Abstract
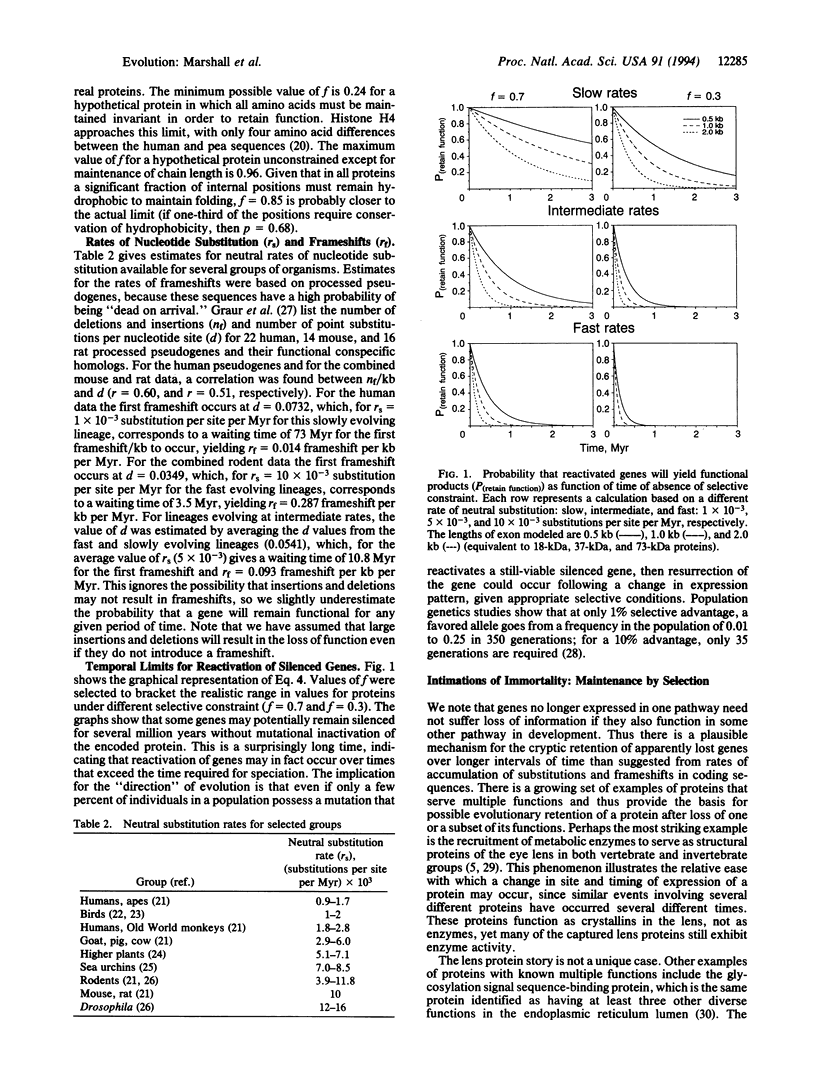
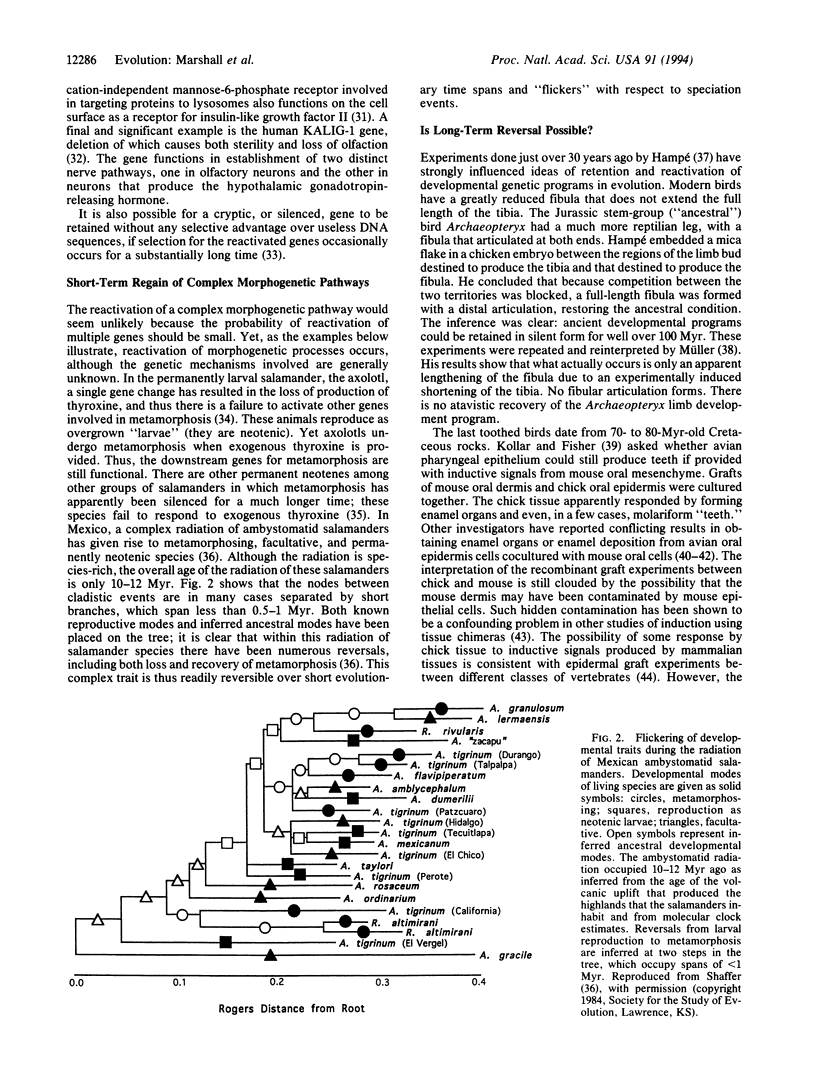
Dollo's law, the concept that evolution is not substantively reversible, implies that the degradation of genetic information is sufficiently fast that genes or developmental pathways released from selective pressure will rapidly become nonfunctional. Using empirical data to assess the rate of loss of coding information in genes for proteins with varying degrees of tolerance to mutational change, we show that, in fact, there is a significant probability over evolutionary time scales of 0.5-6 million years for successful reactivation of silenced genes or "lost" developmental programs. Conversely, the reactivation of long (> 10 million years)-unexpressed genes and dormant developmental pathways is not possible unless function is maintained by other selective constraints; the classic example of the resurrection of "hen's teeth" is most likely an experimental artifact, and the experimental reactivation of the Archaeopteryx limb developmental program has been shown to be a misinterpretation. For groups undergoing adaptive radiations, lost features may "flicker" on and off, resulting in a distribution of character states that does not reflect the phylogeny of the group.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bowie J. U., Reidhaar-Olson J. F., Lim W. A., Sauer R. T. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science. 1990 Mar 16;247(4948):1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- Britten R. J. Rates of DNA sequence evolution differ between taxonomic groups. Science. 1986 Mar 21;231(4744):1393–1398. doi: 10.1126/science.3082006. [DOI] [PubMed] [Google Scholar]
- Cummings E. G., Bringas P., Jr, Grodin M. S., Slavkin H. C. Epithelial-directed mesenchyme differentiation in vitro model of murine odontoblast differentiation mediated by quail epithelia. Differentiation. 1981;20(1):1–9. doi: 10.1111/j.1432-0436.1981.tb01149.x. [DOI] [PubMed] [Google Scholar]
- Cuthbertson R. A., Tomarev S. I., Piatigorsky J. Taxon-specific recruitment of enzymes as major soluble proteins in the corneal epithelium of three mammals, chicken, and squid. Proc Natl Acad Sci U S A. 1992 May 1;89(9):4004–4008. doi: 10.1073/pnas.89.9.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E. The structures of cytochrome c and the rates of molecular evolution. J Mol Evol. 1971;1(1):26–45. doi: 10.1007/BF01659392. [DOI] [PubMed] [Google Scholar]
- Dickinson W. J. On the architecture of regulatory systems: evolutionary insights and implications. Bioessays. 1988 Jun;8(6):204–208. doi: 10.1002/bies.950080608. [DOI] [PubMed] [Google Scholar]
- Fackenthal J. D., Turner F. R., Raff E. C. Tissue-specific microtubule functions in Drosophila spermatogenesis require the beta 2-tubulin isotype-specific carboxy terminus. Dev Biol. 1993 Jul;158(1):213–227. doi: 10.1006/dbio.1993.1180. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Markowitz E. An improved method for determining codon variability in a gene and its application to the rate of fixation of mutations in evolution. Biochem Genet. 1970 Oct;4(5):579–593. doi: 10.1007/BF00486096. [DOI] [PubMed] [Google Scholar]
- Fitch W. M. The molecular evolution of cytochrome c in eukaryotes. J Mol Evol. 1976 Jun 23;8(1):13–40. doi: 10.1007/BF01738880. [DOI] [PubMed] [Google Scholar]
- Franco B., Guioli S., Pragliola A., Incerti B., Bardoni B., Tonlorenzi R., Carrozzo R., Maestrini E., Pieretti M., Taillon-Miller P. A gene deleted in Kallmann's syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991 Oct 10;353(6344):529–536. doi: 10.1038/353529a0. [DOI] [PubMed] [Google Scholar]
- Fuenzalida M., Lemus R., Romero S., Fernandez-Valencia R., Lemus D. Behavior of rabbit dental tissues in heterospecific association with embryonic quail ectoderm. J Exp Zool. 1990 Dec;256(3):264–272. doi: 10.1002/jez.1402560305. [DOI] [PubMed] [Google Scholar]
- Geetha-Habib M., Noiva R., Kaplan H. A., Lennarz W. J. Glycosylation site binding protein, a component of oligosaccharyl transferase, is highly similar to three other 57 kd luminal proteins of the ER. Cell. 1988 Sep 23;54(7):1053–1060. doi: 10.1016/0092-8674(88)90120-1. [DOI] [PubMed] [Google Scholar]
- Gould S. J. Dollo on Dollo's law: irreversibility and the status of evolutionary laws. J Hist Biol. 1970 Fall;3:189–212. doi: 10.1007/BF00137351. [DOI] [PubMed] [Google Scholar]
- Graur D., Shuali Y., Li W. H. Deletions in processed pseudogenes accumulate faster in rodents than in humans. J Mol Evol. 1989 Apr;28(4):279–285. doi: 10.1007/BF02103423. [DOI] [PubMed] [Google Scholar]
- HAMPE A. [The competition between the osseous elements of the zeugopod of the chicken]. J Embryol Exp Morphol. 1960 Sep;8:241–245. [PubMed] [Google Scholar]
- Henry J. J., Grainger R. M. Inductive interactions in the spatial and temporal restriction of lens-forming potential in embryonic ectoderm of Xenopus laevis. Dev Biol. 1987 Nov;124(1):200–214. doi: 10.1016/0012-1606(87)90472-6. [DOI] [PubMed] [Google Scholar]
- Kollar E. J., Fisher C. Tooth induction in chick epithelium: expression of quiescent genes for enamel synthesis. Science. 1980 Feb 29;207(4434):993–995. doi: 10.1126/science.7352302. [DOI] [PubMed] [Google Scholar]
- Lemus D., Coloma L., Fuenzalida M., Illanes J., Paz de la Vega Y., Ondarza A., Blanquez M. J. Odontogenesis and amelogenesis in interacting lizard-quail tissue combinations. J Morphol. 1986 Aug;189(2):121–129. doi: 10.1002/jmor.1051890203. [DOI] [PubMed] [Google Scholar]
- Li W. H. Retention of cryptic genes in microbial populations. Mol Biol Evol. 1984 Feb;1(2):213–219. doi: 10.1093/oxfordjournals.molbev.a040312. [DOI] [PubMed] [Google Scholar]
- Li W. H., Tanimura M., Sharp P. M. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol. 1987;25(4):330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- Limbach K. J., Wu R. Characterization of two Drosophila melanogaster cytochrome c genes and their transcripts. Nucleic Acids Res. 1985 Jan 25;13(2):631–644. doi: 10.1093/nar/13.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyngstadaas S. P., Risnes S., Nordbø H., Flønes A. G. Amelogenin gene similarity in vertebrates: DNA sequences encoding amelogenin seem to be conserved during evolution. J Comp Physiol B. 1990;160(5):469–472. doi: 10.1007/BF00258973. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Coulondre C., Hofer M., Schmeissner U., Sommer H., Schmitz A., Lu P. Genetic studies of the lac repressor. IX. Generation of altered proteins by the suppression of nonsence mutations. J Mol Biol. 1979 Jun 25;131(2):191–222. doi: 10.1016/0022-2836(79)90073-1. [DOI] [PubMed] [Google Scholar]
- Quigley F., Brinkmann H., Martin W. F., Cerff R. Strong functional GC pressure in a light-regulated maize gene encoding subunit GAPA of chloroplast glyceraldehyde-3-phosphate dehydrogenase: implications for the evolution of GAPA pseudogenes. J Mol Evol. 1989 Nov;29(5):412–421. doi: 10.1007/BF02602911. [DOI] [PubMed] [Google Scholar]
- Sauer R. T. DNA sequence of the bacteriophage gama cI gene. Nature. 1978 Nov 16;276(5685):301–302. doi: 10.1038/276301a0. [DOI] [PubMed] [Google Scholar]
- Scarpulla R. C. Processed pseudogenes for rat cytochrome c are preferentially derived from one of three alternate mRNAs. Mol Cell Biol. 1984 Nov;4(11):2279–2288. doi: 10.1128/mcb.4.11.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. On the rate of DNA sequence evolution in Drosophila. J Mol Evol. 1989 May;28(5):398–402. doi: 10.1007/BF02603075. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Tong P. Y., Tollefsen S. E., Kornfeld S. The cation-independent mannose 6-phosphate receptor binds insulin-like growth factor II. J Biol Chem. 1988 Feb 25;263(6):2585–2588. [PubMed] [Google Scholar]
- Whitfield L. S., Lovell-Badge R., Goodfellow P. N. Rapid sequence evolution of the mammalian sex-determining gene SRY. Nature. 1993 Aug 19;364(6439):713–715. doi: 10.1038/364713a0. [DOI] [PubMed] [Google Scholar]
- Wistow G. Lens crystallins: gene recruitment and evolutionary dynamism. Trends Biochem Sci. 1993 Aug;18(8):301–306. doi: 10.1016/0968-0004(93)90041-k. [DOI] [PubMed] [Google Scholar]
- Wright S. An Analysis of Variability in Number of Digits in an Inbred Strain of Guinea Pigs. Genetics. 1934 Nov;19(6):506–536. doi: 10.1093/genetics/19.6.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The Results of Crosses between Inbred Strains of Guinea Pigs, Differing in Number of Digits. Genetics. 1934 Nov;19(6):537–551. doi: 10.1093/genetics/19.6.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. I., Li W. H., Shen J. J., Scarpulla R. C., Limbach K. J., Wu R. Evolution of cytochrome c genes and pseudogenes. J Mol Evol. 1986;23(1):61–75. doi: 10.1007/BF02100999. [DOI] [PubMed] [Google Scholar]


