Abstract
The incorporation of the structural elements of thermostable enzymes into their less stable counterparts is generally used to improve enzyme thermostability. However, the process of engineering enzymes with both high thermostability and high activity remains an important challenge. Here, we report that the thermostability and activity of a thermophilic subtilase were simultaneously improved by incorporating structural elements of a psychrophilic subtilase. There were 64 variable regions/residues (VRs) in the alignment of the thermophilic WF146 protease, mesophilic sphericase, and psychrophilic S41. The WF146 protease was subjected to systematic mutagenesis, in which each of its VRs was replaced with those from S41 and sphericase. After successive rounds of combination and screening, we constructed the variant PBL5X with eight amino acid residues from S41. The half-life of PBL5X at 85°C (57.1 min) was approximately 9-fold longer than that of the wild-type (WT) WF146 protease (6.3 min). The substitutions also led to an increase in the apparent thermal denaturation midpoint temperature (Tm) of the enzyme by 5.5°C, as determined by differential scanning calorimetry. Compared to the WT, PBL5X exhibited high caseinolytic activity (25 to 95°C) and high values of Km and kcat (25 to 80°C). Our study may provide a rational basis for developing highly stable and active enzymes, which are highly desired in industrial applications.
INTRODUCTION
Many microorganisms have successfully colonized extreme temperature environments ranging from −20°C to 122°C (1, 2). Enzymes from psychrophiles, mesophiles, and (hyper)thermophiles usually perform efficient catalysis at low, moderate, and high temperatures, respectively. Psychrophilic enzymes are characterized as cold active but heat labile, and these characteristics may arise from an increase in either the global or localized flexibility of enzyme structure (3, 4). Compared to their psychrophilic and mesophilic counterparts, (hyper)thermophilic enzymes generally exhibit enhanced conformational rigidity (5–7). However, some (hyper)thermophilic enzymes may combine local flexibility in their active site with high overall rigidity, thus making them more thermostable and cold active than their mesophilic counterparts (5–7).
Accumulating evidence suggests that the cumulative effect of minor improvements of local interactions enhances the intrinsic stability of (hyper)thermophilic enzymes (5, 8). Identification of protein stabilization mechanisms is normally based on comparative studies of homologous enzymes that are adapted to different temperatures, on mutational analyses, on directed evolution and on computational methods (6, 9). The results of these studies have provided a rational basis for improving enzyme stability by site-directed mutagenesis (SDM) (10). Nevertheless, the process of engineering enzymes for higher thermostability and activity remains important and difficult. One reason for this problem is that structural differences between homologous enzymes that are adapted to different temperatures may result from not only thermal adaptation but also random events or other sources of evolutionary pressure. Therefore, it is crucial but difficult to identify key structural features that are related to temperature adaptation. To minimize phylogenetic noise, it is desirable to compare enzymes with minimal differences in their primary structures (11, 12). In addition, the incorporation of the structural elements of thermostable enzymes into their less stable counterparts has generally been used to engineer enzymes. However, there is evidence showing that structural components that are involved in the catalytic cycle have improved flexibility to increase low-temperature activity of psychrophilic enzymes, while other protein regions that are not implicated in catalysis may be even more rigid than their mesophilic counterparts (13–15). Indeed, thermostable variants of thermophilic and mesophilic proteins have been successfully constructed by incorporating structural elements of their less stable counterparts (16, 17).
Subtilisin-like serine proteases (subtilases) have been extensively studied due to their contributions to the understanding of the mechanism of enzyme catalysis as well as their industrial potential (18). Thermostability is one of the main requirements for commercial enzymes, and enzymes with high catalytic activity at low temperatures offer economic and environmental benefits through energy savings (6, 19, 20). Many efforts have been made to engineer mesophilic and psychrophilic subtilases with both high stability and high low-temperature activity, either by directed evolution or SDM (21–24). However, synthetic substrates have generally been used for activity selection in previous studies, leading to poor or no improvements in activity toward macromolecular substrates, and thus they may miss the biotechnological goal. The WF146 protease is an extracellular subtilase produced by thermophile Bacillus sp. WF146 (25), and its stabilization and maturation mechanisms have been characterized previously (26–30). We previously engineered a cold-adapted variant of the WF146 protease that displays improved low-temperature activity but shows a decrease in thermostability (31). Notably, the WF146 protease shares high amino acid sequence identities (65 to 68%) with psychrophilic subtilisins S41 (32–34) and S39 (35), as well as with mesophilic subtilisin SSII (36, 37) and sphericase (Sph) (38). The crystal structures of S41 (39) and Sph (38) have been determined. The WF146 protease, Sph, and S41 constitute a trio of thermophilic, mesophilic, and psychrophilic representatives with high sequence identity, thus making them ideal for the investigation of the temperature adaptation mechanisms of enzymes. The purpose of this study is to investigate the roles of variable sites or regions (VRs) in the alignment of the trio of enzymes in terms of their thermostability and enzyme activity and to construct variants of the WF146 protease with improved thermostability and low-temperature activity.
MATERIALS AND METHODS
Materials.
Restriction enzymes and T4 DNA ligase were purchased from Fermentas (Burlington, Canada). DpnI was from Thermo Scientific (Rockford, IL, USA), and Fast Pfu DNA polymerase was from TransGen Biotech (Beijing, China). Phenylmethylsulfonyl fluoride (PMSF), N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide (suc-AAPF-pNA), azocasein, casein, and subtilisin A were from Sigma (St. Louis, MO, USA).
Strains and growth conditions.
Escherichia coli DH5α and E. coli BL21(DE3) were used as the hosts for cloning and expression, respectively. Bacteria were grown at 37°C in Luria-Bertani medium supplemented with kanamycin (30 μg/ml), as needed.
Plasmid construction and mutagenesis.
The DNA fragment encoding the proform of the WF146 protease, which comprises the N-terminal propeptide and the catalytic domain, was amplified from the genomic DNA of Bacillus sp. WF146 (25) by PCR using the primer pair PB-F/PX-R (see Table S1 in the supplemental material). Afterwards, the amplified fragment was inserted into pET26b to construct the expression plasmid (pWT) for the proform of the wild-type (WT) WF146 protease. The QuikChange SDM method (40) was employed to construct most of the WF146 protease variants by using the primers listed in Table S1. The plasmid pWT was subjected to single SDM or successive rounds of SDM to generate a series of variants containing one or more structural elements of S41 or Sph (see Table S2 in the supplemental material). The active-site variants of the WT (S/A) and PBL5X (P-S/A; PBL5X contains eight amino acid residues from the S41), in which the catalytic residue Ser249 was mutated to Ala, were also constructed using the SDM method. Meanwhile, some variants were constructed by the overlap extension PCR method, as described previously (26). Briefly, the 5′ and 3′ ends of the WF146 protease gene were amplified from pWT by using the primer pairs listed in Tables S1 and S2 in the supplemental material. The first-round, 5′- and 3′-end PCR products were then used for the second-round PCR without the addition of primers. In the third-round PCR, the intact gene was amplified using the primer pair PB-F/PX-R (see Tables S1 and S2), with the products of the second-round PCR serving as the template. The overlap extension PCR products were then inserted into pET26b to generate expression plasmids for target proteins (see Table S2). The sequences of all recombinant plasmids were confirmed by DNA sequencing.
Expression, activation, and purification.
The expression of the recombinant proteins was carried out as described previously (25). The cells were then harvested and suspended in buffer A (50 mM Tris-HCl, 10 mM CaCl2, 10 mM NaCl, pH 8.0), followed by sonication on ice. After centrifugation at 13,400 × g for 10 min (4°C), the supernatant (cell extract) containing the proform was incubated at 60°C for 1 h to activate the enzyme. The mature enzyme was purified using a bacitracin-Sepharose 4B (Amersham Biosciences, Sweden) column (1.6 cm by 20 cm), as previously described (26). The purity of the enzyme was confirmed by SDS-PAGE analysis, where a single protein band was observed (see Fig. S1 in the supplemental material). The enzyme solution was concentrated with a Micron YM-3 centrifugal filter (Millipore, Bedford, MA, USA), as needed. The protein concentrations of the purified enzyme samples were measured using the Bradford method (41), with bovine serum albumin (BSA) as the standard.
To prepare the sample for differential scanning calorimetry (DSC) analysis, proforms of the active-site variants (S/A and P-S/A) in cell extracts were purified by affinity chromatography on a column containing Ni2+-charged chelating Sepharose Fast Flow resin (GE Healthcare), as previously described (30). The purified proforms (150 μg/ml) were then incubated with subtilisin A (30 μg/ml) at 60°C for 2 h in buffer A, thereby allowing the proforms to be converted into inactive mature forms after the processing and degradation of their N-terminal propeptides by subtilisin A (30). Subsequently, an extension of the incubation period to 5 h at 60°C led to the auto-degradation of subtilisin A molecules. Finally, the samples were supplemented with 10 mM PMSF and incubated at 25°C for 0.5 h to inactivate any residual subtilisin A, followed by dialysis against buffer A at 4°C overnight. Protein concentrations of mature S/A and P-S/A were determined spectrophotometrically at 280 nm using an extinction coefficient of 49,740 M−1 cm−1, which was calculated from their amino acid compositions.
Enzyme activity assay.
Azocaseinolytic activity of the enzyme was determined at 25°C for 3 h or at 60 to 90°C for 15 min in 500 μl of reaction mixture containing 250 μl of enzyme sample and 0.25% (wt/vol) azocasein in buffer A. Caseinolytic activity of the enzyme was measured at 25°C for 3 h, 35°C for 100 min, 50°C for 30 min, or 60 to 95°C for 15 min in 500 μl of reaction mixture containing 250 μl of enzyme sample and 1% (wt/vol) casein in buffer A. The proteolytic activity of the enzyme against BSA was determined at 25°C for 3 h or at 90°C for 15 min in 500 μl of reaction mixture containing 250 μl of enzyme sample and 1% (wt/vol) BSA in buffer A. The reaction was terminated by the addition of 40% (wt/vol) trichloroacetic acid (TCA) to the reaction mixture. After incubation at room temperature for 15 min, the mixture was centrifuged at 13,400 × g for 10 min, and the absorbance of the supernatant was measured at 335 nm (azocasein) or 280 nm (casein and BSA) in a 1-cm cell. One unit (U) of activity was defined as the amount of enzyme that was required to increase the corresponding absorbance value by 0.01 units per min under the conditions described above.
The activity of the enzyme toward suc-AAPF-pNA (0.2 mM) was measured at 25 or 90°C in buffer A. The activity was recorded in a thermostated spectrophotometer (Cintra 10e; GBC, Australia) by monitoring the initial velocity of suc-AAPF-pNA hydrolysis with an extinction coefficient of 8,480 M−1 cm−1 for p-nitroaniline (pNA) at 410 nm (42). One unit (U) of activity was defined as the amount of enzyme that produced 1 μmol of pNA per min under assay conditions. Using suc-AAPF-pNA as the substrate, kinetic constants for enzymes were determined, as described previously (27).
Half-lives of thermal inactivation.
Purified enzymes (1.5 μg/ml) were incubated in buffer A at 80 or 85°C. Aliquots were taken at different time periods, and the residual activity was measured at 60°C using azocasein as the substrate. The experimental data were fitted to the following equation: ln(A) = ln(A0) − kt, where A and A0 represent the residual activity at time t and the original activity, respectively, and k is the inactivation rate constant (k). Half-lives (t1/2s) were calculated as t1/2 = ln2/k.
SDS-PAGE analysis.
SDS-PAGE was carried out with the glycine-Tris system (43). To prevent self-degradation of the protease during sample preparation (boiling) or electrophoresis, the protease was precipitated by TCA at a final concentration of 20%. After incubation at room temperature for 15 min, the precipitated proteins were recovered by centrifugation at 13,400 × g for 10 min and then resuspended in ice-cold acetone. Afterwards, the proteins were recovered by centrifugation at 13,400 × g for 10 min, air dried, boiled in loading buffer, and then subjected to SDS-PAGE.
N-terminal sequencing and mass spectrometry.
The proteins separated by SDS-PAGE were electroblotted onto a polyvinylidene difluoride membrane and then stained with Coomassie brilliant blue R-250. The target protein bands were excised and subjected to N-terminal amino acid sequence analysis using a PPSQ-33A protein sequencer (Shimadzu, Japan). For mass spectrometry, purified protein samples were analyzed by using a matrix-assisted laser desorption ionization–two-stage time of flight (MALDI-TOF/TOF) system(5800 MALDI-TOF/TOF mass spectrometer; AB Sciex, USA).
DSC analysis.
Calorimetric measurements were carried out with a capillary cell Nano DSC microcalorimeter (TA Instruments, North Lindon, UT, USA), as described by Feng et al. (44). Briefly, protein samples (1 mg/ml) in buffer A were degassed prior to use. A heating rate of 1°C/min was used, and the scanning was performed from 25 to 120°C. A baseline was obtained by scanning the buffer with an identical heating rate, which was subtracted from the experimental runs. Data analysis was performed using NanoAnalyze software.
Homology modeling.
The structure models of the WF146 protease and PBL5X were generated using SWISS-MODEL (45), with sphericase (PDB accession number 1EA7) (38) and subtilisin S41 (PDB accession number 2GKO) (39) as templates. The PyMOL molecular graphics system (46) was used to prepare the figure.
RESULTS
Systematic mutational analysis of the VRs of the WF146 protease.
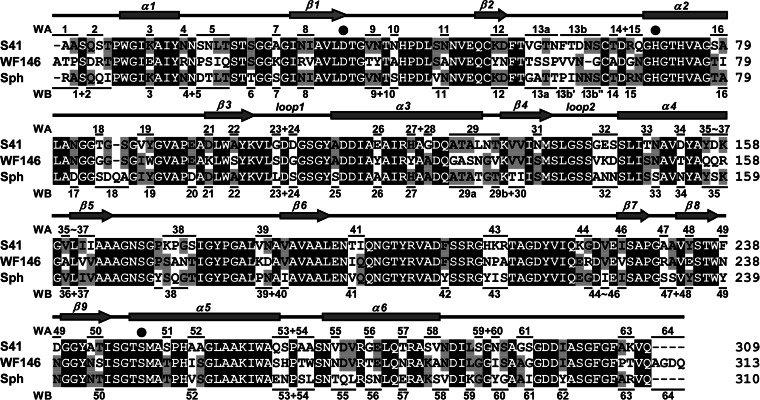
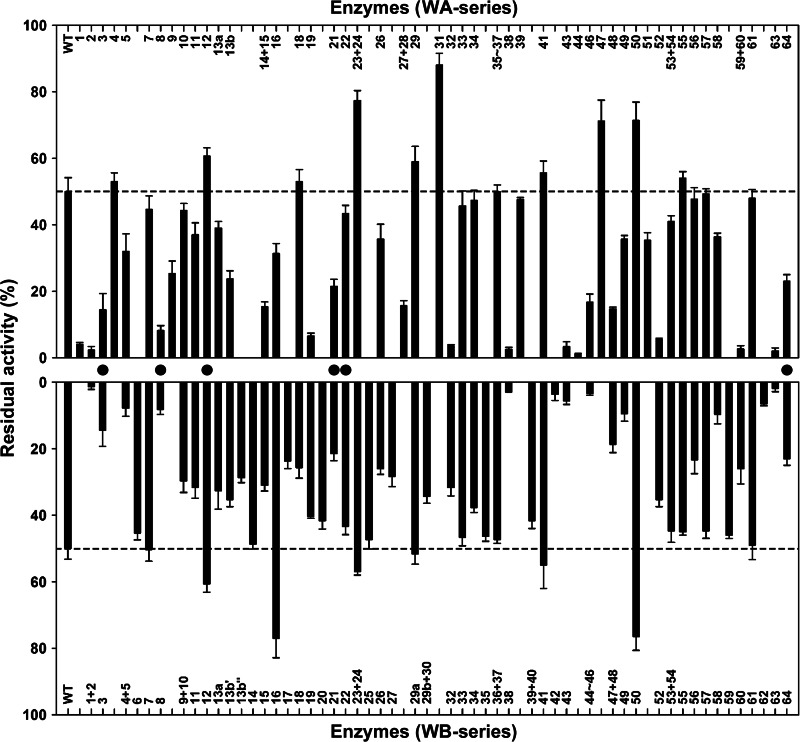
Sixty-four VRs were located between or outside conserved regions/residues in the alignment of S41, Sph, and the WF146 protease (Fig. 1). To probe the contributions of these VRs to enzyme stability and activity, each VR of the WF146 protease was replaced with the corresponding VR from S41 or Sph. In some cases, neighboring VRs were mutated simultaneously. In addition, some long VRs (e.g., VR13 and VR19) were divided into two or three parts and mutated individually. A total of 96 WF146 protease variants were obtained, including 43 variants with VRs from S41 (WA series), 47 variants with VRs from Sph (WB series), and six variants with VRs that were identical in S41 and Sph (for simplicity, these variants are referred to as WA series) (Fig. 1). In comparison with the WT, roughly 60% of the variants showed low residual activities after heat treatment at 80°C for 1 h (Fig. 2), suggesting that the majority of the VRs from S41 or Sph confer thermolability to the WF146 protease variants. Interestingly, six variants (WA23-WA24, WA31, WA47, WA50, WB16, and WB50) retained significantly (P < 0.05) higher residual activities (∼70 to 90%) than WT (∼50%) (Fig. 2). This suggests that the incorporation of certain VRs (stabilizing VRs) from the less stable S41 or Sph into the WF146 protease can further stabilize the thermophilic enzyme.
FIG 1.
Amino acid sequence alignment (Clustal X) (61) of the mature forms of the WF146 protease (GenBank accession number AAQ82911), subtilisin S41 (PDB accession number 2GKO), and sphericase (Sph; PDB accession number 1EA7). Filled circles mark the catalytic triad residues. Conserved regions/residues are shaded in black. The VRs that are marked by horizontal lines above or below the sequence of S41 or Sph represent those that were used to replace corresponding VRs of the WF146 protease. The numbers above and below the sequence alignments represent the resulting variants of the WF146 protease containing VRs of S41 (WA series) and Sph (WB series), respectively. The α-helices (α1 to α6) and β-strands (β1 to β9) in the WF146 protease, as well as the two loops (1 and 2) that form the S4 substrate-binding subsite of the enzyme, are shown above the sequence alignment.
FIG 2.
Heat inactivation profiles of the WT and its variants. The enzymes (1.5 μg/ml) in buffer A were incubated at 80°C for 1 h and then subjected to an azocaseinolytic activity assay at 60°C. The residual activity is expressed as a percentage of the original activity, and the values are means ± standard deviations (bars) of the results of three independent experiments. Filled circles indicate the variants with VRs that are identical in S41 and Sph. The dashed lines mark the levels of residual activity of the WT.
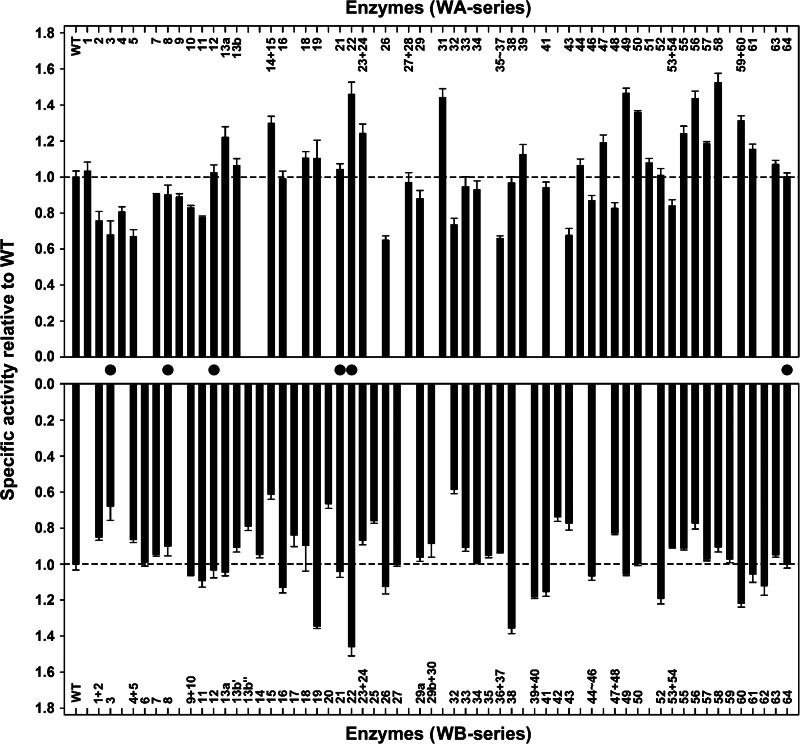
The effects of VR substitutions on the low-temperature activity of the enzymes were investigated by measuring their azocaseinolytic activities at 25°C. Several variants containing VRs from either S41 or Sph displayed improved activity (Fig. 3). The number of activity-enhancing variants with S41-derived VRs was larger than that of the variants with Sph-derived VRs (Fig. 3). In addition, the five most active variants (WA22, WA31, WA49, WA56, and WA58), which exhibited roughly 40 to 60% increases in azocaseinolytic activity, contained S41-derived VRs (Fig. 3). These results demonstrate that the low-temperature activity of the thermophilic WF146 protease could be improved by incorporating structural elements of its less stable counterparts, particularly those from psychrophilic S41.
FIG 3.
Low-temperature activities of the WT and its variants. The azocaseinolytic activities of the enzymes (0.75 μg/ml) were determined at 25°C. The values are means ± standard deviations (bars) of three independent experiments. The specific activities of the variants relative to the WT were calculated. Filled circles indicate the variants with VRs that are identical in S41 and Sph. The dashed lines mark the activity levels of the WT.
Most variants showed a change in thermostability and/or low-temperature activity (Fig. 2 and 3), suggesting that most VRs of the WF146 protease, S41, and Sph are related to temperature adaptation. In comparison with the WT, some variants (e.g., WA13a, WA14-WA15, WA19, WA49, WA58, WA59-WA60, WB19, WB38, WB52, and WB60) showed low residual activities after heat treatment at 80°C (Fig. 2) but high activities at 25°C (Fig. 3). This shows that these variants had lost their thermostability in order to gain higher low-temperature activity; this process is known as the trade-off between stability and activity (3, 47, 48). However, some other variants showed an increase (e.g., WA23-WA24, WA31, WA47, WA50, and WB16) or decrease (e.g., WA2, WA3, WA5, WA11, WA26, WA32, WA43, WB2, WB13b″, WB15, WB42, WB43, and WB56) in both thermostability and low-temperature activity (Fig. 2 and 3), suggesting that there is no intrinsic correlation between stability and activity. Additionally, we noticed that a small number of variants (e.g., WA33, WA34, WB7, WB14, WB29a, and WB35) did not show a significant change in either thermostability or low-temperature activity, thereby implying that the VRs incorporated into these variants are unlikely to be related to temperature adaptation but may result from random events or other sources of evolutionary pressures.
Combination of VRs and screening of variants with improved thermostability and activity.
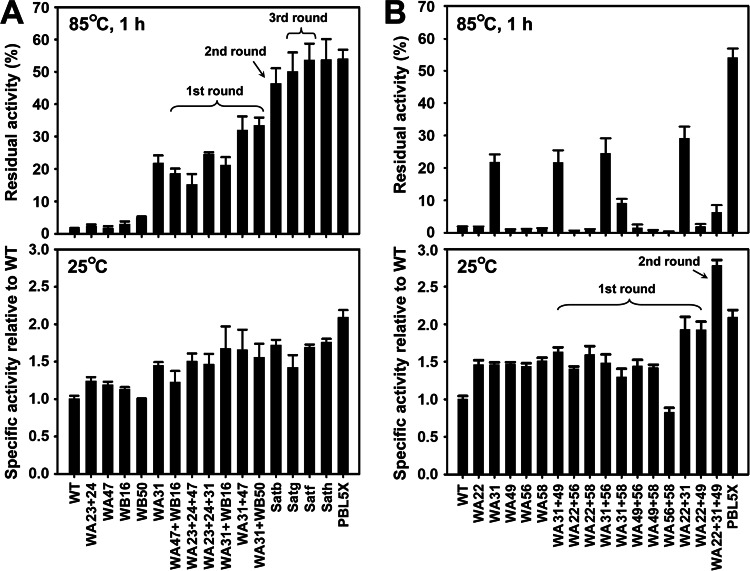
To investigate whether the structural elements of S41 and Sph can stabilize the WF146 protease in a cumulative manner, VRs that led to an increase in variant thermostability were subjected to sequential rounds of combination and screening. Five stability-enhancing variants, which were generated by systematic mutational analysis (WA23-WA24, WA31, WA47, WB16, and WB50) (Fig. 2), were subjected to heat treatment at 85°C for 1 h, followed by the azocaseinolytic activity assay. WA31 retained the highest residual activities (Fig. 4A). In the first round of combination, the stabilizing VRs of WA23-WA24, WA47, WB16, and WB50 were individually combined with that of WA31. The resulting variants were compared with WA31 by measuring their residual activity after heat treatment at 85°C for 1 h. The results show that the WA31-WA47 and WA31-WB50 variants retained higher residual activities than the WA31 variant (Fig. 4A). Subsequently, the stabilizing VRs of WA31-WA47 and WA31-WB50 were combined (second-round combination) to generate Satb (WA31-WA47-WB50), which showed a higher residual activity than the two parental enzymes (Fig. 4A) and demonstrated that these stabilizing VRs had a cumulative effect on enzyme stability. We noticed that two variants (WA23-WA24-WA31 and WA31-WB16) that were generated by the second round of combination exhibited a level of residual activity similar to that of WA31 (Fig. 4A), indicating that the combination of the stabilizing VRs of WA23-WA24 and WB16 with that of WA31 does not cumulatively enhance enzyme stability. Nevertheless, it is possible that the stabilizing VRs of WA23-WA24 and WB16 may act cumulatively with the stabilizing VRs of other stability-enhancing variants (e.g., WA47 and WB50). To test this possibility, two additional variants (WA23-WA24-WA47 and WA47-WB16) were constructed and subjected to heat treatment and the activity assay. Indeed, these two variants were more stable than their parental enzymes (Fig. 4A). Based on this evidence, we performed a third round of combination in which the stabilizing VRs of WA23-WA24 and WB16 were individually introduced into Satb to generate Satg (Satb-WA23-WA24) and Satf (Satb-WB16), respectively. As expected, Satg and Satf showed further increases in their residual activities after heat treatment at 85°C for 1 h, with Satf being slightly more stable than Satg (Fig. 4A). Finally, the stabilizing VRs of Satg and Satf were combined, but no effects on the residual activity of the resulting variant Sath (Satf-WA23-W24) were observed (Fig. 4A). Additionally, low-temperature activities (25°C) of all variants were comparable to or higher than those of their parental enzymes (Fig. 4A). Overall, three sequential rounds of combination and screening yielded two variants (Satg and Satf) with significantly enhanced stability at high temperatures. These findings suggest that some structural elements of less stable S41 and Sph can act cumulatively to further stabilize their thermophilic counterpart without causing a decrease in low-temperature activity.
FIG 4.
Thermal resistance (upper panels) and low-temperature activity (lower panels) of stability-enhancing (A) and activity-enhancing (B) variants generated by combinations of VRs. The enzymes (1.5 μg/ml) in buffer A were incubated at 85°C for 1 h and then subjected to an azocaseinolytic activity assay at 60°C (upper panels). The residual activity is expressed as a percentage of the original activity, and the values are means ± standard deviations (bars) of the results of three independent experiments. The azocaseinolytic activities of the enzymes (0.75 μg/ml) were determined at 25°C in buffer A (lower panels). The values are means ± standard deviations (bars) of three independent experiments, and the mean value of the WT was defined as 1. The specific activities of the variants relative to the WT were calculated.
To further improve the low-temperature activity of the enzyme, the S41-derived, activity-promoting VRs of the five most active variants, which were generated by systematic mutational analysis (WA22, WA31, WA49, WA56, and WA58) (Fig. 3), were subjected to two rounds of combination and screening. In the first round of combination, the activity-promoting VRs of the five variants were paired with each other, thus generating 10 variants containing two activity-promoting VRs that were derived from S41. The azocaseinolytic activities of these variants were then measured at 25°C. Two variants (WA22-WA31 and WA22-WA49) exhibited higher activities than their parental enzymes (Fig. 4B). Subsequently, the activity-promoting VRs of WA22-WA31 and WA22-WA49 were combined to generate WA22-WA31-WA49, which exhibited an increase in azocaseinolytic activity at 25°C to a level that was ∼2-fold higher than that of the WT (Fig. 4B). These results suggest that some structural elements of psychrophilic S41 can act cumulatively to improve the low-temperature activity of its thermophilic counterpart.
Although the WA22-WA31-WA49 variant was more active and stable than the WT, it was less stable than one of its parental enzymes, WA31 (Fig. 4B). Next, we constructed a variant that coupled high stability with high low-temperature activity by incorporating activity-promoting VRs into Satf. Satf represented one of the most thermostable variants of the WF146 protease (Fig. 4A). As shown in Fig. 3, the activity-promoting VR of WA31 is already present in Satf. Furthermore, the activity-promoting VR of WA58 was not selected because combining it with the VR of WA31 led to a significantly reduced thermostability of the variant WA31-WA58 compared to that of WA31 (Fig. 4B). Considering that the combination of the activity-promoting VR of WA31 with that of WA22 (WA22-WA31), WA49 (WA31-WA49), or WA56 (WA31-WA56) did not lead to a decrease in the thermostability of the enzyme (Fig. 4B), the activity-promoting VRs of WA22, WA49, and WA56 were selected and simultaneously combined into Satf to yield the variant PBL5X. The azocaseinolytic activity of PBL5X at 25°C was higher than that of Satf (Fig. 4A). Meanwhile, PBL5X showed residual activity that was similar to that of Satf after heat treatment at 85°C for 1 h (Fig. 4A). These results demonstrate that the thermostability and low-temperature activity of a thermophilic enzyme (the WF146 protease) were cumulatively improved by incorporating selected structural elements that were derived from its less stable counterparts (S41 and Sph).
Properties of PBL5X.
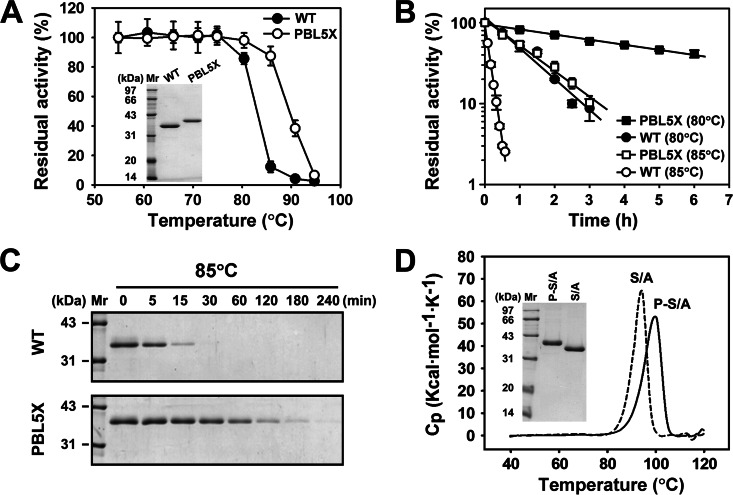
Although the variant PBL5X was shown by SDS-PAGE analysis to have a higher apparent molecular size than the WT (Fig. 5A), N-terminal sequencing of PBL5X revealed that its N terminus is identical to that of the WT, with the first four amino acid residues identified as ATPS. Moreover, PBL5X and the WT were analyzed by mass spectrometry, and their molecular masses were determined to be 31738.2 Da and 31841.1 Da, respectively, which were consistent with their theoretical molecular masses (31,781.5 Da and 31,893.6 Da). The variant PBL5X was compared with the WT in terms of their thermostabilities, enzymatic activities, and kinetic parameters. When the thermostabilities of the two enzymes were tested at various temperatures, PBL5X and the WT were stable until the temperature reached 80 and 75°C, respectively (Fig. 5A). Meanwhile, the variant PBL5X displayed half-lives of 285.6 and 57.1 min at 80 and 85°C, respectively, which were much longer than those of the WT (47.4 and 6.3 min, respectively) (Fig. 5B), suggesting that PBL5X is more stable than the WT. SDS-PAGE analysis revealed that the WT auto-degraded at 85°C more easily than PBL5X as the incubation time increased (Fig. 5C). This indicates that the WT is more sensitive to thermal denaturation. DSC was also carried out to investigate the contribution of the VRs of S41 and Sph to the structural stability of PBL5X. It is known that replacing the catalytic residue Ser with Ala abolishes the activity of the WF146 protease but does not affect enzyme folding (30). Here, the active-site variants of the WT (S/A) and PBL5X (P-S/A) were prepared to prevent enzyme auto-degradation and subjected to DSC analysis. The apparent thermal denaturation midpoint temperature (Tm) of P-S/A (99.7°C) was higher than that of S/A (94.2°C) (Fig. 5D). These results demonstrate that the VRs from S41 and Sph significantly stabilized the global structure of PBL5X.
FIG 5.
Comparison of the thermostabilities of PBL5X and the WT. (A) Effects of temperature on enzyme stability. After the enzymes (1.5 μg/ml) were incubated in buffer A at different temperatures for 15 min, the remaining activities were measured at 60°C using azocasein as the substrate. The residual activity is expressed as a percentage of the original activity, and the values are means ± standard deviations (bars) of the results of three independent experiments. The inset depicts the SDS-PAGE analysis of purified PBL5X and the WT. (B and C) Heat inactivation profiles. The enzymes (1.5 μg/ml) were incubated in buffer A at 80 or 85°C for the indicated time periods and then subjected to an azocaseinolytic activity assay at 60°C (B) and SDS-PAGE analysis (C). In panel B the residual activity (log scale) is plotted versus incubation time and expressed as a percentage of the original activity. The values are means ± standard deviations (bars) of the results of three independent experiments. (D) Typical DSC curves for the mature forms of the active-site variants of the WT (S/A) and PBL5X (P-S/A). Protein samples (1 mg/ml) in buffer A were heated at a rate of 1°C/min, and excess heat capacity was recorded by DSC. The inset depicts the SDS-PAGE analysis of protein samples. Cp, heat capacity at constant pressure.
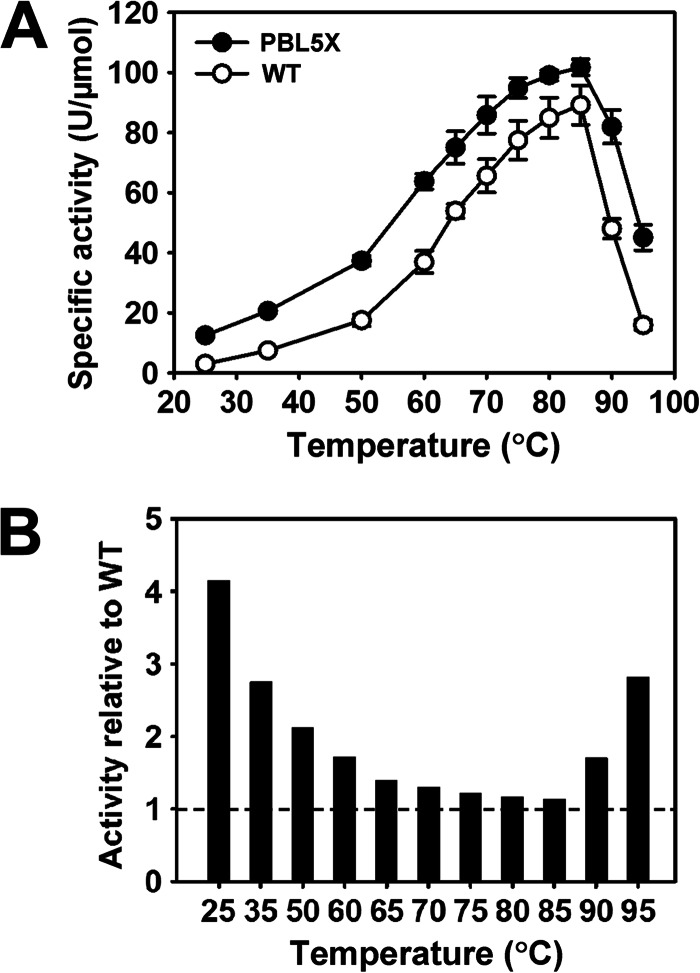
In comparison with the WT, PBL5X exhibited high activities toward azocasein, casein, BSA, and suc-AAPF-pNA at 25 and 90°C (Table 1), thus suggesting an improvement in the activity of PBL5X under both low- and high-temperature conditions. In addition, the caseinolytic activity of PBL5X was higher than that of WT over the tested temperature range of 25 to 95°C (Fig. 6A). The caseinolytic activity of PBL5X relative to that of the WT was temperature dependent, and the relative superiority of its activity over that of the WT increased not only with decreasing temperatures at a lower temperature range (25 to 85°C) but also with increasing temperatures at a higher temperature range (85 to 95°C) (Fig. 6B). This increase in the low-temperature activity of PBL5X suggests that incorporating structural elements of psychrophilic S41 and mesophilic Sph into the thermophilic WF146 protease can facilitate enzymatic catalysis at low temperatures. In contrast, the increase in the high-temperature activity of PBL5X was most likely attributed to its improved thermostability, which rendered the enzyme less prone to the loss of structural integrity, as required for catalysis at high temperatures. Furthermore, using suc-AAPF-pNA as the substrate, kinetic parameters of PBL5X and the WT were determined at different temperatures. In comparison with the WT, PBL5X displayed high values of Km and kcat at 25, 60, and 80°C (Table 2). This suggests that incorporating the VRs of S41 and Sph into the WF146 protease leads to a decrease in substrate affinity and an increase in the turnover rate of the enzyme. Overall, these results demonstrate that certain structural elements of S41 and Sph in PBL5X were involved in modifying the structure around the active site and/or substrate-binding region, which led to a change in the catalytic behavior of the enzyme.
TABLE 1.
Specific activities of the wild-type WF146 protease and PBL5X toward various substrates
| Substrate | Specific activity (U/μmol)a |
|||
|---|---|---|---|---|
| WT protease at: |
PBL5X at: |
|||
| 25°C | 90°C | 25°C | 90°C | |
| Casein | 3.0 ± 0.2 | 48.1 ± 3.3 | 12.5 ± 0.5 | 81.9 ± 5.6 |
| Azocasein | 14.3 ± 2.5 | 207.5 ± 7.2 | 31.5 ± 2.61 | 397.6 ± 14.4 |
| BSA | 4.3 ± 0.3 | 30.7 ± 2.9 | 6.1 ± 0.9 | 82.2 ± 15.3 |
| Suc-AAPF-pNa | 600.1 ± 20.0 | 2003.4 ± 108.7 | 763.9 ± 38.5 | 2411.8 ± 83.0 |
The values are means ± standard deviations of three independent experiments.
FIG 6.
Comparison of the activities of PBL5X and the WT. (A) Temperature dependence of the specific activities of the WT and PBL5X. The caseinolytic activities of the enzymes were determined at the indicated temperatures in buffer A. The values are means ± standard deviations (bars) of the results of three independent experiments. (B) Ratio of the specific activities of PBL5X and the WT. The activity of PBL5X relative to that of the WT was calculated, with that of WT at the respective temperatures defined as 1.
TABLE 2.
Kinetic parameters of the WT protease and PBL5Xa
| Temperature |
Km (mM) |
kcat (s−1) |
||
|---|---|---|---|---|
| WT protease | PBL5X | WT protease | PBL5X | |
| 25°C | 0.3 ± 0.03b | 0.45 ± 0.06 | 51 ± 5b | 113 ± 10 |
| 60°C | 0.61 ± 0.04b | 0.88 ± 0.07 | 348 ± 27b | 606 ± 69 |
| 80°C | 0.95 ± 0.07 | 1.77 ± 0.27 | 508 ± 16 | 975 ± 91 |
The kinetic parameters were determined at different temperatures using suc-AAPF-pNA as the substrate. The values are means ± standard deviations of three independent experiments.
Data obtained from Yang et al. (27).
DISCUSSION
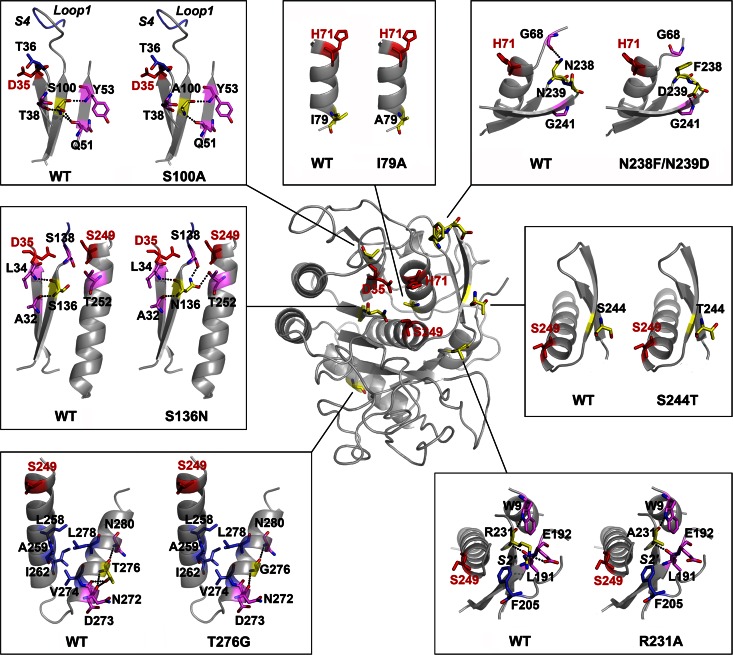
Our results demonstrate that the thermostability of a thermophilic subtilase could be improved by incorporating structural elements from its less stable counterparts. The variant PBL5X contains eight amino acid substitutions (I79A, S100A, S136N, R231A, N238F, N239D, S244T, and T276G), and its high thermostability relative to that of the WT was achieved by cumulative effects of the stabilizing substitutions I79A, S136N, R231A, and S244T. This is consistent with the proposal that the enhanced intrinsic stability of thermostable enzymes is the cumulative effect of minor improvements of local interactions (5). The substitution S136N is of particular interest because it had the strongest stabilizing effect on the enzyme (Fig. 2 and 4A, WA31). This substitution resulted in the formation of two new hydrogen bonds with Ser138 and Thr252 that resided in the α5 helix (Fig. 1 and 7). Meanwhile, Asn136 also formed two hydrogen bonds with Ala32 and Leu34 that resided in the β1 strand (Fig. 1 and 7). Therefore, the substitution S136N appeared to improve the thermostability of the enzyme by locking the secondary structure elements. The substitutions I79A and S244T resided in the α2 helix and the β9 strand that formed an antiparallel β-sheet with the β8 strand, respectively (Fig. 1 and 7). Because Ala is known to be a strong helix former (49, 50) and because Thr exhibits a higher propensity to form β-sheets than Ser (51), the substitutions I79A and S244T may stabilize the enzyme by reinforcing the secondary structure elements. The substitution R231A was adjacent to the substrate-binding subsite S2′ (Fig. 7), which is a hydrophobic pocket in subtilases (18). In the WT, the side chain of Arg231 protruded into the hydrophobic S2′ pocket (Fig. 7). The replacement of the positively charged side chain (Arg) with a hydrophobic methyl group (Ala) may increase the local hydrophobicity of the S2′ pocket and thus contribute to the improvement of the thermostability of the enzyme. We noticed that the accumulative effects in PBL5X depended on the locations of the stabilizing substitutions. For instance, the combination of I79A (WB16) with R231A (WA47) resulted in a further increase in the thermostability of the variant WA47-WB16. However, the combination of I79A with S136N (WA31) did not cumulatively enhance the thermostability of the variant WA31-WB16. Additionally, PBL5X also contained destabilizing substitutions (S100A and N238F/N239D). Nevertheless, PBL5X was as stable as the variant Satf (containing only I79A, S136N, R231A, and S244T) (Fig. 4A), which clearly indicates that the effect of the destabilizing substitutions is buffered by that of the stabilizing substitutions, thus rendering the global structure of PBL5X more resistant to thermal denaturation than that of the WT.
FIG 7.
The effects of substitutions on enzyme structure. An overview of the positions of substituted residues (in yellow) and catalytic triad residues (D35, H71, and Ser249; in red) in the homology model of PBL5X is shown in the center of the picture. Local structures around the original residues in the wild-type (WT) WF146 protease and the substituted residues in PBL5X are shown in boxes. Dotted lines represent hydrogen bonds that are formed by the original and substituted residues in the WT and PBL5X, respectively. The substrate-binding subsites, S4 and S2′, as well as loop 1 of the substrate-binding subsite S4 are indicated in the corresponding boxes.
PBL5X exhibited higher activities than the WT over a wide temperature range, and all substitutions except for S244T contributed positively to enzymatic activity. It is well known that the catalytic action of serine proteases depends on the interplay of the catalytic triad comprising Asp, His, and Ser, and a highly precise arrangement of the catalytic triad residues is required for them to attack and cleave peptide bonds in substrates (52, 53). Indeed, the catalytic triad residues are among the least mobile residues in serine proteases (54, 55). In PBL5X, the substitution S136N resulted in the formation of additional hydrogen bonds, thereby locking the secondary structure elements where the catalytic triad residues Asp35 and Ser249 were situated, respectively (Fig. 7). The stabilizing substitution I79A may reinforce the α2 helix containing the catalytic triad residue His71 (Fig. 7). The substitution T276G resided in the amphipathic α6 helix, which interacted with the α5 helix containing the catalytic residue Ser249 through a hydrophobic patch at the interface (Fig. 1 and 7). The replacement of Thr276 with Gly, which has a very low helix propensity (50), resulted in the disruption of two hydrogen bonds formed by Thr276 with Asp273 and Val274 (Fig. 7). Therefore, the substitution T276G may induce a subtle rearrangement of Ser249 by influencing the interaction between the α5 and α6 helices. It is very likely that the substitutions I79A, S136N, and T276G can lead to a relatively stable and proper arrangement of the catalytic triad, which facilitates the interplay of the catalytic triad residues to improve enzymatic activity. In addition to the action of the catalytic triad, the enzymatic activities of serine proteases rely on their abilities to bind to substrates and release products (53). In PBL5X, the substitution S100A caused the disruption of two hydrogen bonds that were formed by Ser100 with Thr38 and Gln51 and perhaps increased the mobilities of the β3 strand and the neighboring loop 1 that formed the substrate-binding subsite S4 with loop 2 (Fig. 1 and 7). The substitution R231A enlarged the size of substrate-binding subsite S2′ by replacing the large side chain of Arg with a small methyl group of Ala (Fig. 7). The substitution N238F/N239D was situated at the turn that formed an antiparallel β-hairpin loop connecting the β8 and β9 strands (Fig. 1 and 7). While the residue Asp239 formed a hydrogen bond with Gly241 on the β9 strand, the substitution N238F resulted in the disruption of a hydrogen bond between Asn238 and Gly68 near the catalytic residue His71 (Fig. 7). In this context, the substitutions S100A, R231A, and N238F/N239D led to the removal of some hydrogen bonds and bulky side chains, which may have resulted in an increase in the conformational mobility and/or structural adjustment of the substrate-binding site. Supporting evidence comes from the finding that PBL5X showed an increase in both Km and kcat values for suc-AAPF-pNA compared to those of the WT. The subtilisin BPN′ variant (Sbt 140) has been reported to have a lower kcat against tight-binding substrates, leading to the proposal that natural evolution appears to have optimized catalytic activity against a range of sequences by achieving a balance between substrate binding and the rate of release of the N-terminal product (56). Accordingly, a higher conformational mobility of the binding site of PBL5X would allow the binding site to adapt to multiple scissile peptide bonds of substrates and improve the release of the product.
Interestingly, the eight substitutions in PBL5X were all present in S41, and three of them (I79A, S100A, and S244T) were also found in Sph. Therefore, the fact that PBL5X exhibited cold-adapted features (e.g., improved low-temperature activity and higher Km and kcat values) was not unexpected. Our results support the hypothesis that a temperature-adaptive increase in kcat will occur concomitantly with an increase in Km, which resulted from the higher conformational mobility around the active site required for rapid catalysis by lowering the activation energy in cold-adapted enzymes (4, 57). Notably, PBL5X was not only more active but also more thermostable than the WT. The stabilizing forces observed in PBL5X, such as the additional hydrogen bonds (S136N), were conserved in S41 and/or Sph, as elucidated from their crystal structures (38, 39). This raises an interesting question as to why these stabilizing forces were preserved in a heat-labile enzyme, such as psychrophilic S41, but were missing in a thermostable enzyme, such as the WF146 protease. Again, this question may be addressed from the perspective of different adaptative strategies of enzymes in response to different temperature stresses. Psychrophilic enzymes are known to exhibit high activity to compensate for the exponential decrease in chemical reaction rates when temperatures decrease (3, 48). This high low-temperature activity is achieved mainly by destabilization of the active site or the whole protein, which allows the catalytic center to be more mobile or flexible at temperatures that tend to freeze molecular motions (3, 48). However, the active site of psychrophilic enzymes should be stable enough to maintain the favorable positioning accuracy of catalytic triad residues. We speculate that the simplest way to solve this problem is to reinforce the catalytic triad geometry by adding stabilizing forces, such as the hydrogen bonds created by the residue Asn136 in S41, and meanwhile to increase the conformational mobility of the substrate-binding site. Conversely, because the chemical reaction rates increase as temperatures are increased, thermophilic enzymes benefit from high temperatures to boost activity. Therefore, a strong selective pressure for high low-temperature activity should be less essential for these enzymes (48, 58). For the WF146 protease, the effect of the absence of the above-mentioned stabilizing forces could be compensated by the presence of other stabilizing interactions around the active-site region. As a result, the geometric arrangement of the catalytic triad is less likely to be affected, thereby rendering the enzyme stable and active for catalysis at high temperatures. Due to a lack of strong selective pressure for highly active thermophilic enzymes that benefit from thermally induced activity in nature (58), the low-temperature activity of the thermophilic WF146 protease can be artificially improved by incorporating structural elements of psychrophilic S41. This enhancement in low-temperature activity will then be accompanied by an increase in enzyme thermostability if the structural elements (e.g., Asn136 in S41) contribute to enzymatic activity by stabilizing key components that are involved in the catalytic cycle (e.g., the catalytic triad). Recently, lipase variants with an increase in both thermostability and activity have been engineered by rigidifying the catalytic residues (59, 60). In this context, stabilizing the geometry of catalytic residues is a valuable strategy to engineer enzymes that couple high thermostability with high activity.
In summary, the successful construction of PBL5X benefited from the high amino acid sequence identity among the WF146 protease, S41, and Sph. This allowed us to efficiently identify key structural elements that were involved in enzyme stability and activity and to improve enzyme properties by combination of the structural elements with stabilizing or/and activity-enhancing effects. As whole-genome sequence data have become more available for microorganisms that grow in different temperature environments, an increasing number of thermophilic, mesophilic, and psychrophilic enzymes with high sequence identity have been identified. This greatly facilitates the identification of key structural elements that are responsible for temperature adaptation and will accelerate the effort to improve the stability and/or activity of enzymes. Our results not only contribute to a better understanding of the temperature adaptation mechanisms of enzymes in terms of the structure-stability-activity relationship but may also provide a rational basis for developing highly stable and active subtilases, which are highly desired in industrial applications.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gengfu Xiao and Zheng Zhou (Wuhan Institute of Virology, Chinese Academy of Sciences, China) for their help with DSC analyses.
This work was supported in part by the National Natural Science Foundation of China (31270099 and 31470185) and the National Infrastructure of Natural Resources for the Science and Technology Program of China (NIMR-2014-8).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01478-15.
REFERENCES
- 1.Deming JW. 2002. Psychrophiles and polar regions. Curr Opin Microbiol 5:301–309. doi: 10.1016/S1369-5274(02)00329-6. [DOI] [PubMed] [Google Scholar]
- 2.Takai K, Nakamura K, Toki T, Tsunogai U, Miyazaki M, Miyazaki J, Hirayama H, Nakagawa S, Nunoura T, Horikoshi K. 2008. Cell proliferation at 122°C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc Natl Acad Sci U S A 105:10949–10954. doi: 10.1073/pnas.0712334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feller G, Gerday C. 2003. Psychrophilic enzymes: hot topics in cold adaptation. Nat Rev Microbiol 1:200–208. doi: 10.1038/nrmicro773. [DOI] [PubMed] [Google Scholar]
- 4.Fields PA, Somero GN. 1998. Hot spots in cold adaptation: localized increases in conformational flexibility in lactate dehydrogenase A4 orthologs of Antarctic notothenioid fishes. Proc Natl Acad Sci U S A 95:11476–11481. doi: 10.1073/pnas.95.19.11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaenicke R, Sterner R. 2013. Life at high temperatures, p 337–374. In Rosenberg E, DeLong E, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes. Springer, Berlin, Germany. [Google Scholar]
- 6.Vieille C, Zeikus GJ. 2001. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev 65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radestock S, Gohlke H. 2011. Protein rigidity and thermophilic adaptation. Proteins 79:1089–1108. doi: 10.1002/prot.22946. [DOI] [PubMed] [Google Scholar]
- 8.Jaenicke R. 2000. Do ultrastable proteins from hyperthermophiles have high or low conformational rigidity? Proc Natl Acad Sci U S A 97:2962–2964. doi: 10.1073/pnas.97.7.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnold FH, Wintrode PL, Miyazaki K, Gershenson A. 2001. How enzymes adapt: lessons from directed evolution. Trends Biochem Sci 26:100–106. doi: 10.1016/S0968-0004(00)01755-2. [DOI] [PubMed] [Google Scholar]
- 10.Van den Burg B, Vriend G, Veltman OR, Venema G, Eijsink VG. 1998. Engineering an enzyme to resist boiling. Proc Natl Acad Sci U S A 95:2056–2060. doi: 10.1073/pnas.95.5.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheridan PP, Panasik N, Coombs JM, Brenchley JE. 2000. Approaches for deciphering the structural basis of low temperature enzyme activity. Biochim Biophys Acta 1543:417–433. doi: 10.1016/S0167-4838(00)00237-5. [DOI] [PubMed] [Google Scholar]
- 12.Bae E, Phillips GN Jr. 2004. Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases. J Biol Chem 279:28202–28208. doi: 10.1074/jbc.M401865200. [DOI] [PubMed] [Google Scholar]
- 13.Lonhienne T, Gerday C, Feller G. 2000. Psychrophilic enzymes: revisiting the thermodynamic parameters of activation may explain local flexibility. Biochim Biophys Acta 1543:1–10. doi: 10.1016/S0167-4838(00)00210-7. [DOI] [PubMed] [Google Scholar]
- 14.Georlette D, Damien B, Blaise V, Depiereux E, Uversky VN, Gerday C, Feller G. 2003. Structural and functional adaptations to extreme temperatures in psychrophilic, mesophilic, and thermophilic DNA ligases. J Biol Chem 278:37015–37023. doi: 10.1074/jbc.M305142200. [DOI] [PubMed] [Google Scholar]
- 15.Chiuri R, Maiorano G, Rizzello A, del Mercato LL, Cingolani R, Rinaldi R, Maffia M, Pompa PP. 2009. Exploring local flexibility/rigidity in psychrophilic and mesophilic carbonic anhydrases. Biophys J 96:1586–1596. doi: 10.1016/j.bpj.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perl D, Mueller U, Heinemann U, Schmid FX. 2000. Two exposed amino acid residues confer thermostability on a cold shock protein. Nat Struct Biol 7:380–383. doi: 10.1038/75151. [DOI] [PubMed] [Google Scholar]
- 17.Bae E, Bannen RM, Phillips GN. 2008. Bioinformatic method for protein thermal stabilization by structural entropy optimization. Proc Natl Acad Sci U S A 105:9594–9597. doi: 10.1073/pnas.0800938105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siezen RJ, Leunissen JAM. 1997. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci 6:501–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavicchioli R, Siddiqui KS, Andrews D, Sowers KR. 2002. Low-temperature extremophiles and their applications. Curr Opin Biotechnol 13:253–261. doi: 10.1016/S0958-1669(02)00317-8. [DOI] [PubMed] [Google Scholar]
- 20.Atomi H, Sato T, Kanai T. 2011. Application of hyperthermophiles and their enzymes. Curr Opin Biotechnol 22:618–626. doi: 10.1016/j.copbio.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Taguchi S, Ozaki A, Momose H. 1998. Engineering of a cold-adapted protease by sequential random mutagenesis and a screening system. Appl Environ Microbiol 64:492–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao H, Arnold FH. 1999. Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Eng 12:47–53. doi: 10.1093/protein/12.1.47. [DOI] [PubMed] [Google Scholar]
- 23.Miyazaki K, Wintrode PL, Grayling RA, Rubingh DN, Arnold FH. 2000. Directed evolution study of temperature adaptation in a psychrophilic enzyme. J Mol Biol 297:1015–1026. doi: 10.1006/jmbi.2000.3612. [DOI] [PubMed] [Google Scholar]
- 24.Taguchi S, Komada S, Momose H. 2000. The complete amino acid substitutions at position 131 that are positively involved in cold adaptation of subtilisin BPN′. Appl Environ Microbiol 66:1410–1415. doi: 10.1128/AEM.66.4.1410-1415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Bian Y, Tang B, Chen X, Shen P, Peng Z. 2004. Cloning and analysis of WF146 protease, a novel thermophilic subtilisin-like protease with four inserted surface loops. FEMS Microbiol Lett 230:251–258. doi: 10.1016/S0378-1097(03)00914-5. [DOI] [PubMed] [Google Scholar]
- 26.Bian Y, Liang X, Fang N, Tang XF, Tang B, Shen P, Peng Z. 2006. The roles of surface loop insertions and disulfide bond in the stabilization of thermophilic WF146 protease. FEBS Lett 580:6007–6014. doi: 10.1016/j.febslet.2006.09.068. [DOI] [PubMed] [Google Scholar]
- 27.Yang YR, Zhu H, Fang N, Liang X, Zhong CQ, Tang XF, Shen P, Tang B. 2008. Cold-adapted maturation of thermophilic WF146 protease by mimicking the propeptide binding interactions of psychrophilic subtilisin S41. FEBS Lett 582:2620–2626. doi: 10.1016/j.febslet.2008.06.041. [DOI] [PubMed] [Google Scholar]
- 28.Liang X, Bian Y, Tang XF, Xiao G, Tang B. 2010. Enhancement of keratinolytic activity of a thermophilic subtilase by improving its autolysis resistance and thermostability under reducing conditions. Appl Microbiol Biotechnol 87:999–1006. doi: 10.1007/s00253-010-2534-2. [DOI] [PubMed] [Google Scholar]
- 29.Fang N, Zhong CQ, Liang X, Tang XF, Tang B. 2010. Improvement of extracellular production of a thermophilic subtilase expressed in Escherichia coli by random mutagenesis of its N-terminal propeptide. Appl Microbiol Biotechnol 85:1473–1481. doi: 10.1007/s00253-009-2183-5. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H, Xu BL, Liang X, Yang YR, Tang XF, Tang B. 2013. Molecular basis for auto- and hetero-catalytic maturation of a thermostable subtilase from thermophilic Bacillus sp. WF146. J Biol Chem 288:34826–34838. doi: 10.1074/jbc.M113.498774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong CQ, Song S, Fang N, Liang X, Zhu H, Tang XF, Tang B. 2009. Improvement of low-temperature caseinolytic activity of a thermophilic subtilase by directed evolution and site-directed mutagenesis. Biotechnol Bioeng 104:862–870. doi: 10.1002/bit.22473. [DOI] [PubMed] [Google Scholar]
- 32.Davail S, Feller G, Narinx E, Gerday C. 1994. Cold adaptation of proteins. Purification, characterization, and sequence of the heat-labile subtilisin from the antarctic psychrophile Bacillus TA41. J Biol Chem 269:17448–17453. [PubMed] [Google Scholar]
- 33.Wintrode PL, Miyazaki K, Arnold FH. 2001. Patterns of adaptation in a laboratory evolved thermophilic enzyme. Biochim Biophys Acta 1549:1–8. doi: 10.1016/S0167-4838(01)00226-6. [DOI] [PubMed] [Google Scholar]
- 34.Martinez R, Schwaneberg U, Roccatano D. 2011. Temperature effects on structure and dynamics of the psychrophilic protease subtilisin S41 and its thermostable mutants in solution. Protein Eng Des Sel 24:533–544. doi: 10.1093/protein/gzr014. [DOI] [PubMed] [Google Scholar]
- 35.Narinx E, Davail S, Feller G, Gerday C. 1992. Nucleotide and derived amino acid sequence of the subtilisin from the Antarctic psychrotroph Bacillus TA39. Biochim Biophys Acta 1131:111–113. doi: 10.1016/0167-4781(92)90108-C. [DOI] [PubMed] [Google Scholar]
- 36.Wati MR, Thanabalu T, Porter AG. 1997. Gene from tropical Bacillus sphaericus encoding a protease closely related to subtilisins from Antarctic bacilli. Biochim Biophys Acta 1352:56–62. doi: 10.1016/S0167-4781(97)00023-7. [DOI] [PubMed] [Google Scholar]
- 37.Wintrode PL, Miyazaki K, Arnold FH. 2000. Cold adaptation of a mesophilic subtilisin-like protease by laboratory evolution. J Biol Chem 275:31635–31640. doi: 10.1074/jbc.M004503200. [DOI] [PubMed] [Google Scholar]
- 38.Almog O, González A, Klein D, Greenblatt HM, Braun S, Shoham G. 2003. The 0.93Å crystal structure of sphericase: a calcium-loaded serine protease from Bacillus sphaericus. J Mol Biol 332:1071–1082. doi: 10.1016/j.jmb.2003.07.011. [DOI] [PubMed] [Google Scholar]
- 39.Almog O, Gonzalez A, Godin N, de Leeuw M, Mekel MJ, Klein D, Braun S, Shoham G, Walter RL. 2009. The crystal structures of the psychrophilic subtilisin S41 and the mesophilic subtilisin Sph reveal the same calcium-loaded state. Proteins 74:489–496. doi: 10.1002/prot.22175. [DOI] [PubMed] [Google Scholar]
- 40.Papworth C, Bauer JC, Braman J, Wright DA. 1996. Site-directed mutagenesis in one day with >80% efficiency. Strategies 9:3–4. doi: 10.1080/08924562.1996.11000299. [DOI] [Google Scholar]
- 41.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 42.DelMar EG, Largman C, Brodrick JW, Geokas MC. 1979. A sensitive new substrate for chymotrypsin. Anal Biochem 99:316–320. doi: 10.1016/S0003-2697(79)80013-5. [DOI] [PubMed] [Google Scholar]
- 43.King J, Laemmli UK. 1971. Polypeptides of the tail fibres of bacteriophage T4. J Mol Biol 62:465–477. doi: 10.1016/0022-2836(71)90148-3. [DOI] [PubMed] [Google Scholar]
- 44.Feng B, Wang Z, Liu T, Jin R, Wang S, Wang W, Xiao G, Zhou Z. 2014. Methionine oxidation accelerates the aggregation and enhances the neurotoxicity of the D178N variant of the human prion protein. Biochim Biophys Acta 1842:2345–2356. doi: 10.1016/j.bbadis.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 45.Schwede T. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeLano WL. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA. [Google Scholar]
- 47.Jaenicke R. 1981. Enzymes under extremes of physical conditions. Annu Rev Biophys Bioeng 10:1–67. doi: 10.1146/annurev.bb.10.060181.000245. [DOI] [PubMed] [Google Scholar]
- 48.Siddiqui KS, Cavicchioli R. 2006. Cold-adapted enzymes. Annu Rev Biochem 75:403–433. doi: 10.1146/annurev.biochem.75.103004.142723. [DOI] [PubMed] [Google Scholar]
- 49.Scholtz JM, Baldwin RL. 1992. The mechanism of alpha-helix formation by peptides. Annu Rev Biophys Biomol Struct 21:95–118. doi: 10.1146/annurev.bb.21.060192.000523. [DOI] [PubMed] [Google Scholar]
- 50.Pace CN, Scholtz JM. 1998. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J 75:422–427. doi: 10.1016/S0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim CA, Berg JM. 1993. Thermodynamic β-sheet propensities measured using a zinc-finger host peptide. Nature 362:267–270. doi: 10.1038/362267a0. [DOI] [PubMed] [Google Scholar]
- 52.Polgar L. 2005. The catalytic triad of serine peptidases. Cell Mol Life Sci 62:2161–2172. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hedstrom L. 2002. Serine protease mechanism and specificity. Chem Rev 102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 54.Nakagawa S, Yu HA, Karplus M, Urneyama H. 1993. Active site dynamics of acyl-chymotrypsin. Proteins 16:172–194. doi: 10.1002/prot.340160205. [DOI] [PubMed] [Google Scholar]
- 55.Bartlett GJ, Porter CT, Borkakoti N, Thornton JM. 2002. Analysis of catalytic residues in enzyme active sites. J Mol Biol 324:105–121. doi: 10.1016/S0022-2836(02)01036-7. [DOI] [PubMed] [Google Scholar]
- 56.Strausberg SL, Ruan B, Fisher KE, Alexander PA, Bryan PN. 2005. Directed coevolution of stability and catalytic activity in calcium-free subtilisin. Biochemistry 44:3272–3279. doi: 10.1021/bi047806m. [DOI] [PubMed] [Google Scholar]
- 57.Tindbaek N, Svendsen A, Oestergaard PR, Draborg H. 2004. Engineering a substrate-specific cold-adapted subtilisin. Protein Eng Des Sel 17:149–156. doi: 10.1093/protein/gzh019. [DOI] [PubMed] [Google Scholar]
- 58.Cipolla A, Delbrassine F, Da Lage J-L, Feller G. 2012. Temperature adaptations in psychrophilic, mesophilic and thermophilic chloride-dependent alpha-amylases. Biochimie 94:1943–1950. doi: 10.1016/j.biochi.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 59.Kamal MZ, Mohammad TAS, Krishnamoorthy G, Rao NM. 2012. Role of active site rigidity in activity: MD simulation and fluorescence study on a lipase mutant. PLoS One 7:e35188. doi: 10.1371/journal.pone.0035188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xie Y, An J, Yang G, Wu G, Zhang Y, Cui L, Feng Y. 2014. Enhanced enzyme kinetic stability by increasing rigidity within the active site. J Biol Chem 289:7994–8006. doi: 10.1074/jbc.M113.536045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.