activity by different cofactors, e.g., Mg2+ and Ca2+, may cause formation of the active ATP-containing hRad51-ssDNA nucleoprotein filaments, or induce their conversion into an inactive ADP-containing form. This modulation of the ATPase provides a means of regulation of the DNA strand exchange activity of hRad51 protein, a basic activity of HR in human cells.
Supplementary Material
Acknowledgments
We thank P. Sung (Yale University) and M. Wold (University of Iowa) for hRad51 and hRPA expression vectors and S. Kowalczykowski (University of California, Davis) and M. Spies (University of California, Davis) for RecA protein. We thank J. Azizkhan-Clifford, P. Bianco, D. Ferrier, M. Jorns, S. Kowalczykowski, O. Mazina, and E. Zaitsev for comments and discussion. This work was supported by Drexel University College of Medicine Start-up Funds, Pennsylvania Health Research Formula Funds from the Tobacco Settlement Act, and National Institutes of Health Grant CA100839 (to A.V.M.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: hRad51, human Rad51; yRad51, yeast Rad51; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; HR, homologous recombination; JM, joint molecule; NC, nicked-circular; hRPA, human replication protein A.
References
- 1.West, S. C. (2003) Nat. Rev. Mol. Cell. Biol. 4, 435-445. [DOI] [PubMed] [Google Scholar]
- 2.Sung, P., Krejci, L., Van Komen, S. & Sehorn, M. G. (2003) J. Biol. Chem. 278, 42729-42732. [DOI] [PubMed] [Google Scholar]
- 3.Kowalczykowski, S. C. (2002) Nat. Struct. Biol. 9, 897-899. [DOI] [PubMed] [Google Scholar]
- 4.Hassold, T. & Sherman, S. (2000) Clin. Genet. 57, 95-100. [DOI] [PubMed] [Google Scholar]
- 5.Masson, J. Y. & West, S. C. (2001) Trends Biochem. Sci. 26, 131-136. [DOI] [PubMed] [Google Scholar]
- 6.Kolodner, R. D., Putnam, C. D. & Myung, K. (2002) Science 297, 552-557. [DOI] [PubMed] [Google Scholar]
- 7.Benson, F. E., Stasiak, A. & West, S. C. (1994) EMBO J. 13, 5764-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumann, P. & West, S. C. (1998) Trends Biochem. Sci. 23, 247-251. [DOI] [PubMed] [Google Scholar]
- 9.Kowalczykowski, S. C., Dixon, D. A., Eggleston, A. K., Lauder, S. D. & Rehrauer, W. M. (1994) Microbiol. Rev. 58, 401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lusetti, S. L. & Cox, M. M. (2002) Annu. Rev. Biochem. 71, 71-100. [DOI] [PubMed] [Google Scholar]
- 11.Gupta, R. C., Bazemore, L. R., Golub, E. I. & Radding, C. M. (1997) Proc. Natl. Acad. Sci. USA 94, 463-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumann, P. & West, S. C. (1997) EMBO J. 16, 5198-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sigurdsson, S., Trujillo, K., Song, B., Stratton, S. & Sung, P. (2001) J. Biol. Chem. 276, 8798-8806. [DOI] [PubMed] [Google Scholar]
- 14.Berridge, M. J., Lipp, P. & Bootman, M. D. (2000) Nat. Rev. Mol. Cell. Biol. 1, 11-21. [DOI] [PubMed] [Google Scholar]
- 15.Spielberg, H., June, C. H., Blair, O. C., Nystrom-Rosander, C., Cereb, N. & Deeg, H. J. (1991) Exp. Hematol. 19, 742-748. [PubMed] [Google Scholar]
- 16.Schieven, G. L., Kirihara, J. M., Gilliland, L. K., Uckun, F. M. & Ledbetter, J. A. (1993) Mol. Biol. Cell 4, 523-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gafter, U., Malachi, T., Ori, Y. & Breitbart, H. (1997) J. Lab. Clin. Med. 130, 33-41. [DOI] [PubMed] [Google Scholar]
- 18.Sakai, H., Ito, E., Cai, R. X., Yoshioka, T., Kubota, Y., Hashimoto, K. & Fujishima, A. (1994) Biochim. Biophys. Acta 1201, 259-265. [DOI] [PubMed] [Google Scholar]
- 19.Negre-Salvayre, A. & Salvayre, R. (1992) Biochim. Biophys. Acta 1123, 207-215. [DOI] [PubMed] [Google Scholar]
- 20.Tombes, R. M., Simerly, C., Borisy, G. G. & Schatten, G. (1992) J. Cell Biol. 117, 799-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll, J., Swann, K., Whittingham, D. & Whitaker, M. (1994) Development (Cambridge, U.K.) 120, 3507-3517. [DOI] [PubMed] [Google Scholar]
- 22.Zaitseva, E. M., Zaitsev, E. N. & Kowalczykowski, S. C. (1999) J. Biol. Chem. 274, 2907-2915. [DOI] [PubMed] [Google Scholar]
- 23.Henricksen, L. A., Umbricht, C. B. & Wold, M. S. (1994) J. Biol. Chem. 269, 11121-11132. [PubMed] [Google Scholar]
- 24.Mazin, A. V., Zaitseva, E., Sung, P. & Kowalczykowski, S. C. (2000) EMBO J. 19, 1148-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menetski, J. P. & Kowalczykowski, S. C. (1985) J. Mol. Biol. 181, 281-295. [DOI] [PubMed] [Google Scholar]
- 26.Menetski, J. P., Varghese, A. & Kowalczykowski, S. C. (1992) J. Biol. Chem. 267, 10400-10404. [PubMed] [Google Scholar]
- 27.Schmitz, C., Perraud, A. L., Johnson, C. O., Inabe, K., Smith, M. K., Penner, R., Kurosaki, T., Fleig, A. & Scharenberg, A. M. (2003) Cell 114, 191-200. [DOI] [PubMed] [Google Scholar]
- 28.Cox, M. M. & Lehman, I. R. (1982) J. Biol. Chem. 257, 8523-8532. [PubMed] [Google Scholar]
- 29.Yu, X., Jacobs, S. A., West, S. C., Ogawa, T. & Egelman, E. H. (2001) Proc. Natl. Acad. Sci. USA 98, 8419-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stasiak, A. & Egelman, E. H. (1988) in Genetic Recombination, eds. Kucherlapati, R. & Smith, G. R. (Am. Soc. Microbiol., Washington, DC), pp. 265-308.
- 31.Cazenave, C., Toulme, J. J. & Helene, C. (1983) EMBO J. 2, 2247-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silver, M. S. & Fersht, A. R. (1982) Biochemistry 21, 6066-6072. [DOI] [PubMed] [Google Scholar]
- 33.Kim, H. K., Morimatsu, K., Norden, B., Ardhammar, M. & Takahashi, M. (2002) Genes Cells 7, 1125-1134. [DOI] [PubMed] [Google Scholar]
- 34.Menetski, J. P., Varghese, A. & Kowalczykowski, S. C. (1988) Biochemistry 27, 1205-1212. [DOI] [PubMed] [Google Scholar]
- 35.Tombline, G., Shim, K. S. & Fishel, R. (2002) J. Biol. Chem. 277, 14426-14433. [DOI] [PubMed] [Google Scholar]
- 36.Liu, Y., Stasiak, A. Z., Masson, J. Y., McIlwraith, M. J., Stasiak, A. & West, S. C. (2004) J. Mol. Biol. 337, 817-827. [DOI] [PubMed] [Google Scholar]
- 37.Bourne, H. R. (1997) Curr. Opin. Cell Biol. 9, 134-142. [DOI] [PubMed] [Google Scholar]
- 38.Kowalczykowski, S. C. (1991) Annu. Rev. Biophys. Biophys. Chem. 20, 539-575. [DOI] [PubMed] [Google Scholar]
- 39.Howard-Flanders, P., West, S. C. & Stasiak, A. (1984) Nature 309, 215-219. [DOI] [PubMed] [Google Scholar]
- 40.Radding, C. M. (1993) Curr. Biol. 3, 358-360. [DOI] [PubMed] [Google Scholar]
- 41.Mazin, A. V. & Kowalczykowski, S. C. (1998) EMBO J. 17, 1161-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugiyama, T., Zaitseva, E. M. & Kowalczykowski, S. C. (1997) J. Biol. Chem. 272, 7940-7945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.